Abstract
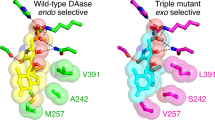
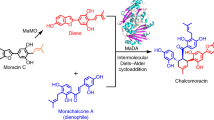
Natural Diels–Alderases that selectively form endo or exo products are increasingly well known but generally form one stereoisomer with limited substrate scope. Here we report the discovery of two homologous groups of flavin-adenine-dinucleotide-dependent enzymes that catalyse intermolecular Diels–Alder reactions on the same substrates with opposite endo/exo selectivity and high enantioselectivity. We show that these enzymes are effective biocatalysts with a wide range of diene and dienophile substrates. The crystal structure of an exo-selective Diels–Alderase was determined at 2.94 Å resolution. Based on the structure and computational investigation of the catalytic mechanism, we designed and prepared mutant enzymes that reverse the stereoselectivity from exo to endo. A combination of structure-based comparison, computational and mutational studies have revealed two different catalytic mechanisms that control the endo/exo selectivity in these enzymatic Diels–Alder reactions.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available with this article and its Supplementary Information. The gene sequences of MaDA-1, MaDA-2 and MaDA-3 are deposited in GenBank under accession nos MW620986, MW620984 and MW620985, respectively. The structural factors and coordinates of MaDA-3 are deposited in the PDB under no. 7E2V. All other data are available from the authors upon reasonable request. Source data are provided with this paper.
References
Diels, O. & Alder, K. Synthesen in der hydroaromatischen Reihe. Justus Liebigs Ann. Chem. 460, 98–122 (1928).
Nicolaou, K. C., Snyder, S. A., Montagnon, T. & Vassilikogiannakis, G. The Diels–Alder reaction in total synthesis. Angew. Chem. Int. Ed. 41, 1668–1698 (2002).
Kagan, H. B. & Riant, O. Catalytic asymmetric Diels Alder reactions. Chem. Rev. 92, 1007–1019 (1992).
Corey, E. J. Catalytic enantioselective Diels–Alder reactions: methods, mechanistic fundamentals, pathways, and applications. Angew. Chem. Int. Ed. 41, 1650–1667 (2002).
Corey, E. J., Imwinkelried, R., Pikul, S. & Xiang, Y. B. Practical enantioselective Diels–Alder and aldol reactions using a new chiral controller system. J. Am. Chem. Soc. 111, 5493–5495 (1989).
Narasaka, K. et al. Asymmetric Diels–Alder reaction catalysed by a chiral titanium reagent. J. Am. Chem. Soc. 111, 5340–5345 (1989).
Han, J. et al. Enantioselective biomimetic total syntheses of kuwanons I and J and brosimones A and B. Angew. Chem. Int. Ed. 53, 9257–9261 (2014).
Gatzenmeier, T. et al. Asymmetric Lewis acid organocatalysis of the Diels–Alder reaction by a silylated C–H acid. Science 351, 949–952 (2016).
Nakashima, D. & Yamamoto, H. Design of chiral N-triflyl phosphoramide as a strong chiral Brønsted acid and its application to asymmetric Diels–Alder reaction. J. Am. Chem. Soc. 128, 9626–9627 (2006).
Ahrendt, K. A., Borths, C. J. & MacMillan, D. W. C. New strategies for organic catalysis: the first highly enantioselective organocatalytic Diels–Alder reaction. J. Am. Chem. Soc. 122, 4243–4244 (2000).
Corey, E. J. & Loh, T.-P. First application of attractive intramolecular interactions to the design of chiral catalysts for highly enantioselective Diels–Alder reactions. J. Am. Chem. Soc. 113, 8966–8967 (1991).
Hayashi, Y., Rohde, J. J. & Corey, E. J. A novel chiral super-Lewis acidic catalyst for enantioselective synthesis. J. Am. Chem. Soc. 118, 5502–5503 (1996).
