Abstract
Widespread antibiotic usage in apiculture contributes substantially to the global dissemination of antimicrobial resistance and has the potential to negatively influence bacterial symbionts of honey bees (Apis mellifera). Here, we show that routine antibiotic administration with oxytetracycline selectively increased tetB (efflux pump resistance gene) abundance in the gut microbiota of adult workers while concurrently depleting several key symbionts known to regulate immune function and nutrient metabolism such as Frischella perrera and Lactobacillus Firm-5 strains. These microbial changes were functionally characterized by decreased capped brood counts (marker of hive nutritional status and productivity) and reduced antimicrobial capacity of adult hemolymph (indicator of immune competence). Importantly, combination therapy with three immunostimulatory Lactobacillus strains could mitigate antibiotic-associated microbiota dysbiosis and immune deficits in adult workers, as well as maximize the intended benefit of oxytetracycline by suppressing larval pathogen loads to near-undetectable levels. We conclude that microbial-based therapeutics may offer a simple but effective solution to reduce honey bee disease burden, environmental xenobiotic exposure, and spread of antimicrobial resistance.
Similar content being viewed by others
Introduction
Pathogens are considered one of the largest contributing factors in the global decline of honey bees (Apis mellifera) and other wild pollinators, which together help support agricultural economies and international food supplies1,2,3. Managed honey bees represent a substantial fraction of total pollinators and are an important reservoir of enzootic pathogens that can affect disease epidemiology in various animal communities4. To address this issue, beekeepers frequently administer antibiotics to their hives in an attempt to prevent or reduce disease occurrence and control intraspecies pathogen transmission between nearby apiaries.
Oxytetracycline (OTC), tylosin, and fumagillin are the most commonly used antibiotics in apiculture5. Tetracycline-based agents are of particular concern given their extensive usage in human medicine and as bacteriostatic feed additives in livestock. Though indispensable under certain circumstances, overuse of tetracyclines pollutes the environment and inadvertently leads to the accumulation of antibiotic-resistance genes in many bacteria, including a multitude of pathogens relevant to human and honey bee health6. Antibiotic exposure can also negatively impact key symbiotic bacteria within the microbiota (community of microorganisms residing on or within a multicellular organism) of honey bees7. This compromises overall health status in hives as the distinct microbiota in healthy bees is crucial to host metabolic competency, immune regulation, growth and development, and resistance towards parasites and pathogens8.
In many parts of the world, prophylactic usage of OTC is recommended under best management practices for beekeepers in the prevention of foulbrood diseases caused by Paenibacillus larvae and Melissococcus plutonius9. In the case of P. larvae, which causes American foulbrood (AFB), there have been reports of widespread tetracycline resistance occurring since the early 2000’s10. Identical sequence homology of tetracycline-resistance loci found in P. larvae and core members of the honey bee microbiota suggests that there is horizontal gene transfer between commensals and pathogens via mobile genetic elements such as transposons and plasmids11. Importantly, despite susceptibility or resistance, OTC is unable to treat P. larvae spores in the comb, which can act as a source of reinfection. To address these issues and guard against the potentially rapid evolution of resistance, it is prudent to test alternative control measures, which so far include hygienic breeding12, use of bioactive essential oils13, bacteriophage therapy14, synthetic indoles with anti-germination properties15, and supplementation of hives with lactic acid bacteria16,17,18. The promise of this latter approach is up-held by studies demonstrating that lactobacilli can promote insect innate immunity and detoxification19,20,21, extend longevity of adult honey bees22, and stimulate queen brood production23,24.
In our previous work, we demonstrated that pollen patty hive supplements infused with three select strains of lactobacilli (Lactobacillus rhamnosus GR-1, Lactobacillus plantarum Lp39, and Lactobacillus kunkeei BR-1; LX3) could reduce P. larvae loads in honey bee hives experiencing active AFB outbreak and improve honey bee survival towards P. larvae infection in vitro25. Here, we evaluated OTC treatment efficacy in subclinically infected hives and characterized how routine exposure to this common antibiotic impacts immune and microbiota dynamics in honey bees. In addition, we evaluated how treatment augmentation with LX3 following OTC exposure influences antibiotic recovery rates as evaluated by immune functionality, microbial homeostasis, and hive productivity.
Results
LX3 enhances larval pathogen eradication by antibiotics
Prophylactic administration of OTC to honey bees is a common practice in beekeeping for the prevention of AFB. To evaluate the efficacy of this long-standing apiculture management strategy, we monitored a 2-week treatment regimen with OTC under natural field conditions in honey bee hives experiencing low-grade chronic infection with P. larvae (Fig. 1a). Using a qPCR-based approach to enumerate pathogen load, P. larvae abundance was found to be significantly lower in honey bee larvae (primary target of AFB) at week 1 and week 2 of OTC treatment (Kruskal–Wallis with Dunn’s multiple comparisons, P = 0.0071 and P = 0.0005, respectively) compared to baseline levels at day 0 (Fig. 1b). In contrast, no observable differences in P. larvae abundance were found in adult honey bees (active vector of AFB) at any time point during this treatment (Kruskal–Wallis with Dunn’s multiple comparisons, P = 0.9999, P = 0.6367, respectively; Fig. 1c).
Experimental hives were subjected to standard antibiotic treatment with oxytetracycline (OTC) for 2 weeks and then supplemented for 4 weeks with either pollen patties containing LX3 (LX3) or pollen patties containing vehicle (VEH). No treatment control (NTC) hives received no further treatment after OTC. a Schematic diagram outlining the experimental design. b, c Molecular-based quantification of P. larvae in honey bee larvae (whole body) and adults (dissected abdomen) collected just prior to the start of OTC exposure (A.0), and then after 1 (A.1) and 2 (A.2) weeks of exposure. Data are depicted as median ± 95% confidence intervals (Kruskal–Wallis with Dunn’s multiple comparisons) at different time points. Each data point represents either one individual (adults) or three pooled individuals (larvae) sampled equally from a total of n = 6 hives. d, e Molecular-based quantification of P. larvae in larvae (whole body) and adults (dissected whole abdomens) at the start of the supplementation period (S.0; corresponding to 3 days post A.2 time point), and then after 2 (S.2) and 4 (S.4) weeks. Data are depicted as mean ± standard deviation (two-way ANOVA with Sidak’s multiple comparisons) at different time points with each data point representing either one individual (adults) or three pooled individuals (larvae) sampled equally from n = 4 hives per treatment group. f, g Capped brood counts during OTC treatment (n = 6 hives) and subsequent supplementation period (n = 4 hives per treatment group). Data represents the median (line in box), IQR (box), and minimum/maximum (whiskers) of relative change in brood counts normalized by hive. Statistics shown for one-way and two-way ANOVA, respectively, with Sidak’s multiple comparisons for both. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns not significant.
A robust body of scientific evidence shows that supplementation with probiotic Lactobacillus spp. can augment the effects of certain antibiotics and attenuate antibiotic-induced dysbiosis in humans and other animals26,27,28,29. Testing this in honey bees, it was found that larval samples from LX3-treated hives exhibited significantly lower levels of P. larvae at week 2 and week 4 compared to both no treatment control (NTC; P < 0.0001 for both) and vehicle-treated (P = 0.0011 and P = 0.0014) hives, respectively (two-way analysis of variance [ANOVA] with Sidak’s multiple comparisons; Fig. 1d). The NTC group also demonstrated a trend towards higher P. larvae loads in larval samples compared to the vehicle group at week 4 (two-way ANOVA with Sidak’s multiple comparisons, P = 0.0568). Similar results were observed in adults with samples from LX3-supplemented hives demonstrating a significantly lower P. larvae load compared to both the NTC group (P = 0.0002 and P = 0.0003) and vehicle-treated group (P < 0.0001 for both) at 2 and 4 weeks, respectively (two-way ANOVA with Sidak’s multiple comparisons; Fig. 1e).
To compare the effects of OTC and LX3 treatments on overall hive health, the coverage of capped brood on hive frames (an established metric for assessing colony strength and reproduction30) was measured weekly during experimentation. No observable differences were found in capped brood counts following 1 week of OTC treatment, whereas a significant reduction was found after 2 weeks (one-way ANOVA with Sidak’s multiple comparisons, P < 0.9999 and P = 0.0041, respectively) compared to pretreatment baseline measurements (Fig. 1f). In contrast, capped brood counts were significantly higher in LX3-treated hives at week 3 and 4 of the supplementation period (two-way ANOVA with Sidak’s multiple comparisons, P = 0.0071 and P = 0.0055, respectively), while no differences were found in NTC and vehicle-treated hives at comparable time points (Fig. 1g).
