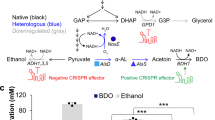
Abstract
Metabolic engineering holds the promise to transform the chemical industry and to support the transition into a circular bioeconomy, by engineering cellular biocatalysts that efficiently convert sustainable substrates into desired products. However, despite decades of research, the potential of metabolic engineering has only been realized to a limited extent at the industrial level. To further realize its potential, it is essential to optimize the synthetic and native metabolic networks of cell factories at a system and genome-wide level. Here we discuss the tools and strategies enabling system-wide (semi-) rational engineering. Recent advances in genome-editing technologies enable directed genome-wide engineering in a growing number of relevant microorganisms. Such system-wide engineering can benefit from machine learning and other in silico design methods, and it needs to be integrated with efficient screening or selection approaches. These approaches are expected to realize the promise of next-generation cell factories for efficient, sustainable production of a wide range of products.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bailey, J. E. Toward a science of metabolic engineering. Science 252, 1668–1675 (1991).
Casini, A., Storch, M., Baldwin, G. S. & Ellis, T. Bricks and blueprints: methods and standards for DNA assembly. Nat. Rev. Mol. Cell Biol. 16, 568–576 (2015).
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016).
Wu, W. Y., Lebbink, J. H. G., Kanaar, R., Geijsen, N. & van der Oost, J. Genome editing by natural and engineered CRISPR-associated nucleases. Nat. Chem. Biol. 14, 642–651 (2018).
Mougiakos, I., Bosma, E. F., Ganguly, J., van der Oost, J. & van Kranenburg, R. Hijacking CRISPR-Cas for high-throughput bacterial metabolic engineering: advances and prospects. Curr. Opin. Biotechnol. 50, 146–157 (2018).
Wannier, T. et al. Recombineering and MAGE. Nat. Rev. Methods Prim. 1, 7 (2021). This review provides an overview on the mechanism, applications and optimal experimental strategies of recombineering and specifically MAGE, in diverse organisms.
Liao, J. C., Mi, L., Pontrelli, S. & Luo, S. Fuelling the future: microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 14, 288–304 (2016).
Claassens, N. J. et al. Harnessing the power of microbial autotrophy. Nat. Rev. Microbiol. 14, 692–706 (2016).
Lee, S. Y. et al. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2, 18–33 (2019). This review article gives a comprehensive overview of industrial production for many bio-based products including their metabolic routes, current performances and challenges.
Choi, K. R. et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 37, 817–837 (2019).
de Jong, E., Stichnothe, H., Bell, G. & Jørgensen, H. Bio-Based Chemicals: a 2020 Update (IEA Bioenergy, 2020).
Oxford Economics. The Global Chemical Industry: Catalyzing Growth and Addressing Our World’s Sustainability Challenges (Oxford Economics, 2019); https://www.oxfordeconomics.com/recent-releases/the-global-chemical-industry-catalyzing-growth-and-addressing-our-world-sustainability-challenges
Blanco, J., Iglesias, J., Morales, G., Melero, J. A. & Moreno, J. Comparative life cycle assessment of glucose production from maize starch and woody biomass residues as a feedstock. Appl. Sci. 10, 2946 (2020).
Naz, T. et al. Transformation of lignocellulosic biomass into sustainable biofuels: major challenges and bioprocessing technologies. Am. J. Biochem. Biotechnol. 16, 308–327 (2020).
Fackler, N. et al. Stepping on the gas to a circular economy: accelerating development of carbon-negative chemical production from gas fermentation. Annu. Rev. Chem. Biomol. Eng. 12, 439–470 (2021).
Cotton, C. A. R., Claassens, N. J., Benito-Vaquerizo, S. & Bar-even, A. Renewable methanol and formate as microbial feedstocks. Curr. Opin. Biotechnol. 62, 168–180 (2020).