Ishihara, K. & Nakano, K. Design of an organocatalyst for the enantioselective Diels–Alder reaction with α-acyloxyacroleins. J. Am. Chem. Soc. 127, 10504–10505 (2005).
Kano, T., Tanaka, Y. & Maruoka, K. exo-Selective asymmetric Diels–Alder reaction catalysed by diamine salts as organocatalysts. Org. Lett. 8, 2687–2689 (2006).
Klas, K., Tsukamoto, S., Sherman, D. H. & Williams, R. M. Natural Diels–Alderases: elusive and irresistable. J. Org. Chem. 80, 11672–11685 (2015).
Jeon, B. S., Wang, S.-A., Ruszczycky, M. W. & Liu, H. W. Natural [4+2]-cyclases. Chem. Rev. 117, 5367–5388 (2016).
Jamieson, C. S., Ohashi, M., Liu, F., Tang, Y. & Houk, K. N. The expanding world of biosynthetic pericyclase: cooperation of experiment and theory for discovery. Nat. Prod. Rep. 36, 698–713 (2019).
Kim, H. J., Ruszczycky, M. W., Choi, S. H., Liu, Y. N. & Liu, H. W. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature 473, 109–112 (2011).
Tian, Z. et al. An enzymatic [4+2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol. 11, 259–265 (2015).
Byrne, M. J. et al. The catalytic mechanism of a natural Diels–Alderase revealed in molecular detail. J. Am. Chem. Soc. 138, 6095–6098 (2016).
Li, L. et al. Biochemical characterization of a eukaryotic decalin-forming Diels–Alderase. J. Am. Chem. Soc. 138, 15837–15840 (2016).
Li, L. et al. Genome mining and assembly-line biosynthesis of the UCS1025A pyrrolizidinone family of fungal alkaloids. J. Am. Chem. Soc. 140, 2067–2071 (2018).
Kato, N. et al. Control of the stereochemical course of [4+2] cycloaddition during trans-decalin formation by Fsa2-family enzymes. Angew. Chem. Int. Ed. 57, 9754–9758 (2018).
Li, Q. et al. Nonspecific heme-binding cyclase, AbmU, catalyses [4+2] cycloaddition during neoabyssomicin biosynthesis. ACS Omega 5, 20548–20557 (2020).
Hantke, V., Skellam, E. J. & Cox, R. J. Evidence for enzyme catalysed intramolecular [4+2] Diels–Alder cyclization during the biosynthesis of pyrichalasin H. Chem. Commun. 56, 2925–2598 (2020).
Caputi, L. et al. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 360, 1235–1239 (2018).
Gao, L. et al. FAD-dependent enzyme-catalysed intermolecular [4+2] cycloaddition in natural product biosynthesis. Nat. Chem. 12, 620–628 (2020).
Siegel, J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science 329, 309–313 (2010).
Basler, S. et al. Efficient Lewis acid catalysis of an abiological reaction in a de novo protein scaffold. Nat. Chem. 13, 231–235 (2021).
Hilvert, D., Hill, K. W., Nared, K. D. & Auditor, M.-T. M. Antibody catalysis of the Diels–Alder reaction. J. Am. Chem. Soc. 111, 9261–9262 (1989).
Gouverneur, V. E. et al. Control of the exo and endo pathways of the Diels-Alder reaction by antibody catalysis. Science 262, 204–208 (1993).
Tan, D. et al. Genome-mined Diels–Alderase catalyses formation of the cis-octahydrodecalins of varicidin A and B. J. Am. Chem. Soc. 141, 769–773 (2019).
Hashimoto, T. et al. Biosynthesis of versipelostatin: identification of an enzyme-catalysed [4+2]-cycloaddition required for macrocyclization of spirotetronate-containing polyketides. J. Am. Chem. Soc. 137, 572–575 (2015).
Zou, Y. et al. Computational investigation of the mechanism of Diels–Alderase PyrI4. J. Am. Chem. Soc. 142, 20232–20239 (2020).