Antibiotics reduce key immune regulators in the adult gut microbiota
Given the broad-spectrum activity of tetracyclines, we evaluated how OTC exposure might influence the symbiotic bacterial communities associated with honey bees. Total bacterial loads, as determined by qPCR-based molecular quantification, were significantly reduced in adult bees following 1 week of OTC treatment (P < 0.0001) and in larvae (P = 0.0421) after 2 weeks of treatment (Kruskal–Wallis tests with Dunn’s multiple comparisons; Supplementary Fig. 2). The nurse-aged adult bees sampled at the experimental start point (pre-OTC exposure) and on the final day of OTC treatment (post-OTC exposure) were chosen for 16S rRNA gene sequencing-based microbiota analysis due to their close physical proximity with brood, passive carriage of P. larvae, and well-balanced representation of overall hive microbial diversity31.
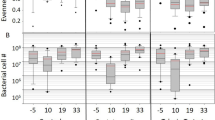
Bar plots shown in Fig. 2a, b visually represent the relative proportion and absolute abundance (adjusted according to qPCR-based quantification of total bacteria) of taxa in samples from pre- and post-OTC exposure, respectively. Adult gut samples collected post-OTC exposure revealed a significant reduction in a single amplicon sequence variant (SV) of Frischella perrera (SV19; Wilcoxon test with Benjamini–Hochberg [BH] multiple comparisons, P = 0.0043) and three unique SVs of Lactobacillus Firm-5 (SV01, SV08, and SV10; Wilcoxon tests with BH multiple comparisons, P = 0.0151, P = 0.0295, and P = 0.04217, respectively) compared to samples collected pre-OTC exposure (absolute effect >0.5; Fig. 2c). SV74 (Lactobacillus Firm-4), SV63 (Bartonella apis), and SV54 (Lactobacillus Firm-4) showed a trend towards decreased relative abundance following OTC exposure (absolute effect <0.5; Fig. 2c).
The gut microbiota of adult honey bees was analyzed before (Pre-OTC) and after (Post-OTC) hive administration with oxytetracycline. a, b Bar plots illustrating the relative and absolute abundance of bacterial species in the gut microbiota of adult honey bee samples as determined by V4 region 16S rRNA gene sequencing. Each bar represents a pooled sample of three adult guts collected from n = 6 hives, with two replicates performed for each hive. Absolute abundance of bacterial taxa was estimated by quantifying total 16S rRNA gene copy number via qPCR. c Strip chart showing differentially abundant taxa in the gut microbiota of OTC-exposed adult honey bees. Positive values indicate an increased relative abundance in response to OTC and negative values indicate a decreased relative abundance. Statistical inference was performed on centered log-ratio transformed read counts of sequence variants using ALDEx2 software in R. Features exceeding absolute effect size (>0.5) and P value (<0.05) thresholds are shown as red. d, e Alpha diversity (measured via Shannon’s H Index) and Beta diversity (measured via Aitchison’s distance between samples at different time points) of adult gut microbiota samples. Data represents the median (line in box), IQR (box), and minimum/maximum (whiskers) of respective microbiota diversity metrics with statistical comparisons shown for separate Wilcoxon tests. f Abundance of seven tetracycline-resistance loci in adult gut samples relative to the total number of 16S rRNA gene copies present. **P < 0.01 and ****P < 0.0001.
The compositional 16S rRNA gene sequencing dataset also demonstrated differences in alpha diversity (intraindividual) and beta diversity (interindividual) metrics of the adult gut microbiota following OTC exposure (Fig. 2d, e). Specifically, Shannon’s H index (balanced alpha diversity metric taking species abundance and evenness into account) was determined to be significantly lower following 2 weeks of OTC treatment (Wilcoxon test, P = 0.0273; Fig. 2d). These findings also corresponded with a decrease in microbiota stability as demonstrated by a significant increase in Aitchison distance (i.e. Euclidean distance after center log-ratio transforming SV read counts) between adults within the same treatment group (Wilcoxon test, P < 0.0001; Fig. 2e).
Next, to determine how the standard practice of treating hives with OTC may influence accumulation of antibiotic-resistance genes in the honey bee gut microbiota, we screened seven tetracycline-resistance loci that have been repeatedly detected in honey bee guts6. These loci included five tetracycline efflux pump genes (tetB, tetC, tetD, tetH, and tetY) and two ribosomal protection protein-encoding genes (tetM and tetW). The abundance of tetB was significantly higher (two-tailed t test, P = 0.0092) whereas tetY showed a trend towards increased abundance (two-tailed Mann–Whitney test, P = 0.0887) in post-OTC adult gut samples (two-tailed Mann–Whitney tests; Fig. 2f). No observable change in abundance were found for any of the remaining five tetracycline-resistance genes examined in this study (Fig. 2f). A positive association was identified between total Gammaproteobacteria (Pearson correlation, r = 0.888, P = 9.25 × 10−17) and Gilliamella apicola (Pearson correlation, r = 0.719, P = 1.23 × 10−08), but not Frischella perrera (Pearson correlation, r = −0.228, P = 0.124), and the presence of tetB (Supplementary Fig. 3).
LX3 improves adult microbiota recovery post-OTC exposure
In humans, probiotic therapy helps to encourage healthy remodeling of the microbiota and can improve recovery following antibiotics32. Here, we tested the ability of LX3 supplementation in honey bees to reduce P. larvae levels and restore microbiota homeostasis in adults and larvae following OTC exposure. As expected, principal component analysis showed clear separation between the microbiota composition of adult and larval samples (Fig. 3a). The largest influencers of separation that were positively associated with adult samples included core microbiota members such as G. apicola, Snodgrassella alvi, F. perrara, Commensalibacter, Lactobacillus Firm-4, and Lactobacillus Firm-5. In contrast, larval samples showed a positive association with mostly opportunistic bacteria including Escherichia/Shigella, Staphylococcus, Enterococcus, Pseudomonas, and P. larvae (Fig. 3a).
a Principle component analysis (PCA) plot of the honey bee microbiota from adult and larval samples before (Pre-supp) and after (Post-supp) the supplementation period. Sequence variants were collapsed at species-level identification, with clr-transformed Aitchison distances used as input values for PCA analysis. Distance between individual samples (points) represent the difference in microbiota composition between samples, with 40.8% of variance explained by the first two principle components shown. Strength of association for taxa are depicted by length of corresponding arrows. b, c ALDEx2 effect plots comparing differences in relative abundance of SVs between groups (ΔA) plotted against the variance, or within-group difference, in relative abundance for each SV (Δw). Low variance SVs that cluster tightly together in adult microbiota samples largely represent well-established core microbiota members (see Supplementary Data 1 for list of corresponding SVs). d Alpha diversity determined by Shannon’s H Index (accounting for abundance and evenness), e Beta diversity measured via Aitchison’s distance (representing within-group microbiota differences), f species dominance (or unevenness) measured via Strong’s Dw Index, and g species richness as determined using the abundance-based coverage estimator (ACE) metric in QIIME2. h, i Differential abundance analysis on adult gut samples between the relative abundance of all core cluster SVs grouped together compared to all noncore SVs grouped together. Data represents median (line in box), IQR(box), and minimum/maximum (whiskers) of clr-transformed relative abundances with statistical comparisons performed by Kruskal–Wallis test with Benjamini–Hochberg multiple comparisons. *P < 0.05, **P < 0.01, ns not significant.
Using ALDEx233 software, Bland–Altmann-like effect plots were generated to investigate the relationship between differences in relative abundance for each SV between treatment groups and the within-group variance of each SV. While no discernible differences were observed within larval samples with an underdeveloped microbiota composition, a tightly clustered group of 13 SVs with low variance were identified in adult samples (Fig. 3b, c). Notably, 12 out of 13 of these SVs trended towards increased abundance in the microbiota of LX3-supplemented adults (Fig. 3b) and consisted of several well-characterized honey bee core microbiota members including five SVs of Lactobacillus Firm-5, two SVs of S. alvi, two SVs of G. apicola, one SV of Lactobacillus Firm-4, one SV of F. perrara, and one SV of Bifidobacterium (see Supplementary Data 1 for full list of SVs). Corroborating these results, LX3 supplementation demonstrated a rescuing effect on microbiota stability as demonstrated by a significant decrease in compositional differences between adult microbiota samples (measured via Aitchison distance; P = 0.0012), whereas no changes were observed in vehicle-treated adults (Kruskal–Wallis test with BH multiple comparisons, P = 0.7570; Fig. 3e).