Wehrs, M. et al. Engineering robust production microbes for large-scale cultivation. Trends Microbiol. 27, 524–537 (2019).
Lee, J. W. et al. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat. Chem. Biol. 8, 536–546 (2012).
Lee, S. Y. & Kim, H. U. Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 33, 1061–1072 (2015).
Sandberg, T. E., Salazar, M. J., Weng, L. L., Palsson, B. O. & Feist, A. M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 56, 1–16 (2019).
Wu, Y., Jameel, A., Xing, X. H. & Zhang, C. Advanced strategies and tools to facilitate and streamline microbial adaptive laboratory evolution. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2021.04.002 (2021).
Biz, A. et al. Systems biology based metabolic engineering for non-natural chemicals. Biotechnol. Adv. 37, 107379 (2019).
Alper, H. & Stephanopoulos, G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab. Eng. 9, 258–267 (2007).
Alper, H., Moxley, J., Nevoigt, E., Fink, G. R. & Stephanopoulos, G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314, 1565–1568 (2006).
Liu, R. et al. Directed combinatorial mutagenesis of Escherichia coli for complex phenotype engineering. Metab. Eng. 47, 10–20 (2018). A demonstration of the use of iCREATE to target ~40,000 mutations across 30 diverse genes in parallel, to optimized C5/C6 sugar co-utilization and tolerance to furfural and acetate in E. coli, which occur in lignocellulose hydrolyates.
Liang, L. et al. Genome engineering of E. coli for improved styrene production. Metab. Eng. 57, 74–84 (2020).
Watson, E., Yilmaz, L. S. & Walhout, A. J. M. Understanding metabolic regulation at a systems level: metabolite sensing, mathematical predictions and model organisms. Annu. Rev. Genet. 49, 553–575 (2015).
Banos, D. T., Trébulle, P. & Elati, M. Integrating transcriptional activity in genome-scale models of metabolism. BMC Syst. Biol. 11, 81–90 (2017).
Nielsen, J. Systems biology of metabolism. Annu. Rev. Biochem. 86, 245–275 (2017).
Stalidzans, E., Seiman, A., Peebo, K., Komasilovs, V. & Pentjuss, A. Model-based metabolism design: constraints for kinetic and stoichiometric models. Biochem. Soc. Trans. 46, 261–267 (2018).
O’Brien, E. J., Monk, J. M. & Palsson, B. O. Using genome-scale models to predict biological capabilities. Cell 161, 971–987 (2015).
Burgard, A. P., Pharkya, P. & Maranas, C. D. OptKnock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84, 647–657 (2003).
Von Kamp, A. & Klamt, S. Growth-coupled overproduction is feasible for almost all metabolites in five major production organisms. Nat. Commun. 8, 15956 (2017).
Banerjee, D. et al. Genome-scale metabolic rewiring improves titers rates and yields of the non-native product indigoidine at scale. Nat. Commun. 11, 5385 (2020).
Sánchez, B. J. et al. Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017).
Kim, O. D., Rocha, M. & Maia, P. A review of dynamic modeling approaches and their application in computational strain optimization for metabolic engineering. Front. Microbiol. 9, 1690 (2018).
Lee, Y., Lafontaine Rivera, J. G. & Liao, J. C. Ensemble modeling for robustness analysis in engineering non-native metabolic pathways. Metab. Eng. 25, 63–71 (2014).
Chen, F. Y. H., Jung, H. W., Tsuei, C. Y. & Liao, J. C. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell 182, 933–946 (2020). This article reports the kinetic-modelling informed design, rational construction, ALE and realization of full growth on methanol in E. coli via the RuMP pathway.
Zhou, H., Vonk, B., Roubos, J. A., Bovenberg, R. A. L. & Voigt, C. A. Algorithmic co-optimization of genetic constructs and growth conditions: application to 6-ACA, a potential nylon-6 precursor. Nucleic Acids Res. 43, 10560–10570 (2015).
