Abstract
DNA can be preserved in marine and freshwater sediments both in bulk sediment and in intact, viable resting stages. Here, we assess the potential for combined use of ancient, environmental, DNA and timeseries of resurrected long-term dormant organisms, to reconstruct trophic interactions and evolutionary adaptation to changing environments. These new methods, coupled with independent evidence of biotic and abiotic forcing factors, can provide a holistic view of past ecosystems beyond that offered by standard palaeoecology, help us assess implications of ecological and molecular change for contemporary ecosystem functioning and services, and improve our ability to predict adaptation to environmental stress.
Similar content being viewed by others
Introduction
Undisturbed lake and marine sediments are natural archives of past changes in biota and their environment, and when dated, they offer the opportunity of reconstructing past changes in e.g. both primary and secondary production and community composition1,2. Analysing organismal remains in freshwater and marine sediment cores provides a long-term perspective of ecological change and has a long history in both pure science and applied contexts3 (Table 1). In the more traditional approaches to the palaeoecology of aquatic systems, microfossil analysis is accompanied by a number of geochemical proxies including lipid biomarkers, pigments and isotope composition (Table 1). The interpretation of these archives, i.e. the science of palaeoecology, is dependent on understanding contemporary ecological controls as well as the sedimentation environment and its context. The remains of a diverse range of organisms, from viruses to mammals, can be preserved in lake and marine sediments (Fig. 1). However, the degree to which they faithfully reflect changing abundance and community composition varies enormously depending on taxon preservation capacity4, the depositional environment, including sedimentation rate, and distance to the depositional site (Fig. 2).
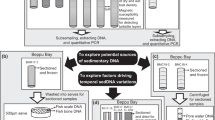
This figure shows where we have data on resurrection ecology series and indicates the food-web and interaction gaps in in the palaeoecological record, which sed-eDNA has the potential to fill in, to reconstruct food-webs, possibly through association networks, as suggested by16 for lake ecosystems. Extracting, respectively, DNA and live propagules from dated sediment core has the potential to greatly enhance both the amount and types of information that we can gain about evolutionary and adaptive processes, by filling different information gaps in the paleo-ecological record. Green stars indicate the organism types for which genetic and phenotypic time-series have already been established from resurrected resting stages; the more stars, the larger the existing dataset. A green star in parenthesis indicates that viable resting stages have been recorded from old sediment layers, but no timeseries data published. Red circles indicate organism types for which there is information based on morphological remains (“traditional” palaeoecology); the more circles, the larger the existing dataset.
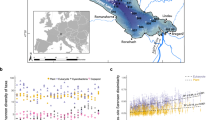
This figure illustrates the processes affecting DNA distribution, degradation and/or preferential preservation during the transitions from the pelagic to the benthic zones, and from the surface sediment to the deeper sediment. The approximate timescales of preservation of different fractions of the sediment record is also illustrated.
In the past decade, the fields of resurrection ecology and environmental DNA (eDNA; Box 1) have developed to a degree which now enables us to complement these traditional analyses with analyses of temporal change at the genetic and genomic levels. This has the potential to enhance our understanding of evolutionary processes in aquatic systems and organisms. We can now evaluate impacts of environmental stressors on both genotypic and phenotypic responses of individual species, as well as on interactions between species and on whole communities. These new approaches can thereby expand our capacity for predictive modelling to project future change, and for impact assessments.
Resurrection ecology is the study of temporal series of revived resting stages from dated sediment layers. Many planktic (as well as benthic) species form dormant or resting stages (propagules), which accumulate over time in aquatic sediments. Such sediment propagule banks contain eggs, spores, cysts and other resilient structures from all domains of life (Bacteria, Archaea and Eukarya) as well as viruses. Importantly, resting stages from many of these taxa can remain viable for centuries5,6 (Fig. 3). From terrestrial environments, a similar phenomenon is seen in seeds, but in most terrestrial environments, time series and reliable chronologies are difficult to achieve, as soil disturbance, due to bioturbation for example, will obscure the age of different depths and oxygenation will enhance organic matter mineralisation. However, a few studies have been able to follow depth series in specialised environmental settings such as marginal water bodies (e.g., in solifluction lobe; ca. 150 years max. age)7 and cedar glades8. Nonetheless, continuous, temporally structured, well-preserved archives covering multiple trophic levels and with strict age-control are unique to aquatic sites with undisturbed sedimentation. Temporal genetic signals of change can also be analysed by extracting total sedimentary DNA (Box 1). Similar to other biomarkers, DNA may be archived in aquatic sediments, but as described below (see Fig. 2), the degree to which this occurs depends heavily on deposition and preservation conditions. Thus, sediment-archived DNA may be either extracellular, in dead tissue/cells, or inside living organisms (within dormant propagules or active microbes). Each of these sources of DNA can be used for detecting genetic change that reflects responses to environmental change in natural populations and communities at a range of trophic levels.
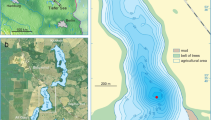
The same strains, from which the DNA was extracted, can be used for side-by-side, or common-garden, tests of phenotypic/physiological response to the same stressors. Here we show modified versions of figures from Lundholm et al.29 (a) and Frisch et al.5 (b), both showing population structure plots. a Analysis of population genetic response of the phytoplankton Pentapharsodinium dalei in a Swedish fjord to environmental change associated with changes in the index of the North Atlantic Oscillation (NAO), which affects, among other things, salinity and water-column stability. b Analysis of population genetic response of the herbivore Daphnia pulicaria in a lake to changing phosphorus concentrations through time.
Combining approaches, and moving from single species to interactions
So far, almost all studies based on resurrection ecology have targeted single species (see the compilation in a special issue of Evolutionary Applications vol. 1, 11, 2018). Here, we present the potential for moving this field toward the more complex level of species interactions by documenting how we can achieve resurrection time-series for new groups of organisms (to fill gaps at different levels of the food-web; Fig. 1).
Recent reviews9 have covered the state-of-the-art on both environmental and ancient DNA, and the body of work on resurrection ecology of single species (see summary below) was presented in a recent special issue of Evolutionary Applications (vol. 1, 11, 2018). Therefore, here we focus on how to develop the real, and mainly untapped, potential benefits of combining the two approaches. We show how the two lines of evidence from resurrection ecology and temporal series of sedimentary eDNA complement each other, and how analysing both in the same sediment record can give synergistic effects and facilitate much deeper insights into past evolutionary and adaptive trajectories and interactions.