No differences in overall alpha diversity, as determined by Shannon’s H Index (accounting for species abundance and evenness), were detectable in either vehicle or LX3 treatment groups (Kruskal–Wallis test with BH multiple comparisons, P = 0.2985 and P = 0.2348, respectively; Fig. 3d). However, further investigation demonstrated that species richness alone (determined using the abundance-based coverage estimator [ACE] algorithm in QIIME2) was significantly lower (P = 0.0428) and that compositional dominance (or unevenness, as determined by Strong’s Dw Index) trended towards being higher (P = 0.0535) in LX3-treated adults compared to vehicle-treated adults (Kruskal–Wallis test with BH multiple comparisons; Fig. 3f, g). Since these data did not provide conclusive evidence as to whether core SVs (dominant microbiota members) or noncore SVs (rare species and transient opportunists) were responsible for the observed differences in diversity indices, a nested compositional analysis was performed on total relative abundance for each group. LX3-treated adults demonstrated a significant enrichment in core SVs (effect size = 1.3631) and a reduction in noncore SVs (effect size = −1.3860; Kruskal–Wallis test with BH multiple comparisons, P = 0.0225; Fig. 3h). In contrast, no change was seen in the total relative abundance of core SVs (effect size = 0.1579) or noncore SVs (effect size = −0.1654) in the vehicle-treated controls (Kruskal–Wallis test with BH multiple comparisons, P = 0.2046; Fig. 3i).
LX3 strains are detectable in-hive members post-supplementation
Since 16S rRNA gene sequencing is unable to resolve bacterial taxonomy at the species level, we performed qPCR-based quantification of Lp39, GR-1, and BR-1 using established primer sets25 to confirm that LX3 strains were being effectively dispersed throughout the hive as intended. LX3-supplemented adults and larvae were found to contain significantly higher levels of L. plantarum (multiple t tests, P = 0.0087 and P = 0.0035, respectively) and L. rhamnosus (multiple t tests, P = 0.0002 for both) compared to vehicle-treated control groups (Fig. 4a). In addition, evaluation of bacterial compositions in honey bee larval samples showed that abundance of P. larvae correlated inversely with L. plantarum (r = −0.442, P = 0.006) and L. rhamnosus (r = −0.456, P = 0.006), but not with L. kunkeei (r = 0.060, P = 0.724, Pearson correlations; Fig. 4b).
a Quantification of LX3 lactobacilli strains in honey bee adults and larvae before and after supplementation period. Data are depicted as mean ± standard deviation (two-tailed t tests) of bacterial abundances (determined via qPCR with species-specific primers) at different time points with each data point representing a single individual (n = 18 adult guts per treatment group at each time point) or a pooled sample of three larvae (n = 12 pooled samples for each treatment group at each time point). b Pearson correlation analysis between P. larvae and LX3 lactobacilli strains (quantified via qPCR) in honey bee larval samples. VEH = Pollen patty supplementation with vehicle, LX3 = Pollen patty supplementation with LX3. *P < 0.05, **P < 0.01 and ****P < 0.0001.
LX3 upregulates both head and gut immunity in adult bees
Repeated exposure to antibiotics can weaken immune defenses in honey bees—a phenomenon thought to be facilitated through the reduction of bacterial species important to immunoregulation34. Using an established zone-of-inhibition (ZOI) assay as a crude measure of immune function35, we assessed the inhibitory potential of honey bee-derived hemolymph against Arthrobacter globiformis. The antimicrobial capacity of adult hemolymph was found to be reduced by 31.27% (95% CI = 8.19–54.34%, one-way ANOVA with Sidak’s multiple comparisons, P = 0.0150) following 2 weeks of OTC treatment (Fig. 5a). In contrast, the antimicrobial capacity of hemolymph from LX3-supplemented adults was significantly increased by 121.30% (95% CI = 22.34–220.30%, two-way ANOVA with Sidak’s multiple comparisons, P = 0.0443) after 2 weeks in comparison to vehicle supplemented controls (Fig. 5b).
Adult hemolymph killing capacity against A. globiformis during a antibiotic treatment and b supplementation periods. Data represents the median (line in box), IQR (box), and minimum/maximum (whiskers) of hemolymph killing capacity for n = 6 hives (during antibiotic treatment) and n = 4 hives per treatment group (during supplementation period), respectively. Representative measurements for each hive at each time point were derived from a total of five randomly sampled adult nurse bees. Statistical analysis shown for one-way and two-way ANOVAs, respectively, with Sidak’s multiple comparisons. c Intraindividual head-to-gut gene expression ratios of nine innate immune- or antioxidant-related genes in adult honey bees. Gene expression was quantified by RT-qPCR with gut gene expression shown as relative to head gene expression. Data shown represents the mean ± standard deviation (one-way ANOVA with Sidak’s multiple comparisons) for n = 24 adults. PCA plots and heatmaps demonstrating innate immune- or antioxidant-related gene expression in d, e head and f, g gut samples of adult honey bees before (Pre-supp) and after (Post-supp) the supplementation period. Log2-transformed relative gene expression estimates (determined via qPCR) were used as input values for PCA analyses. Distance between individual samples (points) represent the difference in gene expression profiles for the nine immune or antioxidant genes shown, with 61.6% (heads) and 65.3% (guts) of variance explained by the first two principle components. Strengths of association for each gene are depicted by the length of corresponding arrows. Ellipses indicate 95% confidence intervals for each treatment group. NTC = no treatment control, VEH = pollen patty supplementation with vehicle, LX3 = pollen patty supplementation with LX3. *P < 0.05, ****P < 0.0001, ns not significant. Capped brood counts during OTC treatment (n = 6 hives) and subsequent supplementation period (n = 4 hives per treatment group). Data represents the median (line in box), IQR (box), and minimum/maximum (whiskers) of relative change in brood counts normalized by hive. Statistics shown for one-way and two-way ANOVA, respectively, with Sidak’s multiple comparisons for both. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns not significant.
Previous work has shown that ex situ supplementation of LX3 to honey bee larvae can increase innate immune gene expression of several antimicrobial peptides (AMPs) that control susceptibility to P. larvae infection25. Here, we tested how LX3 supplemented directly to the hive impacted adult bee immunity. Expression of nine well-characterized innate immune- and antioxidant-related genes (defensin-1, defensin-2, hymenoptaecin, apismin, apidaecin, VgMC, catalase, and lysozyme-1) were measured in adult head and gut tissue as their respective anatomical sites are known to play a major role in social and individual immunity36. Exploratory analysis showed that basal gene expression levels in pre-supplemented adult bees demonstrated a vast enrichment of defensin-1 (900.0 ± 163.6-fold higher) and apismin (297,292 ± 63,498-fold higher) in adults heads relative to gut expression, as compared to the other AMP genes examined (one-way ANOVA with Sidak’s multiple comparisons, P < 0.0001 for both; Fig. 5c)—suggesting that changes to the expression of these genes in the head, as opposed to the gut, might produce a more potent immune response and protective benefit at the colony level.
During the supplementation period, head expression of defensin-1, apismin, and apidaecin showed a significant increase over time in only the LX3-supplemented group (two-way ANOVAs with Sidak’s multiple comparisons, P = 0.0406, P < 0.0001, and P = 0.0004, respectively) whereas expression of hymenoptaecin increased in both LX3 and vehicle supplemented groups (two-way ANOVAs with Sidak’s multiple comparisons, P = 0.0240 and P = 0.0365, respectively; Supplementary Fig. 4A, C–E). All experimental groups showed an increase in head expression level of catalase (two-way ANOVA with Sidak’s multiple comparisons, P = 0.0050, P < 0.0001, and P < 0.0001, respectively) over time with no changes in expression of defensin-2 or lysozyme-1 (Supplementary Fig. 4B, H, I). Gut expression of defensin-1, hymenoptaecin, apidaecin, and VgMC were exclusively increased by LX3 supplementation (two-way ANOVAs with Sidak’s multiple comparisons, P = 0.0004, P = 0.0201, P < 0.0001, and P < 0.0001, respectively), whereas lysozyme-1 and catalase expression increased in both vehicle (P = 0.0013 and P < 0.0001, respectively) and LX3 (P < 0.0001 for both) treatment groups (two-way ANOVA with Sidak’s multiple comparisons; Supplementary Fig. 4J, M, N, P–R and Supplementary Data 2). Relative expression of immune- and antioxidant genes in head and gut samples are visually summarized by the PCA plots and heatmaps in Fig. 5d–g.