Xu, P., Rizzoni, E. A., Sul, S. Y. & Stephanopoulos, G. Improving metabolic pathway efficiency by statistical model-based multivariate regulatory metabolic engineering. ACS Synth. Biol. 6, 148–158 (2017).
Young, E. M. et al. Iterative algorithm-guided design of massive strain libraries, applied to itaconic acid production in yeast. Metab. Eng. 48, 33–43 (2018).
Jervis, A. J. et al. Machine learning of designed translational control allows predictive pathway optimization in Escherichia coli. ACS Synth. Biol. 8, 127–136 (2019).
Zhou, Y. et al. MiYA, an efficient machine-learning workflow in conjunction with the YeastFab assembly strategy for combinatorial optimization of heterologous metabolic pathways in Saccharomyces cerevisiae. Metab. Eng. 47, 294–302 (2018).
Zhang, J. et al. Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism. Nat. Commun. 11, 4880 (2020). Good illustration of integrated system metabolic engineering approaches in yeast, based on modeling to identify five gene targets, Cas9-based editing of promoters variants for these genes, recording data with a tryptophan biosensor and using generated data for machine learning for further improve tryptophan production.
Lawson, C. E. et al. Machine learning for metabolic engineering: a review. Metab. Eng. 63, 34–60 (2021).
Volk, M. J. et al. Biosystems design by machine learning. ACS Synth. Biol. 9, 1514–1533 (2020).
Kim, G. B., Kim, W. J., Kim, H. U. & Lee, S. Y. Machine learning applications in systems metabolic engineering. Curr. Opin. Biotechnol. 64, 1–9 (2020).
Li, F. et al. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction. Nat. Catal. 5, 662–672 (2022).
Urtecho, G. et al. Genome-wide functional characterization of Escherichia coli promoters and regulatory elements responsible for their function. Preprint at https://www.biorxiv.org/content/10.1101/2020.01.04.894907v1 (2020).
La Fleur, T., Hossain, A. & Salis, H. M. Automated model-predictive design of synthetic promoters to control transcriptional profiles in bacteria. Preprint at https://www.biorxiv.org/content/10.1101/2021.09.01.458561v1 (2021).
Wang, H. H. et al. Genome-scale promoter engineering by coselection MAGE. Nat. Methods 9, 591–593 (2012).
Yona, A. H., Alm, E. J. & Gore, J. Random sequences rapidly evolve into de novo promoters. Nat. Commun. 9, 1530 (2018).
Nyerges, Á. et al. Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance. Proc. Natl Acad. Sci. USA 115, E5726–E5735 (2018).
Cetnar, D. P. & Salis, H. M. Systematic quantification of sequence and structural determinants controlling mRNA stability in bacterial operons. ACS Synth. Biol. 10, 318–332 (2021).
Nieuwkoop, T., Finger-Bou, M., van der Oost, J. & Claassens, N. J. The ongoing quest to crack the genetic code for protein production. Mol. Cell 80, 193–209 (2020).
Zelcbuch, L. et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 41, e98 (2013).
Wang, H. H. et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009). This landmark study established the principles of MAGE in E. coli and demonstrated its strong potential for targeted engineering of 24 genes involved in lycopene biosynthesis.
Reis, A. C. & Salis, H. M. An automated model test system for systematic development and improvement of gene expression models. ACS Synth. Biol. 9, 3145–3156 (2020).
Farasat, I. et al. Efficient search, mapping and optimization of multi‐protein genetic systems in diverse bacteria. Mol. Syst. Biol. 10, 731 (2014).
Jeschek, M., Gerngross, D. & Panke, S. Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort. Nat. Commun. 7, 11163 (2016).
Bonde, M. T. et al. Predictable tuning of protein expression in bacteria. Nat. Methods 13, 233–236 (2016).
Höllerer, S. et al. Large-scale DNA-based phenotypic recording and deep learning enable highly accurate sequence-function mapping. Nat. Commun. 11, 3551 (2020).