Finally, we show how insights from more traditional palaeoecology, including preservation and spatio-temporal variability in deposition signals, can help in interpreting the genetic signals from sediment cores. Building on these insights allows the evaluation of the robustness of different chronologies and the validity of the emerging signals—to what extent do they represent real change and to what extent are they affected by preservation and taphonomic processes?
eDNA analyses can greatly expand the taxonomic coverage of traditional palaeoecological reconstructions, and of resurrection series. It can also be used to specifically target signals from resurrection model-species and to expand the range of coverage temporally. Resurrection ecology, on the other hand, adds the dimension of phenotypic and physiological response to the genetic information, and enables tracking of adaptive trait shifts. The two types of data thus offer information on different fractions of past communities.
We argue that judicious application of these emerging methods in resurrection ecology and sedimentary eDNA make it possible to track impacts of environmental change on aquatic ecosystems at centennial, and even millennial, time scales, and to link temporal phenotypic with genotypic responses, thus enabling us to assess adaptibility, resilience and evolvability across whole ecosystems. We highlight the opportunities that are resulting from the rapid development of sophisticated methodology but also discuss the current limitations that need to be addressed in order to achieve the full potential of temporal reconstruction of genetic and phenotypic responses to environmental change.
Resurrection ecology: studies on temporal series of intact cells/propagules
Dormant stages of many species can remain alive in aquatic sediments for decades and even centuries10,11,12 (see examples in Fig. 3), and it is possible to “resurrect” past populations from these resting stages buried in undisturbed sediment via hatching of zooplankton eggs or germination of phytoplankton cysts and spores (Box 1), and potentially spores and other dormant forms of Bacteria, Archaea, Fungi, as well as, potentially, other unicellular heterotrophic organisms. Resurrection ecology, which is the science based on testing temporal, revived, series of strains, now encompasses several trophic levels of the aquatic food web (Figs. 1 and 3) and can vastly increase our understanding of responses to changes at both phenotypic and genotypic levels. In addition, DNA can be obtained from resurrected strains in a quantity and quality that make in-depth analysis of full genome sequences feasible, providing enormous potential for insights into evolutionary genomics13. Genomic information from living propagules will be a key resource for reconstructing marine and freshwater biological community responses to environmental change, e.g. by discovery of loci of adaptation by applying genome-wide association studies. Resurrection studies further allow reconstruction of past phenotypes in the laboratory to directly study historical populations, thereby assessing trait changes over time as well as their transcriptomic basis14. Resurrection ecological studies thus occupy a unique niche by documenting actual processes of century-scale adaptation at both genetic (Fig. 3) and phenotypic levels. So far, almost all studies based on resurrection ecology have targeted single species and, to date, there has been a preference for studying large, identifiable, long-lived resting stages such as Daphnia eggs and dinoflagellate cysts. Below, we document the potential for achieving resurrection time-series for new groups of organisms, to fill gaps in food-chain levels, and discuss the potential for expanding resurrection ecology to include interspecific and trophic-level interactions. We thus argue that reviving resting stages from multiple organism groups and trophic levels from the same site can now make it possible to reconstruct interactions and co-adaptation trajectories over evolutionarily relevant time scales (Fig. 1).
Viruses—potential for resurrection series
Viruses are a ubiquitous component of the biosphere associated with other life forms in all ecosystems. Lakes and their sediment have abundant viral communities which are are dominated by bacteriophages15 and the sediment documents the past microbial populations from the over-lying water body. Importantly, valuable information on virus biology contained within this sediment archive can be revealed from the direct resurrection of the viruses, for example by isolating phages that infected the cyanobacterium Microcystis up to 50 years ago. Recently, Baltic Sea sediments were found to harbour diverse, allochthonous and autochthonously produced viruses down to the deepest point investigated of 37 m, representing about 6000 years since deposition16. The extensive literature documenting phage particles’ morphology from deep-sediment layers suggests that the phages are intact16, and so if the correct host strains are available, these viruses could be cultured. Past populations of viruses and their hosts could thus be resurrected and used to infer co-evolutionary dynamics over time.
Bacteria, Archaea and heterotrophic protists
Longevity of some microbes over millions of years is now widely recognised17,18,19. Resurrection of Bacteria and Archaea is thus not new but differs from studies on larger, morphologically conspicuous organisms in that, usually, enrichment culture targetting a range of taxa is used rather than first separating individual organisms. Even though bacterial palaeoecological resurrection studies started in 1990, using (allochthonous) bacterial spores as palaeo-indicators of agricultural land use20, and resting stages (akinetes) of cyanobacteria have been germinated from old sediment layers21,22, the full potential of this strategy has yet to be realised. Wunderlin et al.23 confirmed the value of spore-formers (using non-resurrection approaches) in that their abundance relative to total bacteria increases with sediment age. Still, our understanding of the potential for DNA preservation in, as well as survival of, resting stages of different bacterial taxa, is inadequate6, and the plethora of techniques to improve microbial cultivation and selectively enrich for specific functional groups has barely been used in palaeoecology. Heterotrophic protists, such as flagellates, cilates and amoebae, may also be present in sediments, both in encysted and actively metabolising states. A few studies have cultivated amoebae found downcore in aquatic sediments24, and other studies have shown that ciliates may survive tens of thousands of years in permafrost soils25. However, to the best of our knowledge, no genetic and/or phenotypic studies of these organisms have been carried out on sediment time-series.
Phytoplankton
Resting stages are found in many species and in most groups of phytoplankton12, but so far resurrection ecological studies on time series have been restricted to a few, marine, species: the dinoflagellates Alexandrium spp., Pentapharsodinium dalei (example in Fig. 3), and Apocalathium malmogiense26 and the diatom Skeletonema marinoi. These have been used to explore the impact of environmental conditions (e.g. salinity6,27,28 and eutrophication29) on population genetic dynamics over multidecadal time scales, as well as phenotypic adaptation to changed environmental conditions (e.g.6,26,27). These examples illustrate that it is possible to trace evolutionary change of genotypes as well as phenotypic reaction norms of different traits in response to environmental state using monoclonal cultures established from revived resting stages and kept in the laboratory side-by-side with modern strains. These phenotypic responses can then be linked to the corresponding genomic data.