Correlations between host gene expression and bacterial loads
It has been known for decades now that probiotic bacteria can induce an immune response in honey bees37. In addition, recent evidence shows that host bacterial communities are selectively regulated by the innate immune system of honey bees, and that core microbiota members demonstrate a higher level of resistance to host AMPs than opportunistic bacterial pathogens38. Here, we examine the simultaneous relationship between bacterial abundances and immune- and antioxidant-related gene expression by using a dual extraction-based method to derive RNA and DNA from adult heads and guts. Experimental end point measurements at week 4 of the supplemental period demonstrated a negative Pearson correlation between total abundance of P. larvae and expression of apidaecin (r = −0.589, P = 0.004), apismin (r = −0.483, P = 0.023), hymenoptaecin (r = −0.460, P = 0.036), defensin-1 (r = −0.599, P = 0.004), and catalase (r = −0.650, P = 0.001) in head samples, and with apidaecin (r = −0.559, P = 0.007), hymenoptaecin (r = −0.467, P = 0.029), and catalase (r = −0.589, P = 0.004) in gut samples (Fig. 6).
At the end of the 6-week experimental period, bacterial abundances in the guts of adult honey bees were compared with head and gut expression of nine immune- or antioxidant-related genes. Scale shown represents Pearson correlation coefficient, r, for n = 20–24 individual adults for each comparison. (G) = Gut gene expression, (H) = Head gene expression. The horizontal dendrogram acts to group host genes that covary in their expression patterns while the vertical dendrogram groups bacteria based on their co-occurring abundance relative to host gene expression. Both dendrograms were calculated using Euclidean distance and the complete ‘hclust’ function in R.
Assessment between the supplemented strains of lactobacilli and immune or antioxidant gene expression showed a positive relationship (Pearson correlations) between L. plantarum abundance and head expression of apidaecin (r = 0.653, P = 0.001), defensin-1 (r = 0.526, P = 0.014), and catalase (r = 0.435, P = 0.043), as well as gut expression of defensin-2 (r = 0.401, P = 0.008) catalase (r = 0.591, P = 0.004), and VgMC (r = 0.468, P = 0.028; Fig. 6). For L. rhamnosus, bacterial abundance was associated with increased head expression of apidaecin (r = 0.454, P = 0.039) and a trend towards increased gut expression of defensin-2 (r = 0.401, P = 0.072). No correlations were observed between gene expression and abundance of L. kunkeei. Core microbiota members demonstrated varying relationships with head and gut expression of immune- and antioxidant-related genes in adult honey bees. Total abundance of Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Bifidobacterium, and F. perrara clustered together (based on Euclidean distance of Pearson correlation coefficients) and were found to be mostly associated with increased AMP gene expression (Fig. 6). Oppositely, total abundance of Bacteroidetes, Firmicutes, and G. apicola clustered together and were mostly associated with an overall decrease in immune- and antioxidant-related gene expression irrespective of body site. A notable exception to these trends was the gut expression of apismin and defensin-1 as well as the head expression of defensin-2 and lysozyme-1, which clustered together (Euclidean distance matrix) based on similarities in gene expression patterns relative to bacterial abundances (Fig. 6). Pearson correlation coefficients (and associated statistics) for all relationships between bacterial abundances and host gene expression are provided in Supplementary Data 3.
Discussion
This study demonstrated that in-hive supplementation with LX3 can augment P. larvae clearance rates during chronic low-grade infection and improve honey bee immunity following standard antibiotic treatment. Notably, the combination of OTC treatment followed by LX3 supplementation suppressed P. larvae more effectively than OTC treatment alone (to nearly undetectable levels) in honey bee brood. This combined treatment approach, but not antibiotics alone, also demonstrated the capacity to lower P. larvae loads in the gut microbiota of adult honey bees. Furthermore, LX3 treatment helped to partially restore antibiotic-induced deficits in-hive productivity, beta diversity of the gut microbiota in adult nurse bees, and immune responsiveness of adult-derived hemolymph. These findings expand on previous work in which Lp39, GR-1, and BR-1 strains of lactobacilli were shown to improve survival of laboratory-reared honey bee larvae in an acute infection model of P. larvae, as well as mitigate disease severity during an active AFB outbreak25.
Tetracycline-derived antibiotics have been shown to reduce the abundance39 and genetic diversity7 of core bacterial species in the gut microbiota of caged honey bee workers. Consistent with these reports, OTC directly administered to the hive altered the gut microbiota diversity of adult bees with significant decreases in relative abundances of F. perrara and Lactobacillus Firm-5 post-treatment (Fig. 2). Interestingly, metagenomic sequencing has demonstrated the genomes of Gammaproteobacteria (e.g. F. perrara) and Firmicutes (e.g. Lactobacillus Firm-5) to be specifically enriched with carbohydrate metabolism enzymes that can degrade mannose40—a prevalent plant-derived sugar known to be toxic to honey bees. In addition to the direct toxicity that antibiotics exert on honey bees41, our results suggest that antibiotic exposure may also indirectly increase mortality via depletion of symbionts crucial for the breakdown of toxic food components. The evaluation of LX3 supplementation in hives following cessation of OTC treatment elucidated a partial restorative effect on the adult gut microbiota (Fig. 3b). This effect was structurally characterized by a relative enrichment of core microbiota members and a reduction of transient opportunists (Fig. 3h, i). Together, these findings validate the negative and long-lasting implications of OTC exposure in honey bees while also highlighting a potential role for probiotics in safeguarding against the blooming effect of low-level pathobionts and transient opportunists following antibiotic exposure42.
Although widespread tetracycline resistance has been identified in honey bees6, there is lacking knowledge on how the adult gut metagenome may facilitate pathogen persistence during in-hive antibiotic treatment. This study demonstrated that OTC could effectively lower P. larvae loads in honey bee brood (unstable microbiota profile characterized by low abundance and diversity43), but not adult nurses (well-characterized microbiota profile with high abundance and diversity44) after 2 weeks of in-hive treatment (Fig. 1b, c). Similarly to the reduced diversity of tetracycline-resistance genes observed in OTC-exposed wax moths (Galleria mellonella)45, the relative abundance of five out of seven resistance genes (tetC, tetD, tetH, tetM, and tetW) in adult nurse bees remained unchanged or was diminished by OTC, while the relative abundance of tetB (and to a lesser extent, tetY) was increased (Fig. 2f). These findings suggest that the adult honey bee gut microbiota may provide P. larvae safe harborage against antibiotic exposure—potentially through polymicrobial synergy of multiple distinct resistance factors46 or via physical exclusion47 within the thick stratified biofilm known to be produced by S. alvi (existing in the bottom layer directly associated with host epithelium) and G. apicola (predominantly in the top layer) in the hindgut of bees48. Supporting this postulation, relative abundance of G. apicola was unaffected by OTC treatment (Fig. 2a–c) yet showed a strong positive association with tetB—an efflux pump-encoding gene which itself was shown to increase during OTC exposure (Fig. 2f). Presence of the tetB gene was confirmed in select G. apicola strains wkB1 and PEB01626, though only ~18% of wild isolates are expected to possess tetracycline-resistance genes. Considering that G. apicola strains vary in their ability to beneficially regulate honey bee nutrition49,50, metabolism40,51, and microbiota composition52,53, the consequences of antibiotic selective pressures on intraindividual strain diversity merits further study.