Kozak, M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13–37 (2005).
De Nijs, Y., De Maeseneire, S. L. & Soetaert, W. K. 5′ untranslated regions: the next regulatory sequence in yeast synthetic biology. Biol. Rev. 95, 517–529 (2020).
Nieuwkoop, T., Claassens, N. J. & van der Oost, J. Improved protein production and codon optimization analyses in Escherichia coli by bicistronic design. Microb. Biotechnol. 12, 173–179 (2018).
Rennig, M. et al. TARSyn: tunable antibiotic resistance devices enabling bacterial synthetic evolution and protein production. ACS Synth. Biol. 7, 432–442 (2018).
Arnold, F. H. Directed evolution: bringing new chemistry to life. Angew. Chem. Int. Ed. 57, 4143–4148 (2018).
Copeland, N. G., Jenkins, N. A. & Court, D. L. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2, 769–779 (2001).
Bassalo, M. C. et al. Rapid and efficient one-step metabolic pathway integration in E. coli. ACS Synth. Biol. 5, 561–568 (2016).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Sharan, S. K., Thomason, L. C., Kuznetsov, S. G. & Court, D. L. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2010).
Gallagher, R. R., Li, Z., Lewis, A. O. & Isaacs, F. J. Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat. Protoc. 9, 2301–2316 (2014).
Nyerges, Á. et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc. Natl Acad. Sci. USA 113, 2502–2507 (2016).
Bonde, M. T. et al. Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides. ACS Synth. Biol. 4, 17–22 (2015).
Aparicio, T., Nyerges, A., Martínez-García, E. & de Lorenzo, V. High-efficiency multi-site genomic editing of Pseudomonas putida through thermoinducible ssDNA recombineering. iScience 23, 100946 (2020).
Chang, Y., Wang, Q., Su, T. & Qi, Q. The efficiency for recombineering is dependent on the source of the phage recombinase function unit. Preprint at https://www.biorxiv.org/content/10.1101/745448v1.full.pdf (2019).
van Pijkeren, J. P., Neoh, K. M., Sirias, D., Findley, A. S. & Britton, R. A. Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered https://doi.org/10.4161/bioe.2104 (2012).
Filsinger, G. T. et al. Characterizing the portability of phage-encoded homologous recombination proteins. Nat. Chem. Biol. 17, 394–402 (2021).
Wannier, T. M. et al. Improved bacterial recombineering by parallelized protein discovery. Proc. Natl Acad. Sci. USA 117, 13689–13698 (2020). This study provides a platform for identifying efficient single-stranded annealing proteins and allowed for more efficient recombineering in E. coli and several other bacteria.
Hueso-Gil, A., Nyerges, Á., Pál, C., Calles, B. & De Lorenzo, V. Multiple-site diversification of regulatory sequences enables interspecies operability of genetic eevices. ACS Synth. Biol. 9, 104–114 (2020).
Barbieri, E. M., Muir, P., Akhuetie-Oni, B. O., Yellman, C. M. & Isaacs, F. J. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell 171, 1453–1467 (2017).
Dicarlo, J. E. et al. Yeast oligo-mediated genome engineering (YOGE). ACS Synth. Biol. 2, 741–749 (2013).
Liang, Z., Metzner, E. & Isaacs, F. J. Advanced eMAGE for highly efficient combinatorial editing of a stable genome. Preprint at https://www.biorxiv.org/content/10.1101/2020.08.30.256743v1 (2020).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337, 816–821 (2012).
Doudna, J. A. & Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014).
Adiego-Pérez, B. et al. Multiplex genome editing of microorganisms using CRISPR-Cas. FEMS Microbiol. Lett. 366, 1–19 (2019). This review provides a comprehensive overview of CRISPR-Cas techniques and their performance for multiplexing in prokaryotic and eukaryotic microorganisms.
Zhang, Z. X. et al. Recent advances in the application of multiplex genome editing in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 105, 3873–3882 (2021).