Metazoa, mainly zooplankton
The most comprehensive dataset of resurrection ecology is probably that from crustaceans with dormant stages. The bulk of these studies have been carried out on the water flea Daphnia, e.g. to identify species invasions30,31 and to track effects of eutrophication32,34,,33 (example in Fig. 3). Many other invertebrates are present in propagule banks but have rarely been exploited for genetic or resurrection studies34. However, Epp et al.35 were able to relate changes in different genotypes of Brachionus rotifers to dramatic environmental change such as water level or the deposition of volcanic ashes over 100 years. Using dormant copepod eggs retrieved from lake sediment, Makino et al.36 recovered 21 haplotypes using 28S ribosomal DNA. The brine shrimp Artemia is also being explored as a model for resurrection ecology in higher salinity environments37.
Resurrection ecological studies on Daphnia spp. have shown how a population evolved (and subsequently lost) its resistance to toxic cyanobacterial blooms over a few decades38, documented a historical change in the phenotypic plasticity of phosphorus physiology in response to anthropogenic eutrophication5 and showed changes in phototactic behaviour39 and other traits40 in response to changes in predation pressure. The comparison of transcriptomic responses of resurrected 10- and 700-year old Daphnia isolates allowed identification of gene networks and key functional drivers involved in the evolutionary adaptation to eutrophication14. Recently, the first attempts to perform whole-genome amplification of DNA from dormant stages have been made41—a method that potentially facilitates whole-genome sequencing even of propagules that cannot be hatched or germinated.
For fish, aDNA has been extracted from remains of otoliths42 or scales43 from museum archives, but, although these remains can accumulate in the sediment, we are not aware of the use of sediment-buried fish remains to reconstruct historic evolutionary adaptation to an environmental change at the genetic or genomic level.
Next steps
Potential pitfalls in the study of resurrected time series regarding e.g. issues of the non-representative nature of revived populations44,45,46 and differential survival can be addressed in the planning phase of a study. Thus, biases and artefacts related to specific phenotypes can be detected by the analysis of multiple sediment cores39,47, cores from greater depths or from anoxic sediments not exposed to early cues for germination/hatching. In the case of phyto- and zooplankton resting stages, rapidly developing single-cell sequencing approaches may help to identify germination and survival biases48, and the application of beneficial bacteria and their (or other) extracts may enhance germination49. Issues of adaptation to culture conditions50,51 can be circumvented by phenotyping the cultures soon after germination.
Currently the main datasets for zooplankton originate from lakes while the main datasets for phytoplankton derive from marine coastal embayments. However, as mentioned above and illustrated in Fig. 1, this bias can be overcome by intensifying the effort to germinate multiple species from different trophic levels from the same site, allowing the evolutionary dynamics of biotic interactions to be studied if both organisms deposit viable resting stages in the same sediment. One of the few studies to adopt this approach revealed co-evolutionary dynamics between D. magna and its microparasites by resurrecting host and parasite populations from different time periods47. Similarly, the influence of viral infections on the changing abundance of cyanobacteria such as Microcystis in eutrophic lakes could be shown by linking pigment concentration with estimates of cyanophage infection (sensu Hargreaves). The multi-faceted aspect of eutrophication on freshwater foodwebs and species interactions has been addressed by coupling resurrected Daphnia with bacterial infections (Pasteuria ramosa) against a background of changing food quality associated with increased nutrient load. Reyserhove et al.52 show that genetic differentiation in Daphnia is affected by food availability, and ultimately influences parasite virulence. Finally, an example from the marine realm: as described earlier, resting stages of the dinoflagellate Pentapharsodinium dalei can survive ca. 100 years in undisturbed sediment cores in temparate and arctic regions27. Parvilucifera is a genus of well-known parasites that are strain-specific for P. dalei53. Germinating infected strains of the dinoflagellate from separated time-slices, isolating the parasite, and attempting to re-infect strains with parasites of a different age can give novel insights into co-evolution in real systems at multidecadal to century time scales.
Going further, adopting a comparative genomics approach will improve our understanding of adaptation to environmental change by targeting specific genes or genomic regions across taxa present in the sediment archive and linking this to specific phenotypic responses.
DNA sedimentary archives and their taphonomy
The potential of using eDNA sediment time series in ecology was recently covered in a thorough review by Balint et al.9. Therefore, here we focus specifically on two points: (1) the potential for linking eDNA data with resurrection ecology, and (2) methological issues that need more attention to tap the full potential for reconstructing planktic communities and interactions.
The taphonomy of DNA
As mentioned in the Introduction, valid interpretations of environmental signals derived from aquatic sediment timeseries rely heavily on an understanding of sedimentological processes (e.g. depositional environment and rates) and process of preservation and degradation (taphonomy). These considerations, perhaps even more critically, apply to the analysis of DNA archived in aquatic sediments. Thus, a thorough understanding of the factors influencing DNA preservation must underpin the field of research into sediment timeseries of eDNA.
The probability of enzymatic and abiotic degradation of DNA increases with time, hence the importance of rapid burial. This particularly applies to extracellular DNA, but even intracellular DNA will be damaged if the cell has ceased active repair. In the dark and anoxic conditions typical of benthic sediments, hydrolysis (depurination and deamination) is likely to be the main abiotic process contributing to DNA decay54. Anoxia55, minimal bioturbation55 and low temperatures56 are conducive to an excellent sedimentary DNA archive. Extreme salinity also has an impact on DNA preservation, as shown for brines from deep-sea anoxic hypersaline lakes57, especially those rich in chaotropic salts which can be effectively sterile and therefore excellent for preserving biomolecules58. Furthermore, hardwater lakes provide good preservation due to calcite formation, which supports rapid sedimentation59.
A large fraction of sediment DNA is extracellular, e.g. 90% in deep-sea sediments60 and 31% in lake sediments61. In fact, extracellular DNA in aquatic sediments is the largest global reservoir of DNA, with implications for ecosystem functioning60 as an important source of carbon, nitrogen and especially phosphorus. Many microbes assimilate these elements from DNA62 and in some cases depolymerised DNA is used as a source of energy62,63.