An auxiliary repercussion of antibiotic-induced dysbiosis is perturbation of innate immunity. Adult bees exhibited a reduction in hemolymph antimicrobial capacity over time during the OTC treatment period (Fig. 5a), suggesting a disturbance in their microbial regulation. These findings support past observations of antibiotic-induced susceptibility to opportunistic infection by Serratia marcescens39—a Gram-negative bacterial pathobiont frequently associated with honey bees. Our current findings show that LX3 supplementation broadly upregulated expression of several AMPs (crucial immune effectors involved in pathogen defense) in the heads and guts of adult honey bees (Fig. 5d–g). Defensin-1 is critical in controlling Gram-positive bacteria54 (including P. larvae55) whereas apidaecin and hymenoptaecin are most active against Gram-negative bacteria (including many opportunistic pathogens)56,57. Notably, expression of apidaecin increased by over fivefold in heads (primarily role in social immunity) and 50-fold in guts (primary role in individual immunity) after 4 weeks of LX3 supplementation, while no discernible changes were observed in vehicle and NTC groups.
In a recent report, Kwong et al. showed expression of apidaecin to be uniquely elevated in healthy adults in comparison to microbiota-depleted counterparts38. Moreover, three core phylotype strains of G. apicola, S. alvi, and Lactobacillus Firm-5 were found to upregulate apidaecin (and to a lesser extent hymenoptaecin) expression in gut tissue yet were largely resistant to both of these purified AMPs in vitro38. This suggests that host AMPs may play a role in shaping the microbiota through pathogen exclusion and selective refinement of evolutionarily adapted bacterial species. Supporting this theory, we found that intraindividual expression of apidaecin and several other key immune genes covaried in their expression patterns based on tissue type as well as the abundance of LX3 strains and core phylotype members in the gut (Fig. 6). Whereas for P. larvae, abundance in the adult gut was negatively associated with the expression of most AMPs tested, which suggests a robust host immune response is critical to suppressing this pathogen in the hive. The findings also suggest that the increase in core SVs and decrease in noncore SVs observed in LX3-treated adults (Fig. 3b) were likely a result of upregulated host AMPs (Fig. 5d–g). This evidence cumulatively supports that timely use of probiotics may help in the re-establishment of a healthy microbiota following OTC exposure and concurs with the proposition that immunostimulatory Lactobacillus spp. can ‘reset’ dysbiotic microbiota phenotypes58. Thus, while it has been long known that probiotics can induce an immune response in honey bees37, the functionality of probiotics in modulating AMPs with microbiota-shaping properties is only now becoming apparent. It is important to note, however, that LX3 supplementation was unable to fully restore microbiota composition to pre-OTC baseline levels in terms of overall diversity and structure during the time frame of this study. Though, a limitation to our findings is that low-level residues of antibiotics can persist in the hive for up to 3 months post-treatment39, which may have functionally impeded the corrective potential of treatment. Nevertheless, it remains plausible that targeted modulation of immune components may facilitate host-guided microbiota restoration following exposure to noxious chemicals distributed across apicultural and agricultural landscapes.
Resolving the mechanisms by which supplemental lactobacilli modulate honey bee immunity under normal field conditions, and the subsequent impact this has on their microbiota, is challenging due to the highly complex social interaction networks of adult workers and convoluted microbial dynamics in the hive. Though, innate immunity of insects is highly conserved59 with peptidoglycan recognition proteins (PGRPs) thought to play an important role in bacterial sensing via cell wall component recognition. Interestingly, some but not all Lactobacillus spp. produce diaminopimelic acid (DAP)-containing peptidoglycan60 which is known to activate Imd pathway signaling (leading to downstream production of AMPs) in insects via select PGRP binding61. Corroborating this, past work has shown that Lp39 (one of the LX3 strains used in this study) produces DAP-type peptidoglycan that can potently upregulate host AMP gene expression via Imd pathway signaling in D. melanogaster19. Alternatively, LGR-1 can modulate other divisions of insect innate immunity (e.g. Duox pathway signaling) and ameliorate neonicotinoid-induced suppression of host-generated microbicidal reactive oxygen species21. Its noteworthy to mention that honey bee-derived core lactobacilli members including L. kunkeei, L. apinorum, L. mellifer, L. mellis, L. melliventris, L. kimbladii, L. helsingborgensis, and L. kullabergensis all lack the ability to produce DAP-type peptidoglycan60,62. Accordingly, the absence of this crucial cell wall immune modulator may potentially explain the multiple recent failures at using honey bee-specific lactobacilli to reduce AFB disease severity63,64. Whether L. kunkeei BR-1 affected the overall immune-boosting performance of LX3 is undetermined, though it would be an interesting avenue for future studies given the opposing immune response to various lactobacilli-derived peptidoglycans observed in mouse models65.
In summary, this study has: (i) recapitulated the deleterious effects of antibiotics on the gut microbiota, immunity, and productivity of honey bees, (ii) substantiated the anti-P. larvae properties of LX3 supplemented to subclinically infected hives, and (iii) characterized the transcriptional response of several important immune- and antioxidant-related genes in response to hive supplementation with LX3. These results are not intended to refute the value of antibiotics or discourage their usage under appropriate circumstances. Instead, we suggest that exploiting the multipronged and evolutionarily adapted immune repertoire of honey bees via microbial-based therapeutics could augment current management strategies and offer a more effective method of controlling hive pathogens.
Methods
Culture conditions of bacterial strains
The three strains of lactobacilli used in this study were L. plantarum Lp39 (American Type Culture Collect [ATCC] 14917), L. rhamnosus GR-1 (ATCC 55826), and L. kunkeei BR-1 (honey bee-derived isolate from a healthy hive)25. Unless otherwise stated, routine culturing of these strains was performed under anaerobic conditions at 37 °C using de Man, Rogosa, and Sharpe (catalog number: 288130, BD Difco) broth or agar supplemented with 10 g/L D-fructose (catalog number: F-3510, Sigma-Aldrich; MRS-F). A. globiformis (ATCC 8010) utilized in hemolymph ZOI assays was routinely cultured aerobically at 37 °C using Luria-Bertani (catalog number: DF0446173, BD Difco; LB) broth or agar.
Apiary conditions and experimental design
Two separate experiments were performed in two distinct apiaries maintained within a single geographic region near Western University (London, ON, Canada). These experimental apiaries were selected based on their geographic inclusion (<1 km away; a distance shown to be at risk of contracting high spore loads66) within a boundary assessed to have a recent increase of AFB incidence denoted through provincial apiary inspection by the Ontario Ministry of Agriculture and Food and Ministry of Rural Affairs (OMAFRA). Apiary A (N = 8 Carniolan hives) and apiary B (N = 8 Buckfast hives) were subjected to similar experimental designs between early summer (June–July) and late summer (August–September), respectively (Supplementary Fig. 1). In each case, all hives received standard dusting treatment with OTC hydrochloride (catalog number: 0223111, MEDIVET Pharmaceuticals) for 2 weeks according to manufacturer instructions. Two hives in each apiary were then left to freely interact with experimental hives in an effort to emulate realistic buffering conditions of undisturbed local neighboring colonies. The remaining six hives in each apiary were longitudinally monitored for an additional 4 weeks after being randomly assorted into the following experimental groups: (i) a NTC group that received no supplementation following administration of OTC, (ii) a vehicle control group (VEH) that received a standard 250 g pollen substitute patty (28.5 g of soy flour, 74.1 g of granulated sucrose, 15.4 g of debittered brewer’s yeast, 132.1 g of a 2:1 [w/v] simple sucrose-based syrup solution) with the addition of 4 mL phosphate-buffered saline (0.01 M) once per week, or (iii) a probiotic supplementation group (LX3) that received a 250 g pollen substitute patty infused with Lp39, GR-1, and BR-1 strains (each at a final concentration of 1 × 109 colony forming units [CFU]/g) once per week. All larval samples in this study represent third-to-fifth instar, whereas adult samples represent nurse-aged workers found in close association with the brood. Sampling was performed in a manner to minimize the number of bees removed from the hive during experimentation. Hive tools were flame sterilized prior to use between each of the hives and sterile latex gloves were employed to prevent cross-contamination of LX3 strains and potential pathogens.