Kim, H., Ji, C., Je, H., Kim, J. & Kang, H. mpCRISTAR: multiple plasmid approach for CRISPR/Cas9 and TAR-mediated multiplexed refactoring of natural product biosynthetic gene clusters. ACS Synth. Biol. 9, 175–180 (2020).
Lian, J., Schultz, C., Cao, M., HamediRad, M. & Zhao, H. Multi-functional genome-wide CRISPR system for high throughput genotype–phenotype mapping. Nat. Commun. 10, 5794 (2019).
Napolitano, M. G. et al. Emergent rules for codon choice elucidated by editing rare arginine codons in Escherichia coli. Proc. Natl Acad. Sci. USA 113, E5588–E5597 (2016).
Ronda, C., Pedersen, L. E., Sommer, M. O. A. & Nielsen, A. T. CRMAGE: CRISPR optimized MAGE recombineering. Sci. Rep. 6, 19452 (2016).
Oesterle, S., Gerngross, D., Schmitt, S., Roberts, T. M. & Panke, S. Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli. Sci. Rep. 7, 12327 (2017).
Asin-Garcia, E., Martin-Pascual, M., Garcia-Morales, L., Van Kranenburg, R. & Martins Dos Santos, V. A. P. ReScribe: an unrestrained tool combining multiplex recombineering and minimal-PAM ScCas9 for genome recoding Pseudomonas putida. ACS Synth. Biol. 10, 2672–2688 (2021).
Jiang, Y. et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514 (2015).
Pyne, M. E., Moo-Young, M., Chung, D. A. & Chou, C. P. Coupling the CRISPR/Cas9 system with lambda red recombineering enables simplified chromosomal gene replacement in Escherichia coli. Appl. Environ. Microbiol. 81, 5103–5114 (2015).
Garst, A. D. et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35, 48–55 (2017). Extensive work that established the CREATE platform for high-throughput targeted and trackable genome engineering in E. coli based on CRISPR-Cas9 and recombineering.
Liu, R. et al. Iterative genome editing of Escherichia coli for 3-hydroxypropionic acid production. Metab. Eng. 47, 303–313 (2018).
Roy, K. R. et al. Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat. Biotechnol. 36, 512–520 (2018).
Gaudelli, N. M. et al. Programmable base editing of A–T to G–C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Banno, S., Nishida, K., Arazoe, T., Mitsunobu, H. & Kondo, A. Deaminase-mediated multiplex genome editing in Escherichia coli. Nat. Microbiol. 3, 423–429 (2018).
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Wang, Y. et al. In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing. Nat. Commun. 12, 678 (2021).
Volke, D. C., Martino, R. A., Kozaeva, E., Smania, A. M. & Nikel, P. I. Modular (de)construction of complex bacterial phenotypes by CRISPR/nCas9-assisted, multiplex cytidine base-editing. Nat. Commun. 13, 3026 (2022). This work established and demonstated an efficient base-editing tool for multiplex editing in the emerging model organism P. putida.
Tong, Y., Jørgensen, T. S., Whitford, C. M., Weber, T. & Lee, S. Y. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat. Commun. 12, 5206 (2021).
Reis, A. C. et al. Simultaneous repression of multiple bacterial genes using nonrepetitive extra-long sgRNA arrays. Nat. Biotechnol. 37, 1294–1301 (2019).
Cui, L. et al. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 9, 1912 (2018).
Si, T., Luo, Y., Bao, Z. & Zhao, H. RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering. ACS Synth. Biol. 4, 283–291 (2015).
Na, D. et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 31, 170–174 (2013).
Xie, W. H., Deng, H. K., Hou, J. & Wang, L. J. Synthetic small regulatory RNAs in microbial metabolic engineering. Appl. Microbiol. Biotechnol. 105, 1–12 (2021).
Kiattisewee, C. et al. Portable bacterial CRISPR transcriptional activation enables metabolic engineering in Pseudomonas putida. Metab. Eng. 66, 283–295 (2021).