Extracellular DNA has differential bioavailability, depending on whether it is free or adsorbed to sediment particles. DNA bound to minerals or organic matter can constitute the majority of extracellular DNA (e.g. >95% in marine sediments64), and attachment can enhance its preservation. Cation bridging of DNA to clay minerals is a major mechanism by which DNA bioavailability is reduced, due to the large surface area to volume ratio of clay minerals and, for some clay minerals, their laminar structure, whereby DNA can be adsorbed between clay layers54,65. In addition to reducing bioavailability of DNA, nucleases adsorb to minerals66, potentially reducing their activity54. Extreme conditions may facilitate preservation of cells/DNA over hundreds, thousands or even millions of years, as found with evaporite minerals, such as halite17. There is still a lot to learn about how mineral type and organic matter composition, coupled with other factors such as ionic composition, influence the early diagenetic processes of extracellular sedimentary DNA54,65.
Temporal extent of DNA archives in aquatic sediments
These preservation issues non-withstanding, DNA signals of a large variety of organism groups have been traced continuously over millenial time scales in aquatic sediment archives. Most studies have extracted bulk DNA from the sediment without distinguishing between DNA inside live cells/resting stages and extracellular DNA. Our focus is on the potential for reconstructing planktic aquatic ecosystems and we refer to other studies for catchment-derived DNA signals (of e.g. trees and other vegetation) stored in lake sediments67.
With regard to eukaryotic aquatic organisms, temporal changes in sed-eDNA have been documented over time scales of thousands to tens of thousands of years. For lakes this was reviewed by Domaizon et al.59. In a marine setting, Lejerzerowicz and co-workers68 recovered DNA from deep-sea sediment cores collected in the South Atlantic, dated to about 32.500 years ago, including DNA from taxa that do not fossilise well and undetermined taxa. A more recent study used sed-eDNA to estimate the colonisation date for white fish in a Swedish lake69. eDNA approaches therefore offer the potential to assess the impact of environmental change across taxonomic groups, over long temporal scales, and potentially with a taxonomic resolution unavailable by traditional microfossil approaches. However, many authors have also highlighted limitations70 (see also Fig. 2) and questioned the reliability of DNA archives from sediments as a stand-alone proxy71. Instead, most researchers advocate a combination of DNA evidence and palaeoecological approaches as the way forward (i.e. using DNA as one proxy within a multiproxy study)70,72,73. Indeed, as indicated in the previous section, the mechanisms of DNA preservation are sometimes unclear and even counterintuitive. This point is illustrated by a sediment core study74 from the stratified Watts Basin in Antarctica, which reported a 10-fold decline in diatom DNA and a 10,000-fold decline in dinoflagellate DNA over 2700 years. However, there was no ecological explanation for this difference, and quantification of the dinoflagellate biomarker, dinosterol, did not support this massive decline. Therefore, these findings were attributed to preferential preservation of diatom DNA within resting stages, which were also found in the sediment record, together with potentially greater lability of dinoflagellate DNA due to their lack of histones74. Applying parallel studies of the two types of temporal genetic signals derived from eDNA and resurrection ecology can further illuminate these issues.
Bacteria, Archaea, Fungi and Viruses
Bacteria, Archaea and to a lesser extent Fungi, pose both an opportunity and a threat to the field of sed-eDNA. The opportunity is that, as for other organisms, their sed-eDNA can be used for temporal reconstruction of communities, with at least two requirements: firstly, that the environment is favourable to preservation (Fig. 2); secondly, that the DNA is from a group that would be present and functional in the water column but not the sediment. The threat comes from the ability of many microorganisms to function in sediments, with two primary, interconnected effects: (1) they increase the bioavailability and degradation of organic matter, including DNA and other biomarkers derived from supposedly archived organisms; (2) they multiply in the sediment, and thus alter the microbial community composition, i.e. mixing up the modern (autochthonous) and ancient sed-eDNA. With increasing depth and time, most DNA derives from autochthonous microbes that are adapted to the present deep-sediment conditions in both marine75 and lacustrine76 environments. However, exceptional preservation may occur as discussed earlier, for example in the case of 217,000-year-old DNA derived from phototrophic Chromatiaceae77 and 2700-year-old DNA from green-sulfur bacteria (Chlorobium)74. Naturally, most bacterial sed-eDNA studies have focussed on phototrophs that are allochthonous to the deep sediment, including cyanobacteria78,79, Chlorobi74 and Chromatiaceae80.
Phylogenetic genes are valuable when there is a near-unambiguous taxonomy-trait relationship, e.g. as found in cyanobacteria, where photosynthesis is a group trait, notwithstanding the capacity for other modes of energy generation in some cyanobacterial lineages such as those detected in the deep subsurface81 and per-alpine lakes82. More recently, genes encoding enzymes with a specific function are being used, such as a cyanobacterial gene (mycA) coding for the synthesis of the toxin microcystin, which has been used to identify where potentially toxic cyanobacterial blooms had occurred in perialpine lakes78. Similarly, the particulate methane monooxygenase gene (pmoA) in anoxic lake sediments has been used to infer past aerobic methane oxidation in the water column83. DNA extracted from Bacillus spores in a lake sediment archive has shown a rise in abundance of antibiotic resistance genes from the 1960s for tetracycline resistance and 1970s for sulfonamide resistance, demonstrating the value of such studies in understanding the historical legacy of antibiotic use84.
Analysis of sed-eDNA has the potential to provide a unique window into host-parasite evolution, such as that between cyanobacterial Planktothrix chemotype hosts and their parasitic chytrid fungi85. The lake sediment record showed stable co-existence of host and parasite, raising questions about spatial avoidance (infection refuges) or evolution of resistance chemotypes in Planktothrix85. The diversity and population dynamics of sediment viruses could also be assessed using metagenomic, culture-independent methods by probing specific groups. Many bacterial taxa are associated with particular phage groups, so their diversity can be explored using primers specific to conserved genes such as the capsid protein or portal protein genes that are currently used as a tool to assess marine cyanophage diversity86. The phage-encoded genes for bacterial metabolism proteins may also provide clues to deciphering past ecological conditions. For example, some cyanobacterial phages harbour the genes for phosphorus uptake proteins87. A third way to explore past viral dynamics is possibly the newly developed method that enables virus sequences to be linked to bacteria using the frequency and abundance of CRISPR sequences in both the viruses and bacteria88.
Next steps
Increasing phylogenetic coverage (including reference sequences) and amount of sequence data and implementing links to phenotypic change by targeting functional genes will help us towards the goal of temporal reconstruction of entire ecosystems and their response to change,
Applying multiproxy approches, and building on insights from palaeoecological studies can give an increased understanding of preservational issues and sampling techniques, leading to more robust and reliable interpretations of past changes.