Hive sampling procedures
For molecular quantification of P. larvae during OTC exposure, adults and larvae were sampled from n = 6 hives in apiary A with several replicates per hive. For adults, four samples from each hive were collected per time point (0, 1, and 2 weeks) with each replicate consisting of an individual dissected abdomen (n = 24 samples for each time point collected equally across the hives). For larvae, five samples from each hive were collected per time point (day 0, day 7, day 14) with each replicate consisting of a pooled sample of three whole larvae (n = 30 samples for each time point collected equally across the hives). Similarly, for quantification of P. larvae during the post-OTC supplementation period, adults and larvae were sampled from a total of n = 12 hives (six hives from apiary A [n = 2 NTC, n = 2 for vehicle treatment, n = 2 for probiotic treatment] and six hives from apiary B [n = 2 NTC, n = 2 for vehicle treatment, n = 2 for probiotic treatment]). For both adults and larvae, four samples from each hive were collected per time point (0, 2, and 4 weeks) with each replicate consisting of either an individual dissected abdomen or a pooled sample of three whole larvae, respectively (n = 48 adult samples and n = 48 larval samples collected equally across 12 hives at each time point).
For experiments evaluating the adult microbiota in response to OTC exposure, n = 6 hives from apiary A were sampled in biological duplicate (i.e. two samples from each hive with each replicate consisting of three nurse-aged adult guts pooled together) at two separate time points (pre-OTC and post-OTC), which represented a total of 72 individual bees sampled randomly and equally across the hives. The same sampling methodology was also implemented in experiments evaluating the adult and larval gut microbiota during the post-OTC supplementation period, for which a total of n = 8 hives were used (four hives from apiary A [n = 2 for vehicle treatment, n = 2 for probiotic treatment] and four hives from apiary B [n = 2 for vehicle treatment, n = 2 for probiotic treatment]).
For experiments characterizing the relationship between intraindividual bacterial abundances and host innate immune or antioxidant gene expression, adults were sampled at the final time point (week 4 of the supplementation period) from a total of n = 12 hives (six hives from apiary A [n = 2 NTC, n = 2 for vehicle treatment, n = 2 for probiotic treatment] and six hives from apiary B [n = 2 NTC, n = 2 for vehicle treatment, n = 2 for probiotic treatment]). Each hive was sampled in duplicate (i.e. two samples from each hive with each replicate consisting of a single individual nurse-aged adult bee) with each adult sample then subcategorized into head and gut groupings (n = 20–24 total paired head and gut samples used as not all variables measured were detectable in all samples).
Molecular-based quantification of P. larvae
Larval (whole body) samples were evaluated as they represent the target developmental stage for P. larvae infection, whereas adults (dissected abdomen) are passive carriers of P. larvae and provide a good estimate of overall microbial diversity31. Respective samples were surface sterilized using 0.25% sodium hypochlorite and then rinsed in ddH2O for 30 s. DNA was then extracted from samples using the previously described CTAB method53. Bacterial loads were determined by qPCR using the Power SYBR Green PCR Master Mix kit (Applied Biosystems) following manufacturer’s instructions. Universal 16S rRNA gene and species-specific primer sets used in this study are listed in Supplementary Table 1. All qPCR reactions were performed in DNase- and RNase-free 384-well microplates on a QuantStudio 5 Real-Time PCR System (Applied Biosystems) and analyzed with associated QuantStudio Design and Analysis software. Copy numbers of target 16S rRNA genes were calculated as previously described using established primer efficiencies and limits of detection25.
Enumeration of capped brood
To evaluate the efficacy of OTC and LX3 supplementation on hive health dynamics, the coverage of capped brood on hive frames (an established metric for assessing colony strength and reproduction30) was measured weekly during experimentation. Briefly, both sides of every frame in all experimental hives were photographed at the specified sampling time points and then semiautomatically counted using image analysis software as previously described24.
Determination of in vitro antimicrobial capacity of adult hemolymph
Antimicrobial activity of adult hemolymph was determined using an established ZOI assay35 Prior to experimentation, the bacterial indicator stock of A. globiformis was grown aerobically in LB broth at 30 °C for 48 h to a final concentration of 1.6 × 109 CFU/mL. Subsequently, 1 mL of broth culture was used to create a lawn of A. globiformis on LB agar plates. Next, flash frozen adult samples were thawed to 4 °C and hemolymph was extracted via centrifugation (1200 × g for 1 min) following the aseptic removal of antennae. Further, antimicrobial capacity was determined by plating 0.75 μL of hemolymph in preconstructed wells on agar plates that had been freshly seeded with a lawn of A. globiformis. The bottom of each plate was marked with a grid consisting of 12 squares, each 2 × 2 cm. Streptomycin (0.1 g/mL in 80% glycerol) was used at 1:200 and 1:300 dilutions as a positive control to account for batch variation among agar plates35. The agar plates were then incubated aerobically at 30 °C for 48 h prior to measurement of ZOI diameters for hemolymph or antibiotic control wells. ZOI was normalized by hive, and data are shown relative to experimental start points for OTC and LX3 treatment periods.
Preparation of the 16S rRNA gene library
Targeted amplification of the V4 region of the bacteria 16S rRNA gene was achieved using established GOLAY-barcoded primers (5′–3′) ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNxxxxxxxxxxxxGTGCCAGCMGCCGCGGTAA and (5′–3′) CGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCTNNNNxxxxxxxxxxxxGGACTACHVGGGTWTCTAAT wherein ‘xxxxxxxxxxxx’ represents the sample-specific 12-mer barcode following the Illumina adapter sequence used for downstream library construction67. A BioMek Automated Workstation (Beckman Coulter) was then used to transfer 2 µL of sample DNA (5 ng/µL) to a 96-well 0.2-mL PCR plate containing 10 µL of each primer per well (3.2 pmol/µL). Next, 20 µL of GoTaq 2X Colorless Master Mix (Promega) was added to each well and plates were sealed using PCR-grade adhesive aluminum foil. PCR was then performed using a Prime Thermal Cycler (Technie) with the following reaction conditions: an initial activation step at 95 °C, followed by 25 cycles of 95 °C for 1 min, 52 °C for 1 min, and 72 °C for 1 min. After completion, the thermocycler was held at 4 °C, and amplicons were stored at −20 °C until further processing.
Sequencing and analysis of the 16S rRNA gene dataset
Processing of amplicon libraries was conducted at the London Regional Genomics Centre (Robarts Research Institute, London, ON, Canada) in which amplicons were quantified using PicoGreen (Quant-It; Life Technologies, Burlington, ON, Canada), pooled at equimolar ratios, and sequenced on the MiSeq paired-end Illumina platform adapted for 2 × 250 bp paired-end chemistry. Sequence reads were then processed, aligned, and categorized using the DADA2 (v1.8) pipeline to infer exact amplicon SVs from the data68. Briefly, sequence reads were filtered (reads truncated after a quality score of ≤2 and forward/reverse reads truncated after 155/110 bases, respectively) and trimmed (10 bases off 5′ end of reverse reads) using optimized parameter settings as recommended. Next, sequence reads were de-replicated, de-noised, and merged using DADA2 default parameters. After omitting PCR blank control samples, the dataset used to assess the effects of OTC on the adult microbiota consisted of 24 nurse bee-derived gut samples and a total of 1,109,022 reads with an average count of 46,209 reads per sample. Following quality assurance measures described in the DADA2 pipeline69, an average of 13.94% reads were removed from each sample, leaving a total of 955,475 filtered reads. The dataset which evaluated the impact of probiotic treatment following antibiotic exposure consisted of 16 nurse bee-derived gut samples (372,198 total reads with an average of 23,262 reads per sample) and 16 larval samples (59,335 total reads with an average of 3708 reads per sample). Following quality control, an average of 11.63% and 31.29% reads were removed from adult and larval datasets, respectively. SV read counts were left in their unadulterated state for analysis (i.e. copy number was not corrected) to prevent additional noise in the dataset70. Taxonomic designations were assigned to SVs using the SILVA nonredundant training set (V132) configured for DADA2. Phylotype identity was determined by cross referencing with a previously established honey bee-specific seed alignment of 276 unique representatives71. Raw sequence reads are uploaded to the NCBI Sequence Read Archive and accessible under BioProject ID PRJNA610196.
Simultaneous extraction of RNA and DNA from individual honey bees
Dissected head and abdomen samples from adult honey bees were surface sterilized using 0.25% sodium hypochlorite and then rinsed in ddH2O for 30 s. Subsequently, samples were homogenized in TRIzol (Invitrogen) by beat beating and RNA extracted following manufacturer’s instructions. Using the same samples, DNA was also extracted from the TRIzol homogenates using a back-extraction buffer consisting of 4 M guanidine thiocyanate, 50 mM sodium citrate dihydrate, and 1 M Tris base as previously described72. Quality of RNA and DNA was assessed using a microvolume spectrophotometer (DS-11 Spectrophotometer; DeNovix) and only samples determined to have A260/280 absorbance ratios between 1.9 and 2.2 were considered for further analyses.