Liu, Y., Wan, X. & Wang, B. Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria. Nat. Commun. 10, 3693 (2019).
Lin, J. L., Wagner, J. M. & Alper, H. S. Enabling tools for high-throughput detection of metabolites: metabolic engineering and directed evolution applications. Biotechnol. Adv. 35, 950–970 (2017).
Leavell, M. D., Singh, A. H. & Kaufmann-Malaga, B. B. High-throughput screening for improved microbial cell factories, perspective and promise. Curr. Opin. Biotechnol. 62, 22–28 (2020).
Sarnaik, A., Liu, A., Nielsen, D. & Varman, A. M. High-throughput screening for efficient microbial biotechnology. Curr. Opin. Biotechnol. 64, 141–150 (2020).
Chen, L., Sun, G.-G., Fang, H.-T. & He, K. in DEStech Transactions on Engineering and Technology Research 110–117 (DEStech Publications, 2019); https://doi.org/10.12783/dtetr/amee2019/33444
Yang, H. et al. Study on fluorescence spectra of thiamine, riboflavin and pyridoxine. In Proc. Seventh International Symposium on Precision Mechanical Measurements 99030H (SPIE, 2016); https://doi.org/10.1117/12.2211248
Sesen, M. & Whyte, G. Image-based single cell sorting automation in droplet microfluidics. Sci. Rep. 10, 8736 (2020).
Fu, L., Zhang, J. & Si, T. Recent advances in high-throughput mass spectrometry that accelerates enzyme engineering for biofuel research. BMC Energy 2, 1 (2020).
Häbe, T. T. et al. Ultrahigh-throughput ESI-MS: sampling pushed to six samples per second by acoustic ejection mass spectrometry. Anal. Chem. 92, 12242–12249 (2020).
Sinclair, I., Davies, G. & Semple, H. Acoustic mist ionization mass spectrometry (AMI-MS) as a drug discovery platform. Expert Opin. Drug Discov. 14, 609–617 (2019).
Wyatt Shields Iv, C., Reyes, C. D. & López, G. P. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 15, 1230–1249 (2015).
Bouzetos, E., Ganar, K. A., Mastrobattista, E., Deshpande, S. & van der Oost, J. (R)evolution-on-a-chip. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2021.04.009 (2021).
Caen, O. et al. High-throughput multiplexed fluorescence-activated droplet sorting. Microsyst. Nanoeng. 4, 33 (2018).
Guo, L., Zeng, W., Xu, S. & Zhou, J. Fluorescence-activated droplet sorting for enhanced pyruvic acid accumulation by Candida glabrata. Bioresour. Technol. 318, 124258 (2020).
Liang, W. F. et al. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply. Metab. Eng. 39, 159–168 (2017). This study nicely demonstrated the high-throughput targeting of a global regulator for improving mevalonate production by using an in vivo sensor for this product in a non-conventional, methylotrophic bacterium.
van Rossum, T., Kengen, S. W. M. & van der Oost, J. Reporter-based screening and selection of enzymes. FEBS J. 280, 2979–2996 (2013).
Rogers, J. K. & Church, G. M. Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl Acad. Sci. USA 113, 2388–2393 (2016).
Fang, M. et al. Intermediate-sensor assisted push-pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli. Metab. Eng. 33, 41–51 (2016).
Xu, P. et al. Design and kinetic analysis of a hybrid promoter-regulator system for malonyl-CoA sensing in Escherichia coli. ACS Chem. Biol. 9, 451–458 (2014).
Tang, S.-Y. & Cirino, P. C. Design and application of a mevalonate-responsive regulatory protein. Angew. Chem. 123, 1116–1118 (2011).
Rogers, J. K., Taylor, N. D. & Church, G. M. Biosensor-based engineering of biosynthetic pathways. Curr. Opin. Biotechnol. 42, 84–91 (2016).