The benefits of synergy: combining sed-eDNA and resurrection ecology
It is apparent from the summaries above of recent work, and other reviews, that these two new fields complement each other, and can greatly enhance traditional palaeoecological information derived from traditional approaches. However, to reach their full potential, knowledge gaps and methodological concerns must be addressed. In this final section we briefly highlight the exciting potential of merging and expanding the analyses of genetic archives to fill the missing ecological links indicated in Fig. 1. By integrating investigations on multiple trophic levels, and combining them with sed-eDNA analyses, the field of resurrection ecology can be taken a step further toward reconstructing whole-ecosystem responses to environmental change. So far, most experiments on eco-evolutionary dynamics spanning different trophic levels have used simplified systems such as prey-predator dynamics in well-controlled experiments89. The methods presented here have the potential to extend this approach to the greater complexity of natural systems. In addition to the examples provided in this paper, here we briefly present examples of such interactions, where temporal trajectories and species interactions, based on existing knowledge, could be reconstructed from sediment sequences.
Most eDNA studies lack even an indirect link to phenotype and therefore cannot connect genetic to phenotypic changes, although functional gene analysis provides this opportunity in some cases. In addition, there may be uncertainty associated with the provenance of the extracted sequences. This problem is especially true for non-model organisms for which many functional pathways are unknown, or gene annotation is based on phylogenetically distant taxa. Furthermore, while palaeogenomics can provide evidence for community changes over time, there is rarely sufficient resolution for population-level analysis (although this might change, see a recent perspective paper90), and it cannot provide a direct link to specific changes at the phenotype level and the organisms’ fitness. Resurrection studies can address these limitations.
One of the benefits of sed-eDNA comes from its ability to provide information about organisms that may not leave an obvious fossil morphological record (Figs. 1 and 3). However, as with the development of other biochemical proxies in palaeoecology, for example, pigments and stable isotopes, without a critical context, sed-eDNA results can simply be another stratigraphic profile to be interpreted subjectively. A more sophisticated approach is to use the molecular results to answer questions that are difficult to address with more traditional palaeoecological approaches. Increased nutrient loading to aquatic ecosystems results in greater production and changed species composition and community structure91. However, high temporal resolution stratigraphic sequences from eutrophic lakes show considerable temporal variability in diatom species and cyanobacteria. Using a multi-proxy approach with relevant statistical methods, it is clear that increasing nutrient load, grazing by zooplankton or climate forcing cannot explain all of this temporal variability in algal abundance. In this context, sed-eDNA can provide supplementary evidence of other trophic links. For example, the role of chytrid infections in controlling phytoplankton abundance has been underestimated in freshwaters (see review by Frenken et al.92). From a longer temporal perspective, combining fossil Asterionella abundance with the sed-eDNA record of the chytrid Zygorhizidium planktocnium could provide evidence of an important mechanism controlling Asterionella abundance at decadal time scales. See also the work of Kyle85 discussed above. A combination of resurrection ecology and sed-eDNA could also be used to test the effect of environmental change on biodiversity, e.g. how genetic diversity changes when one member disappears, and to identify possible corresponding phenotypic responses in the remaining species.
Conclusions: tracing evolution of planktonic food-webs in sediment archives
The science of palaeoecology allows us to reconstruct past changes in biological communities, but has limitations in species coverage and in the type of information that can be inferred about past processes. New developments in resurrection ecology and sedimentary timeseries of eDNA promise to address these gaps and can become powerful tools for predicting futures changes to ecosystem function. Building on our knowledge about preservation and degradation of organic molecules in aquatic sediments, we can develop and optimise sampling and interpretation of these unique historical and ancient continuous time series of genetic and phenotypic adaptation. DNA preserved in dated sediment cores has the potential to increase taxonomic coverage to include key organisms and processes that are missing in the microfossil record. Resurrection ecology further adds the dimension of linking population genetic changes with adaptive trait shifts, and linking both to environmental drivers. Moreover, expanding coverage in terms of species, organism groups and strain numbers will allow us to reconstruct rates and magnitudes of change under both natural and anthropogenic forcing.
We are confident that in a few years we will be able to address, for example, questions about the sources of adaptive variation and their underlying genomic architecture. Using undisturbed aquatic sediment cores is the only way to obtain such continuous records for high-resolution temporal reconstructions. The quantitative data acquired from this approach thus have the potential to illuminate generic adaptive processes and responses in the face of multiple environmental stressors.
Change history
09 July 2020
The original version of this Article was updated shortly after publication following an error that resulted in the ORCID ID of N. John Anderson being omitted.
References
Anderson, N. J., Renberg, I. & Segerstrom, U. Diatom production responses to the development of early agriculture in a boreal forest lake-catchment (Kassjon, Northern Sweden). J. Ecol. 83, 809–822 (1995).
Davidson, T. A. et al. The role of cladocerans in tracking long-term change in shallow lake trophic status. Hydrobiologia 676, 299–315 (2011).
Battarbee, R. W., Anderson, N. J., Jeppesen, E. & Leavitt, P. R. Combining palaeolimnological and limnological approaches in assessing lake ecosystem response to nutrient reduction. Freshw. Biol. 50, 1772–1780 (2005).
Ryves, D. B., Battarbee, R. W., Juggins, S., Fritz, S. C. & Anderson, N. J. Physical and chemical predictors of diatom dissolution in freshwater and saline lake sediments in North America and West Greenland. Limnol. Oceanogr. 51, 1355–1368 (2006).
Frisch, D. et al. A millennial-scale chronicle of evolutionary responses to cultural eutrophication in Daphnia. Ecol. Lett. 17, 360–368 (2014).
Ribeiro, S. et al. Phytoplankton growth after a century of dormancy illuminates past resilience to catastrophic darkness. Nat. Commun. 2, https://doi.org/10.1038/ncomms1314 (2011).
Bennington, C. C., McGraw, J. B. & Vavrek, M. C. Ecological genetic-variation in seed banks: 2. Phenotypic and genetic-differences between young and old subpopulations of luzula-parviflora. J. Ecol. 79, 627–643 (1991).
Morris, A. B., Baucom, R. S. & Cruzan, M. B. Stratified analysis of the soil seed bank in the cedar glade endemic Astragalus bibullatus: evidence for historical changes in genetic structure. Am. J. Bot. 89, 29–36 (2002).
Balint, M. et al. Environmental DNA time series in ecology. Trends Ecol. Evol. 33, 945–957 (2018).