Measurement of host gene expression and molecular quantification of bacterial communities
To determine host gene expression, a total of 1500 ng of the extracted RNA from adult head and gut samples was reverse transcribed to cDNA using a High-Capacity cDNA Reverse Transcription Kit following manufacturer’s instructions (Applied Biosystems, catalog number: 4368813). RT-qPCR reactions were performed with tenfold-diluted cDNA using the Power SYBR Green kit (Applied Biosystems) following manufacturer’s instructions. Oligonucleotide primers used to evaluate host expression of innate immune and antioxidant-related genes are listed in Supplementary Table 2. Honey bee alpha-tubulin was determined to be the most stably expressed endogenous control gene (compared to ribosomal protein S5, microsomal glutathione-S-transferase, and UDP-glucuronyltransferase) under experimental conditions in this study, and thus was chosen as the internal standard for normalization as per MIQE guidelines73. To measure bacterial abundance in samples, qPCR reactions were performed with tenfold-diluted DNA (from back extraction) using the Power SYBR Green kit (Applied Biosystems) following manufacturer’s instructions. The universal 16S rRNA gene, phylotype-specific, and species-specific primer sets that were used for molecule quantification of bacteria are listed in Supplementary Table 1. All qPCR reactions were performed in DNase- and RNase-free 384-well microplates using a QuantStudio 5 Real-Time PCR System (Applied Biosystems) and analyzed with associated QuantStudio Design and Analysis software. Relative gene expression was calculated using the 2−ΔΔ Ct method74 whereas bacterial abundance (measured via copy number of target 16S rRNA genes) was calculated using previously established primer efficiencies and limits of detection44,52,53.
Statistics and reproducibility
All compositional analyses on 16S rRNA gene sequencing datasets were performed using QIIME2 (v2020.2) or in R (v3.6.0) using ALDEx2 software (v1.18.0). Statistics for all other datasets were performed using GraphPad Prism (v8.3.0). Datasets with unique values were tested for normality using the Shapiro–Wilk test, whereas datasets with ties (two or more identical values) were tested for normality using the D’Agostino–Pearson test. Normally distributed data were statistically compared with two-tailed t tests, Pearson correlations, one-way ANOVAs, or two-way ANOVAs as indicated. ANOVA tests were complemented with Sidak’s multiple comparisons when appropriate. Nonparametric datasets were statistically compared using Wilcoxon, Mann–Whitney, or Kruskal–Wallis tests and complemented with Dunn’s multiple comparisons (quantitative datasets) or BH false discovery rate method (compositional datasets) when appropriate. Sample sizes and replicate details are described in the relevant ‘Methods’ sections.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
References
Otterstatter, M. C. & Thomson, J. D. Does pathogen spillover from commercially reared bumble bees threaten wild pollinators? PLoS One 3, e2771 (2008).
Goulson, D., Nicholls, E., Botías, C. & Rotheray, E. L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347, 1255957 (2015).
Smith, K. M. et al. Pathogens, pests, and economics: drivers of honey bee colony declines and losses. EcoHealth 10, 434–445 (2013).
Fürst, M. A., McMahon, D. P., Osborne, J. L., Paxton, R. J. & Brown, M. J. F. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 506, 364–366 (2014).
Piva, S. et al. Could honey bees signal the spread of antimicrobial resistance in the environment? Lett. Appl. Microbiol. 70, 349–355 (2020).
Tian, B., Fadhil, N. H., Powell, J. E., Kwong, W. K. & Moran, N. A. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio. 3, e00377–12 (2012).
Raymann, K., Bobay, L.-M. & Moran, N. A. Antibiotics reduce genetic diversity of core species in the honeybee gut microbiome. Mol. Ecol. 27, 2057–2066 (2018).
Raymann, K. & Moran, N. A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci. 26, 97–104 (2018).
Arbia, A. & Babbay, B. Management strategies of honey bee diseases. J. Entomol. 8, 1–15 (2011).
Murray, K. D. & Aronstein, K. A. Oxytetracycline-resistance in the honey bee pathogen Paenibacillus larvae is encoded on novel plasmid pMA67. J. Apic. Res. 45, 207–214 (2006).
Evans, J. D. Diverse origins of tetracycline resistance in the honey bee bacterial pathogen Paenibacillus larvae. J. Invertebr. Pathol. 83, 46–50 (2003).
Spivak, M. & Reuter, G. S. Resistance to American foulbrood disease by honey bee colonies Apis mellifera bred for hygienic behavior. Apidologie 32, 555–565 (2001).
González, M. J. & Marioli, J. M. Antibacterial activity of water extracts and essential oils of various aromatic plants against Paenibacillus larvae, the causative agent of American Foulbrood. J. Invertebr. Pathol. 104, 209–213 (2010).
Beims, H. et al. Paenibacillus larvae-directed bacteriophage HB10c2 and its application in American Foulbrood-affected honey bee larvae. Appl Env. Microbiol. 81, 5411–5419 (2015).
Alvarado, I. et al. Inhibitory effect of indole analogs against Paenibacillus larvae, the causal agent of American foulbrood disease. J. Insect Sci. 17, 104–112 (2017).
Arredondo, D. et al. Lactobacillus kunkeei strains decreased the infection by honey bee pathogens Paenibacillus larvae and Nosema ceranae. Benef. Microbes. 9, 279–290 (2017).
Chmiel, J. A., Daisley, B. A., Pitek, A. P., Thompson, G. J. & Reid, G. Understanding the effects of sublethal pesticide exposure on honey bees: a role for probiotics as mediators of environmental stress. Front. Ecol. Evol. 8, e00022 (2020).
Mudroňová, D. et al. Lactobacillus sp. as a potential probiotic for the prevention of Paenibacillus larvae infection in honey bees. J. Apic. Res. 50, 323–324 (2011).
Daisley, B. A. et al. Neonicotinoid-induced pathogen susceptibility is mitigated by Lactobacillus plantarum immune stimulation in a Drosophila melanogaster model. Sci. Rep. 7, 2703 (2017).
Trinder, M. et al. Probiotic Lactobacillus rhamnosus reduces organophosphate pesticide absorption and toxicity to Drosophila melanogaster. Appl. Environ. Microbiol. 82, 6204–6213 (2016).
Chmiel, J. A., Daisley, B. A., Burton, J. P. & Reid, G. Deleterious effects of neonicotinoid pesticides on Drosophila melanogaster immune pathways. mBio 10, e01395–19 (2019).
Kaznowski, A. et al. The effects of probiotic supplementation on the content of intestinal microflora and chemical composition of worker honey bees (Apis mellifera). J. Apic. Res. 44, 10–14 (2005).
Alberoni, D. et al. Impact of beneficial bacteria supplementation on the gut microbiota, colony development and productivity of Apis mellifera L. Benef. Microbes. 9, 269–278 (2018).
Audisio, M. & Benítez-Ahrendts, M. Lactobacillus johnsonii CRL1647, isolated from Apis mellifera L. bee-gut, exhibited a beneficial effect on honeybee colonies. Benef. Microbes. 2, 29–34 (2011).
Daisley, B. A. et al. Novel probiotic approach to counter Paenibacillus larvae infection in honey bees. ISME J. 14, 476–491 (2019).
Reid, G. Probiotics to prevent the need for, and augment the use of, antibiotics. Can. J. Infect. Dis. Med. Microbiol. 17, 291–295 (2006).
Du, Y.-Q. et al. Adjuvant probiotics improve the eradication effect of triple therapy for Helicobacter pylori infection. World J. Gastroenterol. WJG 18, 6302–6307 (2012).
Verdú, E. F. et al. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut 55, 182–190 (2006).
Tarasova, E. et al. The influence of probiotic Enterococcus faecium strain L5 on the microbiota and cytokines expression in rats with dysbiosis induced by antibiotics. Benef. Microbes. 1, 265–270 (2010).
Delaplane, K. S., Steen, Jvander & Guzman-Novoa, E. Standard methods for estimating strength parameters of Apis mellifera colonies. J. Apic. Res. 52, 1–12 (2013).