Caputi, L. et al. Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids. Nat. Chem. Biol. 16, 383–386 (2020).
Heine, A. et al. An antibody exo Diels–Alderase inhibitor complex at 1.95 angstrom resolution. Science 279, 1934–1940 (1998).
Yang, Y., Tan, Y.-X., Chen, R.-Y. & Kang, J. The latest review on the polyphenols and their bioactivities of Chinese Morus plants. J. Asian Nat. Prod. Res. 16, 690–702 (2014).
Kumar, K. et al. Cation–π interactions in protein–ligand binding: theory and data-mining reveal different roles for lysine and arginine. Chem. Sci. 9, 2655–2665 (2018).
Tantillo, D. J., Chen, J. & Houk, K. N. Theozymes and compuzymes: theoretical models for biological catalysis. Curr. Opin. Chem. Biol. 2, 743–750 (1998).
Ujaque, G. et al. Catalysis on the coastline: theozyme, molecular dynamics, and free energy perturbation analysis of antibody 21D8 catalysis of the decarboxylation of 5-nitro-3-carboxybenzisoxazole. J. Comput. Chem. 24, 98–110 (2003).
Fujiyama, K. et al. Molecular basis for two stereoselective Diels–Alderases that produce decalin skeletons. Angew. Chem. Int. Ed. 60, 22401–22410 (2021).
Sato, M. et al. Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels–Alderase. Nat. Catal. 4, 223–232 (2021).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Frisch, M. J. et al. Gaussian 16, v.A.03 (Gaussian, 2016).
Grimme, S. Supramolecular binding thermodynamics by dispersion-corrected density functional theory. Chem. Eur. J. 18, 9955–9964 (2012).
Takano, Y. & Houk, K. N. Benchmarking the conductor-like polarizable continuum model (CPCM) for aqueous solvation-free energies of neutral and ionic organic molecules. J. Chem. Theory Comput. 1, 70–77 (2005).
Spartan’18 (Wavefunction, 2018).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Pierce, L. C. et al. Routine access to millisecond time scale events with accelerated molecular dynamics. J. Chem. Theory Comput. 8, 2997–3002 (2012).
Case, D. A. et al. AMBER 2016 (University of California, San Francisco, 2016).
Case, D. A. et al. AMBER 2018 (University of California, San Francisco, 2018).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Bayly, C. I., Cieplak, P., Cornell, W. D. & Kollman, P. A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 97, 10269–10280 (1993).
Besler, B. H., Merz, K. M. & Kollman, P. A. Atomic charges derived from semiempirical methods. J. Comput. Chem. 11, 431–439 (1990).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Kelley, L. A., Gardner, S. P. & Sutcliffe, M. J. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng. Des. Sel. 9, 1063–1065 (1996).
Acknowledgements
We thank J. Zhou (Peking University) for assistance with high-resolution MS analysis; X. D. Su and B. Wang for assistance with data collection and structure determination of MaDA-3; and Q. Ding for bioinformatic analysis. Anton 2 computer time was provided by the Pittsburgh Supercomputing Center through grant no. R01GM116961 from the National Institutes of Health. The Anton 2 machine at the Pittsburgh Supercomputing Center was generously made available by D. E. Shaw Research. L.G. is supported in part by a Postdoctoral Fellowship of Peking-Tsinghua Center for Life Sciences. We are grateful to the staff members of the Shanghai Synchrotron Radiation Facility (beamline BL17U) for their support during X-ray data collection. This project was supported by the National Key Research & Development Plan (2017YFA0505200 to X. Lei), National Natural Science Foundation of China grants (21625201, 21961142010, 21661140001, 91853202 and 21521003 to X. Lei; 22101009 to L.G.; 22177006 to J.F.) and the Beijing Outstanding Young Scientist Program (BJJWZYJH01201910001001 to X. Lei).