Hargreaves, K. R., Anderson, N. J. & Clokie, M. R. J. Recovery of viable cyanophages from the sediments of a eutrophic lake at decadal timescales. Fems Microbiol. Ecol. 83, 450–456 (2013).
Frisch, D. et al. Paleogenetic records of Daphnia pulicaria in two North American lakes reveal the impact of cultural eutrophication. Glob. Change Biol. 23, 708–718 (2017).
Ellegaard, M. & Ribeiro, S. The long-term persistence of phytoplankton resting stages in aquatic ‘seed banks’. Biol. Rev. 93, 166–183 (2018).
Orsini, L. et al. The evolutionary time machine: using dormant propagules to forecast how populations can adapt to changing environments. Trends Ecol. Evol. 28, 274–282 (2013).
Roy Chowdhury, P. et al. Differential transcriptomic responses of ancient and modern Daphnia genotypes to phosphorus supply. Mol. Ecol. 24, 123–135 (2015).
Clokie, M. R. J., Millard, A. D., Letarov, A. V. & Heaphy, S. Phages in nature. Bacteriophage 1, 31–45 (2011).
Cai, L. L. et al. Active and diverse viruses persist in the deep sub-seafloor sediments over thousands of years. ISME J. 13, 1857–1864 (2019).
Gramain, A., Diaz, G. C., Demergasso, C., Lowenstein, T. K. & McGenity, T. J. Archaeal diversity along a subterranean salt core from the Salar Grande (Chile). Environ. Microbiol. 13, 2105–2121 (2011).
Lomstein, B. A., Langerhuus, A. T., D’Hondt, S., Jorgensen, B. B. & Spivack, A. J. Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment. Nature 484, 101–104 (2012).
Inagaki, F. et al. Exploring deep microbial life in coal-bearing sediment down to similar to 2.5 km below the ocean floor. Science 349, 420–424 (2015).
Nilsson, M. & Renberg, I. Viable endospores of thermoactinomyces-vulgaris in lake-sediments as indicators of agricultural history. Appl. Environ. Microbiol. 56, 2025–2028 (1990).
Livingstone, D. & Jaworski, G. H. M. The viability of akinetes of blue-green-algae recovered from the sediments of Rostherne Mere. Br. Phycol. J. 15, 357–364 (1980).
Legrand, B., Miras, Y., Beauger, A., Dussauze, M. & Latour, D. Akinetes and ancient DNA reveal toxic cyanobacterial recurrences and their potential for resurrection in a 6700-year-old core from a eutrophic lake. Sci. Total Environ. 687, 1369–1380 (2019).
Wunderlin, T., Junier, T., Roussel-Delif, L., Jeanneret, N. & Junier, P. Endospore-enriched sequencing approach reveals unprecedented diversity of Firmicutes in sediments. Environ. Microbiol. Rep. 6, 631–639 (2014).
Smirnov, A. V. Vertical distribution and abundance of gymnamoebae (Rhizopoda) in bottom sediments of the Brackish water Niva Bay (Baltic Sea, The Sound). Protist 153, 239–250 (2002).
Shatilovich, A., Stoupin, D. & Rivkina, E. Ciliates from ancient permafrost: assessment of cold resistance of the resting cysts. Eur. J. Protistol. 51, 230–240 (2015).
Kremp, A., Hinners, J., Klais, R., Leppanen, A. P. & Kallio, A. Patterns of vertical cyst distribution and survival in 100-year-old sediment archives of three spring dinoflagellate species from the Northern Baltic Sea. Eur. J. Phycol. 53, 135–145 (2018).
Ribeiro, S., Berge, T., Lundholm, N. & Ellegaard, M. Hundred Years of Environmental Change and Phytoplankton Ecophysiological Variability Archived in Coastal Sediments. Plos ONE 8, https://doi.org/10.1371/journal.pone.0061184 (2013).
Lundholm, N., Ribeiro, S., Godhe, A., Nielsen, L. R. & Ellegaard, M. Exploring the impact of multidecadal environmental changes on the population genetic structure of a marine primary producer. Ecol. Evol. 7, 3132–3142 (2017).
Harnstrom, K., Ellegaard, M., Andersen, T. J. & Godhe, A. Hundred years of genetic structure in a sediment revived diatom population. Proc. Natl Acad. Sci. USA 108, 4252–4257 (2011).
Duffy, M. A., Perry, L. J., Kearns, C. M., Weider, L. J. & Hairston, N. G. Paleogenetic evidence for a past invasion of Onondaga Lake, New York, by exotic Daphnia curvirostris using mtDNA from dormant eggs. Limnol. Oceanogr. 45, 1409–1414 (2000).
Mergeay, J., Verschuren, D. & De Meester, L. Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proc. R. Soc. B: Biol. Sci. 273, 2839–2844 (2006).
Brede, N. et al. The impact of human-made ecological changes on the genetic architecture of Daphnia species. Proc. Natl Acad. Sci. USA 106, 4758–4763 (2009).
Limburg, P. A. & Weider, L. J. ‘Ancient’ DNA in the resting egg bank of a microcrustacean can serve as a palaeolimnological database. Proc. R. Soc. B: Biol. Sci. 269, 281–287 (2002).
Radzikowski, J. Resistance of dormant stages of planktonic invertebrates to adverse environmental conditions. J. Plankton Res. 35, 707–723 (2013).
Epp, L. S., Stoof, K. R., Trauth, M. H. & Tiedemann, R. Historical genetics on a sediment core from a Kenyan lake: intraspecific genotype turnover in a tropical rotifer is related to past environmental changes. J. Paleolimnol. 43, 939–954 (2010).
Makino, W., Ohtsuki, H. & Urabe, J. Finding copepod footprints: a protocol for molecular identification of diapausing eggs in lake sediments. Limnology 14, 269–282 (2013).
Lenormand, T. et al. Resurrection ecology in Artemia. Evolut. Appl. 11, 76–87 (2018).
Hairston, N. G. et al. Lake ecosystems—rapid evolution revealed by dormant eggs. Nature 401, 446–446 (1999).
Cousyn, C. et al. Rapid, local adaptation of zooplankton behavior to changes in predation pressure in the absence of neutral genetic changes. Proc. Natl Acad. Sci. USA 98, 6256–6260 (2001).
Stoks, R., Govaert, L., Pauwels, K., Jansen, B. & De Meester, L. Resurrecting complexity: the interplay of plasticity and rapid evolution in the multiple trait response to strong changes in predation pressure in the water flea Daphnia magna. Ecol. Lett. 19, 180–190 (2016).