Erban, T. et al. Honeybee (Apis mellifera)-associated bacterial community affected by American foulbrood: detection of Paenibacillus larvae via microbiome analysis. Sci. Rep. 7, 5084 (2017).
Reid, G. et al. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38 (2011).
Fernandes, A. D., Macklaim, J. M., Linn, T. G., Reid, G. & Gloor, G. B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-seq. PLoS One 8, e67019 (2013).
Li, J. H. et al. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee’s vulnerability to Nosema infection. PLoS One 12, e0187505 (2017).
Harpur, B. A. et al. No genetic tradeoffs between hygienic behaviour and individual innate immunity in the honey bee, Apis mellifera. PLoS One 9, e104214 (2014).
López-Uribe, M. M., Fitzgerald, A. & Simone-Finstrom, M. Inducible versus constitutive social immunity: examining effects of colony infection on glucose oxidase and defensin-1 production in honeybees. R. Soc. Open Sci. 4, 170224 (2017).
Evans, J. D. & Lopez, D. L. Bacterial probiotics induce an immune response in the honey bee (Hymenoptera: Apidae). J. Econ. Entomol. 97, 752–756 (2004).
Kwong, W. K., Mancenido Amanda, A. L. & Moran, N. A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 4, 170003 (2017).
Raymann, K., Shaffer, Z. & Moran, N. A. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 15, e2001861 (2017).
Engel, P., Martinson, V. G. & Moran, N. A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl Acad. Sci. USA 109, 11002–11007 (2012).
Peng, Y.-S. C., Mussen, E., Fong, A., Montague, M. A. & Tyler, T. Effects of chlortetracycline of honey bee worker larvae reared in vitro. J. Invertebr. Pathol. 60, 127–133 (1992).
Stecher, B., Maier, L. & Hardt, W.-D. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 11, 277–284 (2013).
Mohr, K. I. & Tebbe, C. C. Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ. Microbiol. 8, 258–272 (2006).
Martinson, V. G., Moy, J. & Moran, N. A. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 78, 2830–2840 (2012).
Ignasiak, K. & Maxwell, A. Oxytetracycline reduces the diversity of tetracycline-resistance genes in the Galleria mellonella gut microbiome. BMC Microbiol. 18, 228 (2018).
Burmølle, M. et al. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 72, 3916–3923 (2006).
Davies, D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114–122 (2003).
Koch, H. & Schmid-Hempel, P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl Acad. Sci. 108, 19288–19292 (2011).
Zheng, H. et al. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio. 7, e01326–16 (2016).
Engel, P. & Moran, N. A. Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis. Gut Microbes 4, 60–65 (2013).
Zheng, H., Powell, J. E., Steele, M. I., Dietrich, C. & Moran, N. A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl Acad. Sci. 114, 4775–4780 (2017).
Kešnerová, L. et al. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 15, e2003467 (2017).
Powell, J. E., Martinson, V. G., Urban-Mead, K. & Moran, N. A. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 80, 7378–7387 (2014).
Ilyasov, R., Gaifullina, L., Saltykova, E., Poskryakov, A. & Nikolenko, A. Review of the expression of antimicrobial peptide defensin in honey bees Apis Mellifera L. J. Apic. Sci. 56, 115–124 (2012).
Khilnani, J. C. & Wing, H. J. Protocols to test the activity of antimicrobial peptides against the honey bee pathogen Paenibacillus larvae. J. Microbiol. Methods 117, 54–56 (2015).
Li, W.-F., Ma, G.-X. & Zhou, X.-X. Apidaecin-type peptides: biodiversity, structure–function relationships and mode of action. Peptides 27, 2350–2359 (2006).
Casteels, P., Ampe, C., Jacobs, F. & Tempst, P. Functional and chemical characterization of Hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honeybee (Apis mellifera). J. Biol. Chem. 268, 7044–7054 (1993).
Daisley, B. A., Chmiel, J. A., Pitek, A. P., Thompson, G. J. & Reid, G. Missing microbes in bees: how systematic depletion of key symbionts erodes immunity. Trends Microbiol. https://doi.org/10.1016/j.tim.2020.06.006. (2020)
Wang, Q., Ren, M., Liu, X., Xia, H. & Chen, K. Peptidoglycan recognition proteins in insect immunity. Mol. Immunol. 106, 69–76 (2019).
Felis, G. E. & Dellaglio, F. Taxonomy of lactobacilli and bifidobacteria. Curr. Issues Intest. Microbiol. 8, 44–61 (2007).
Lesperance, D. N. & Broderick, N. A. Microbiomes as modulators of Drosophila melanogaster homeostasis and disease. Curr. Opin. Insect Sci. 39, 84–90 (2020).
Olofsson, T. C., Alsterfjord, M., Nilson, B., Butler, È. & Vásquez, A. Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int. J. Syst. Evol. Microbiol. 64, 3109–3119 (2014).
Stephan, J. G. et al. Honeybee-specific lactic acid bacterium supplements have no effect on American Foulbrood-infected honeybee colonies. Appl. Environ. Microbiol. 85, e00606–e00619 (2019).
Lamei, S. et al. Feeding honeybee colonies with honeybee-specific lactic acid bacteria (Hbs-LAB) does not affect colony-level Hbs-LAB composition or Paenibacillus larvae spore levels, although American Foulbrood affected colonies harbor a more diverse Hbs-LAB community. Microb. Ecol. 79, 743–755 (2020).
Shida, K., Kiyoshima-Shibata, J., Kaji, R., Nagaoka, M. & Nanno, M. Peptidoglycan from lactobacilli inhibits interleukin-12 production by macrophages induced by Lactobacillus casei through Toll-like receptor 2-dependent and independent mechanisms. Immunology 128, e858–e869 (2009).
Lindström, A., Korpela, S. & Fries, I. Horizontal transmission of Paenibacillus larvae spores between honey bee (Apis mellifera) colonies through robbing. Apidologie 39, 515–522 (2008).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Callahan, B. J., McMurdie, P. J. & Holmes, S. P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Louca, S., Doebeli, M. & Parfrey, L. W. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome 6, 41 (2018).
Newton, I. L. & Roeselers, G. The effect of training set on the classification of honey bee gut microbiota using the Naive Bayesian Classifier. BMC Microbiol. 12, 221 (2012).
Triant, D. A. & Whitehead, A. Simultaneous extraction of high-quality RNA and DNA from small tissue samples. J. Hered. 100, 246–250 (2009).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Acknowledgements
This work was funded by the Government of Canada through a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant and the Ontario Ministry of Agriculture, Food and Rural Affairs (OMAFRA) through a New Directions Research Program grant.
Author information
Authors and Affiliations
Contributions
B.D., A.P., G.T., and G.R. conceived the study design. A.P., J.C., A.C., K.F., and G.T. managed the experimental hives, administered treatments, and collected honey bee samples. B.D. performed all RNA/DNA extractions, enumerated P. larvae loads in adult and larval samples, and analyzed the majority of data for all experiments. A.P. performed laboratory assays to measure hemolymph antimicrobial capacity. S.G. performed qPCR experiments to molecularly quantify core bacterial loads and antibiotic-resistance genes in honey bee adults. K.F. and A.C. enumerated brood counts from hive frame photos. K.A. and J.P. provided scientific input and resources for 16S rRNA gene sequencing experiments. K.A. assisted with microbiota data interpretation and figure design. B.D. drafted the manuscript and all authors revised it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Daisley, B.A., Pitek, A.P., Chmiel, J.A. et al. Lactobacillus spp. attenuate antibiotic-induced immune and microbiota dysregulation in honey bees. Commun Biol 3, 534 (2020). https://doi.org/10.1038/s42003-020-01259-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-020-01259-8
This article is cited by
-
A longitudinal field study of commercial honey bees shows that non-native probiotics do not rescue antibiotic treatment, and are generally not beneficial
Scientific Reports (2024)
-
Disentangling the gut bacterial communities of the agave weevil, Scyphophorus acupunctatus (Coleoptera: Curculionidae)
Symbiosis (2024)
-
Prospects of probiotics in beekeeping: a review for sustainable approach to boost honeybee health
Archives of Microbiology (2024)
-
Delivery mechanism can enhance probiotic activity against honey bee pathogens
The ISME Journal (2023)
-
Bacterial and Fungal Symbionts in Pollen Provisions of a Native Solitary Bee in Urban and Rural Environments
Microbial Ecology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.