Author information
Authors and Affiliations
Contributions
X. Lei, K.N.H. and L.G. initiated the project. X. Lei, K.N.H., L.G., Y.Z., X. Liu and J.Y. conceived and designed the experiments, analysed the data and prepared the manuscript with input from all the authors. X. Liu and J.W. synthesized the substrates and racemic D–A adducts. L.G. performed the large-scale enzymatic reactions. L.G. and X.Y. cloned and expressed the MaDA-1, MaDA-2 and MaDA-3 genes and biochemically characterized MaDA-1, MaDA-2 and MaDA-3. X.D. and J.F. performed the X-ray crystallization and data analysis of MaDA-3. Y.Z. performed the docking and MD studies. M.J. and Y.L. performed the DFT calculations with the help of Y.Z.; J.Y. performed the protein mutagenesis and expression. X. Lei and K.N.H. managed the whole project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Catalysis thanks Jiri Damborsky, Bernhard Hauer and Bernhard Loll for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Functional and biochemical characterization of Diels-Alderases using dienophile 7 and diene 10.
a, In vitro analysis of 7 and 10 at 50 °C for 10 min with buffer (i), 137 nM MaDA (ii), 139 nM MaDA-1 (iii), 207 nM MaDA-2 (iv), 206 nM MaDA-3 (v), and standard compounds 2 and 5 (vi). Three experiments were repeated independently with similar results. b, Kinetic analysis of MaDA-1 for dienophile 7. c, Kinetic analysis of MaDA-1 for diene 10. d, Kinetic analysis of MaDA-3 for dienophile 7. e, Kinetic analysis of MaDA-3 for diene 10. KM, kcat, kcat/KM and Ki values represent the mean ± SD. Error bars represent standard deviations of three independent measurements.
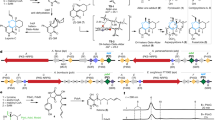
Extended Data Fig. 2 Chemo-enzymatic syntheses of Diels-Alder type natural products using MaDA-1 and MaDA-2/3.
All enzymatic products of MaDA-1 were determined as endo products since their NMR data and optical rotations are consistent with those of the Diels-Alder type natural products with endo configuration. Similarly, all enzymatic products of MaDA-2 and MaDA-3 were determined as exo products. The ee values for all the enzymatic products were determined as >99%. The detailed protocols and chiral HPLC analysis were described in the chemical synthesis part below. Note: chemo-enzymatic syntheses of all the D-A products with exo configuration were achieved using MaDA-3 except mulberrofuran J and albafuran C.
Extended Data Fig. 3 Investigation of the diene scope.
For the endo-selective Diels-Alder reaction, the enzymatic reaction containing 20 mM Tris•HCl and 9.6 μg MaDA-1 was run at pH = 8.0 and 55 °C for 5 min. For the exo-selective Diels-Alder reaction, the enzymatic reaction containing 20 mM Tris•HCl and 9.6 μg MaDA-3 was run at pH = 7.0 and 50 °C for 5 min. The conversion rates of the diene were calculated from the ratios between peak areas of dienes and the internal peak areas in the enzymatic reaction mixture and its negative control. Note: s.i. means only single product was observed with no or trace of its stereoisomer observed in UPLC analysis (endo/exo or exo/endo > 50 :1).
Extended Data Fig. 4 The endo/exo selectivities of MaDA or MaDA-1 variant involved enzymatic reactions.
a, Relative enzymatic activities of MaDA-1 and its variants using dienophile 7 and diene 8 as substrates. The exo/endo ratio of the MaDA-1-G283R variant-catalyzed reaction was about 1:108. b, Relative enzymatic activities of MaDA and its variants of G294 using dienophile 7 and diene 8 as substrates. c, Relative enzymatic activities of MaDA-1 and its G283R variant using dienophile 7 and diene 10 as substrates. The exo/endo ratio of the MaDA-1-G283R variant-catalyzed reaction was about 9 :1 when diene 10 was used. The red and blue columns indicate the exo-activities and endo activities respectively. Enzyme activity values represent mean ± standard deviation (s.d.) of three independent replicates.