Lack, J. B., Weider, L. J. & Jeyasingh, P. D. Whole genome amplification and sequencing of a Daphnia resting egg. Mol. Ecol. Resour. 18, 118–127 (2018).
Schaerlaekens, D. G., Dekker, W., Wickstrom, H., Volckaert, F. A. M. & Maes, G. E. Extracting a century of preserved molecular and population demographic data from archived otoliths in the endangered European eel (Anguilla anguilla L.). J. Exp. Mar. Biol. Ecol. 398, 56–62 (2011).
Iwamoto, E. M., Myers, J. M. & Gustafson, R. G. Resurrecting an extinct salmon evolutionarily significant unit: archived scales, historical DNA and implications for restoration. Mol. Ecol. 21, 1567–1582 (2012).
De Meester, L., Louette, G., Duvivier, C., Van Darnme, C. & Michels, E. Genetic composition of resident populations influences establishment success of immigrant species. Oecologia 153, 431–440 (2007).
Roulin, A. C. et al. High genetic variation in resting-stage production in a metapopulation: is there evidence for local adaptation? Evolution 69, 2747–2756 (2015).
Weis, A. E. Detecting the “invisible fraction” bias in resurrection experiments. Evolut. Appl. 11, 88–95 (2018).
Decaestecker, E. et al. Host-parasite ‘Red Queen’ dynamics archived in pond sediment. Nature 450, 870–U816 (2007).
Blainey, P. C. The future is now: single-cell genomics of bacteria and archaea. Fems Microbiol. Rev. 37, 407–427 (2013).
Bolch, C. J. S., Bejoy, T. A. & Green, D. H. Bacterial associates modify growth dynamics of the Dinoflagellate Gymnodinium catenatum. Frontiers Microbiol. 8, https://doi.org/10.3389/fmicb.2017.00670 (2017).
Flynn, J. M., Chain, F. J. J., Schoen, D. J. & Cristescu, M. E. Spontaneous mutation accumulation in Daphnia pulex in selection-free vs. competitive environments. Mol. Biol. Evol. 34, 160–173 (2017).
Berge, T., Daugbjerg, N. & Hansen, P. J. Isolation and cultivation of microalgae select for low growth rate and tolerance to high pH. Harmful Algae 20, 101–110 (2012).
Reyserhove, L. et al. A historical perspective of nutrient change impact on an infectious disease in Daphnia. Ecology 98, 2784–2798 (2017).
Figueroa, R. I., Garces, E., Massana, R. & Camp, J. Description, host-specificity, and strain selectivity of the dinoflagellate parasite Parvilucifera sinerae sp nov (Perkinsozoa). Protist 159, 563–578 (2008).
Torti, A., Lever, M. A. & Jorgensen, B. B. Origin, dynamics, and implications of extracellular DNA pools in marine sediments. Mar. Genomics 24, 185–196 (2015).
Corinaldesi, C., Barucca, M., Luna, G. M. & Dell’Anno, A. Preservation, origin and genetic imprint of extracellular DNA in permanently anoxic deep-sea sediments. Mol. Ecol. 20, 642–654 (2011).
Xu, Z. H. et al. DNA extraction, amplification and analysis of the 28S rRNA portion in sediment-buried copepod DNA in the Great Wall Bay and Xihu Lake, Antarctica. J. Plankton Res. 33, 917–925 (2011).
Borin, S. et al. DNA is preserved and maintains transforming potential after contact with brines of the deep anoxic hypersaline lakes of the Eastern Mediterranean Sea. Saline Syst. 4, 10. https://doi.org/10.1186/1746-1448-4-10 (2008).
Hallsworth, J. E. et al. Limits of life in MgCl2-containing environments: chaotropicity defines the window. Environ. Microbiol. 9, 801–813 (2007).
Domaizon, I., Winegardner, A., Capo, E., Gauthier, J. & Gregory-Eaves, I. DNA-based methods in paleolimnology: new opportunities for investigating long-term dynamics of lacustrine biodiversity. J. Paleolimnol. 58, 1–21 (2017).
Dell’Anno, A. & Danovaro, R. Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 309, 2179–2179 (2005).
Lennon, J. T., Placella, S. A. & Muscarella, M. E. Relic DNA contributes minimally to estimates of microbial diversity. bioRxiv, https://doi.org/10.1101/131284 (2017).
Mulcahy, H., Charron-Mazenod, L. & Lewenza, S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 12, 1621–1629 (2010).
Pinchuk, G. E. et al. Utilization of DNA as a sole source of phosphorus, carbon, and energy by Shewanella spp.: Ecological and physiological implications for dissimilatory metal reduction. Appl. Environ. Microbiol. 74, 1198–1208 (2008).
Corinaldesi, C., Dell’Anno, A. & Danovaro, R. Early diagenesis and trophic role of extracellular DNA in different benthic ecosystems. Limnol. Oceanogr. 52, 1710–1717 (2007).
Levy-Booth, D. J. et al. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 39, 2977–2991 (2007).
Khanna, M. & Stotzky, G. Transformation Of Bacillus-Subtilis By Dna Bound On Montmorillonite And Effect Of Dnase On The Transforming Ability Of Bound Dna. Appl. Environ. Microbiol. 58, 1930–1939 (1992).
Parducci, L. et al. Ancient plant DNA in lake sediments. N. Phytologist 214, 924–942 (2017).
Lejzerowicz, F. et al. Ancient DNA complements microfossil record in deep-sea subsurface sediments. Biol. Lett. 9, https://doi.org/10.1098/rsbl.2013.0283 (2013).
Olajos, F. et al. Estimating species colonization dates using DNA in lake sediment. Methods Ecol. Evol. 9, 535–543 (2018).
Pedersen, M. W. et al. Ancient and modern environmental DNA. Phil. Trans. Roy. Soc. B: Biol. Sci. 370, https://doi.org/10.1098/rstb.2013.0383 (2015).
Birks, H. J. B. & Birks, H. H. How have studies of ancient DNA from sediments contributed to the reconstruction of Quaternary floras? N. Phytologist 209, 499–506 (2016).
Anderson-Carpenter, L. L. et al. Ancient DNA from lake sediments: bridging the gap between paleoecology and genetics. BMC Evolutionary Biol. 11, https://doi.org/10.1186/1471-2148-11-30 (2011).