Raman, S., Rogers, J. K., Taylor, N. D. & Church, G. M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl Acad. Sci. USA 111, 17803–17808 (2014). This study demontrates the strenght of combining rational genome-wide targetting (MAGE) with product biosensors controlling selectable markers, leading to many-fold improved naringenin and glucaric acid production in E. coli.
Orsi, E., Claassens, N. J., Nikel, P. I. & Lindner, S. N. Growth-coupled selection of synthetic modules to accelerate cell factory development. Nat. Commun. 12, 5295 (2021).
Kim, S. et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat. Chem. Biol. 16, 538–545 (2020).
Antonovsky, N. et al. Sugar synthesis from CO2 in Escherichia coli. Cell 166, 115–125 (2016).
Guzmán, G. I. et al. Enzyme promiscuity shapes adaptation to novel growth substrates. Mol. Syst. Biol. 15, e8462 (2019).
Saleski, T. E. et al. Syntrophic co-culture amplification of production phenotype for high- throughput screening of microbial strain libraries. Metab. Eng. 54, 232–243 (2019).
Patinios, C. et al. Streamlined CRISPR genome engineering in wild-type bacteria using SIBR-Cas. Nucleic Acids Res. 49, 11392–11404 (2021).
Chao, R., Mishra, S., Si, T. & Zhao, H. Engineering biological systems using automated biofoundries. Metab. Eng. 42, 98–108 (2017).
Hillson, N. et al. Building a global alliance of biofoundries. Nat. Commun. 10, 2040 (2019).
Isaacs, F. J. et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science 6040, 348–353 (2017).
Ostrov, N. et al. Design, synthesis and testing toward a 57-codon genome. Science 353, 819–822 (2016).
Szili, P. et al. Rapid evolution of reduced susceptibility against a balanced dual-targeting antibiotic through stepping-stone mutations. Antimicrob. Agents Chemother. 63, e00207–e00219 (2019).
Campa, C. C., Weisbach, N. R., Santinha, A. J., Incarnato, D. & Platt, R. J. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods 16, 887–893 (2019).
Jung, S. W., Yeom, J., Park, J. S. & Yoo, S. M. Recent advances in tuning the expression and regulation of genes for constructing microbial cell factories. Biotechnol. Adv. 50, 107767 (2021).
Acknowledgements
We thank R. van Kranenburg and E. Orsi for critical reading of this manuscript. S.Y., N.J.C. and J.v.d.O. acknowledge the support of the Dutch Research Council (NWO) via the Gravitation Project BaSyC (024.003.019) and Spinoza (SPI 93-537), awarded to J.v.d.O. In addition, N.J.C. acknowledges support from his NWO Veni fellowship (VI.Veni.192.156). Funding for this research was also provided by the US Department of Energy (DOE) under grant no. DE-FG02-02ER63445 and by the National Science Foundation (NSF) award no. 2123243 (both to G.M.C.). A.N. was supported by the EMBO LTF 160-2019 Long-Term fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.v.d.O. is included as inventor on several CRISPR-related patents and is scientific advisor of NTrans Technologies, Scope Biosciences and Hudson River Biotechnology. G.M.C. is a founder of companies in which he has related financial interests: ReadCoor, EnEvolv and 64x Bio. For a complete list of G.M.C.’s financial interests, see also https://arep.med.harvard.edu/gmc/tech.html. A.N. is an inventor on a patent related to directed evolution with random genomic mutations (DIvERGE) (US patent 10669537B2: Mutagenizing intracellular nucleic acids). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Catalysis thanks Sang Yup Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yilmaz, S., Nyerges, A., van der Oost, J. et al. Towards next-generation cell factories by rational genome-scale engineering. Nat Catal 5, 751–765 (2022). https://doi.org/10.1038/s41929-022-00836-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-022-00836-w
This article is cited by
-
Enabling whole pathway reconstruction using artificial chromosomes
Cell Research (2024)