Extended Data Fig. 5 The relative activities of MaDA variants with mutation at E414 or R443 positions.
The enzymatic reactions were performed using dienophile 7 and diene 8 as substrates in the presence of 1 μg MaDA or its variants at the optical conditions for 5 min. No exo product was observed in all aliquots. Enzyme activity values represent mean ± standard deviation (s.d.) of three independent replicates.
Extended Data Fig. 6 The R294/E414/R443 triad is vital for the exo activity in MaDA-3.
All these mutations regardless of synonymous mutation or not led to notable decrease of the exo activity of MaDA-3, indicating the hydrogen bonding interactions among the R294/E414/R443 triad are delicate contributors to form strong cation-π interaction between R294 and dienophile for promoting exo selective reaction. The E414 and R443 in MaDA-3 were found to be indispensable for endo activity of MaDA-3 since mutations at these two residues completely lost endo activities. The red and blue columns indicate the exo-activities and endo activities respectively. Enzyme activity values represent mean ± standard deviation (s.d.) of three independent replicates.
Extended Data Fig. 7 The different substitutions in the dienophiles have an influence on both overall activities and stereoselectivities.
The enzymatic reactions were performed using different dienophiles (200 μM) and diene 8 (200 μM) as substrates in the presence of 3 μg MaDA-3 or its variants at the optical conditions for 10 min. The red and blue columns indicate the exo/endo selectivities and dienophile conversions respectively. Enzyme activity values represent mean ± standard deviation (s.d.) of three independent replicates.
Extended Data Fig. 8 Orbital analysis for the a) LUMOs of the dienophile, b) HOMO of the diene, c) the LUMO of the dienophile bound to R443 (MaDA) and d) the dienophile bound to R294(MaDA-3).
Orbital energies are calculated using ωB97X-D/6-31G(d,p) CPCM (diethyl ether) level of theory. All orbital energies are reported in eV. The reaction proceeds via a normal-electron demand DA reaction, with HOMODiene-LUMOdienophile gap shrinking from 6.7 eV for the uncatalyzed reaction between a and b to 5.9 eV between b and c by 0.8 eV for R443-Dienophile (MaDA). The HOMODiene-LUMOdienophile gap shrunk to 6.0 eV between b and d by 0.7 eV for R294-Dienophile (MaDA-3).
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1 and 2 and Methods.
Supplementary Data 1
The binding pose of the endo transition state in MaDA before MD simulations.
Supplementary Data 2
The binding pose of the endo transition state in MaDA after MD simulations.
Supplementary Data 3
The binding pose of the exo transition state in MaDA before MD simulations.
Supplementary Data 4
The binding pose of the exo transition state in MaDA after MD simulations.
Supplementary Data 5
The binding pose of the endo transition state in MaDA-3 before MD simulations.
Supplementary Data 6
The binding pose of the endo transition state in MaDA-3 after MD simulations.
Supplementary Data 7
The binding pose of the exo transition state in MaDA-3 before MD simulations.
Supplementary Data 8
The binding pose of the exo transition state in MaDA-3 after MD simulations.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
About this article
Cite this article
Gao, L., Zou, Y., Liu, X. et al. Enzymatic control of endo- and exo-stereoselective Diels–Alder reactions with broad substrate scope. Nat Catal 4, 1059–1069 (2021). https://doi.org/10.1038/s41929-021-00717-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-021-00717-8
This article is cited by
-
The evolutionary origin of naturally occurring intermolecular Diels-Alderases from Morus alba
Nature Communications (2024)
-
Computational study on the catalytic control of endo/exo Diels-Alder reactions by cavity quantum vacuum fluctuations
Nature Communications (2023)
-
Altogether changed, and yet the same
Nature Chemistry (2023)
-
Chemoenzymatic approaches for exploring structure–activity relationship studies of bioactive natural products
Nature Synthesis (2023)
-
Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nature Catalysis (2023)