Capo, E., Debroas, D., Arnaud, F. & Domaizon, I. Is planktonic diversity well recorded in sedimentary DNA? Toward the reconstruction of past protistan diversity. Microb. Ecol. 70, 865–875 (2015).
Boere, A. C., Damste, J. S. S., Rijpstra, W. I. C., Volkman, J. K. & Coolen, M. J. L. Source-specific variability in post-depositional DNA preservation with potential implications for DNA based paleoecological records. Org. Geochem. 42, 1216–1225 (2011).
Marshall, I. P. G., Karst, S. M., Nielsen, P. H. & Jorgensen, B. B. Metagenomes from deep Baltic Sea sediments reveal how past and present environmental conditions determine microbial community composition. Mar. Genomics 37, 58–68 (2018).
Vuillemin, A. et al. Microbial community composition along a 50 000-year lacustrine sediment sequence. Fems Microbiol. Ecol. 94, https://doi.org/10.1093/femsec/fiy029 (2018).
Coolen, M. J. L. & Overmann, J. 217 000-year-old DNA sequences of green sulfur bacteria in Mediterranean sapropels and their implications for the reconstruction of the paleoenvironment (vol 9, pg 238, 2007). Environ. Microbiol. 9, 1099–1099 (2007).
Monchamp, M. E., Walser, J. C., Pomati, F. & Spaak, P. Sedimentary DNA reveals cyanobacterial community diversity over 200 Years in two perialpine lakes. Appl. Environ. Microbiol. 82, 6472–6482 (2016).
Monchamp, M. E. et al. Homogenization of lake cyanobacterial communities over a century of climate change and eutrophication. Nat. Ecol. Evol. 2, 317-+ (2018).
Coolen, M. J. L. & Overmann, J. Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment. Appl. Environ. Microbiol. 64, 4513–4521 (1998).
Puente-Sanchez, F. et al. Viable cyanobacteria in the deep continental subsurface. Proc. Natl Acad. Sci. USA 115, 10702–10707 (2018).
Monchamp, M. E., Spaak, P. & Pomati, F. Long term diversity and distribution of non-photosynthetic cyanobacteria in Peri-Alpine Lakes. Frontiers Microbiol. 9, https://doi.org/10.3389/fmicb.2018.03344 (2019).
Belle, S. et al. Temporal changes in the contribution of methane-oxidizing bacteria to the biomass of chironomid larvae determined using stable carbon isotopes and ancient DNA. J. Paleolimnol. 52, 215–228 (2014).
Madueno, L. et al. A historical legacy of antibiotic utilization on bacterial seed banks in sediments. Peerj 6, https://doi.org/10.7717/peerj.4197 (2018).
Kyle, M., Haande, S., Ostermaier, V. & Rohrlack, T. The red queen race between parasitic chytrids and their host, planktothrix: a test using a time series reconstructed from sediment DNA. Plos ONE 10, https://doi.org/10.1371/journal.pone.0118738 (2015).
Dutilh, B. E. et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 5, https://doi.org/10.1038/ncomms5498 (2014).
Sepulveda, B. P. et al. Marine phage genomics: the tip of the iceberg. FEMS Microbiol. Lett. 363, https://doi.org/10.1093/femsle/fnw158 (2016).
Sabehi, G. et al. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc. Natl Acad. Sci. USA 109, 2037–2042 (2012).
Becks, L., Ellner, S. P., Jones, L. E. & Hairston, N. G. The functional genomics of an eco-evolutionary feedback loop: linking gene expression, trait evolution, and community dynamics. Ecol. Lett. 15, 492–501 (2012).
Adams, C. I. M. et al. Beyond biodiversity: can environmental DNA (eDNA) cut it as a population genetics tool? Genes 10, https://doi.org/10.3390/genes10030192 (2019).
McGowan, S. et al. Controls of algal abundance and community composition during ecosystem state change. Ecology 86, 2200–2211 (2005).
Frenken, T. et al. Integrating chytrid fungal parasites into plankton ecology: research gaps and needs. Environ. Microbiol. 19, 3802–3822 (2017).
Lewis, J. P. et al. The shellfish enigma across the Mesolithic-Neolithic transition in southern Scandinavia. Quat. Sci. Rev. 151, 315–320 (2016).
Saros, J. E., Northington, R. M., Anderson, D. S. & Anderson, N. J. A whole-lake experiment confirms a small centric diatom species as an indicator of changing lake thermal structure. Limnol. Oceanogr. Lett. 1, 27–35 (2016).
Weckstrom, K. in Applications of Paleoenvironmental Techniques in Estuarine Studies Vol. 20 (eds Weckström, K., Gell Saunders, P. & Skilbeck, G.) 615–662 (Kluwer, 2012).
Thomsen, P. F. & Willerslev, E. Environmental DNA—an emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18 (2015).
Allentoft, M. E. et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc. R. Soc. B: Biol. Sci. 279, 4724–4733 (2012).
Author information
Authors and Affiliations
Contributions
All authors were participants in a workshop at Loughborough University in March 2017, led by M.E. and N.J.A., and aimed at debating new possibilities and challenges in using genetic signals stored in aquatic sediment archives as tools in evolutionary ecology. M.E., M.C., T.C., D.F., A.G., A.K., A.L., T.M., S.R., and N.J.A. have subsequently contributed to writing this position paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ellegaard, M., Clokie, M.R.J., Czypionka, T. et al. Dead or alive: sediment DNA archives as tools for tracking aquatic evolution and adaptation. Commun Biol 3, 169 (2020). https://doi.org/10.1038/s42003-020-0899-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-020-0899-z
This article is cited by
-
Millennial-scale variations in Arctic sea ice are recorded in sedimentary ancient DNA of the microalga Polarella glacialis
Communications Earth & Environment (2024)
-
Choice of primer pairs and PCR polymerase affect the detection of fish eDNA
Environmental Sciences Europe (2023)
-
Paleoreconstructions of ciliate communities reveal long-term ecological changes in temperate lakes
Scientific Reports (2022)
-
Reconstruction of 100-year dynamics in Daphnia spawning activity revealed by sedimentary DNA
Scientific Reports (2022)
-
Reviving and characterizing three species of dinoflagellate cysts dormant for about 70 years in the East China Sea: Biecheleria brevisulcata, Biecheleriopsis adriatica, and Scrippsiella donghaienis
Journal of Oceanology and Limnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.