Abstract
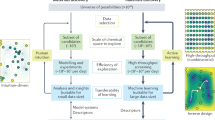
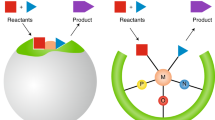
The organic chemist’s toolbox is vast, with technologies to accelerate the synthesis of novel chemical matter. The field of asymmetric catalysis is one approach to accessing new areas of chemical space and computational power is today sufficient to assist in this exploration. Unfortunately, existing techniques generally require computational expertise and are therefore underutilized in synthetic chemistry. Here we present our platform Virtual Chemist, which allows bench chemists to predict outcomes of asymmetric chemical reactions ahead of testing in the laboratory, in just a few clicks. Modular workflows facilitate the simulation of various sets of experiments, including the four realistic scenarios discussed: one-by-one design, library screening, hit optimization and substrate-scope evaluation. Catalyst candidates are screened within hours and the enantioselectivity predictions provide substantial enrichments compared to random testing. The achieved accuracies within ~1 kcal mol–1 provide opportunities for computational chemistry in the field of asymmetric catalyst design, allowing bench chemists to guide the design and discovery of asymmetric catalysts.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The sets of molecules used in this study (Supplementary Tables 1–8) and representative computed data (Supplementary Tables 9–18) are available as Supplementary Information. A tutorial for the use of this platform is provided as Supplementary Data 1. The programs are available (free of charge for academic research) at www.molecularforecaster.com. All the data, parameter files and structures are available on moitessier-group.mcgill.ca/software.html.
All other data is available from the authors upon reasonable request.
Code availability
Description and pseudocode of all the programs used in this study are provided in the Supplementary Methods. The programs are available for download upon request from the authors (www.molecularforecaster.com).
References
ACS. Where Is Organic Chemistry Used? ACS Chemistry for life www.acs.org/content/acs/en/careers/college-to-career/areas-of-chemistry/organic-chemistry.html (accessed 30 June 2020).
Wang, A. et al. Unraveling the mysterious failure of Cu/SAPO-34 selective catalytic reduction catalysts. Nat. Commun. 10, 1137 (2019).
Wang, X.-G. et al. Three-component ruthenium-catalyzed direct meta-selective C–H activation of arenes: a new approach to the alkylarylation of alkenes. J. Am. Chem. Soc. 141, 13914–13922 (2019).
Meucci, E. A. et al. Nickel(IV)-catalyzed C–H trifluoromethylation of (hetero)arenes. J. Am. Chem. Soc. 141, 12872–12879 (2019).
Durrant, J. D. & McCammon, J. A. Molecular dynamics simulations and drug discovery. BMC Biol. 9, 71–71 (2011).
Borhani, D. W. & Shaw, D. E. The future of molecular dynamics simulations in drug discovery. J. Comput.-Aided Mol. Des. 26, 15–26 (2012).
Kuntz, I. D., Blaney, J. M., Oatley, S. J., Langridge, R. & Ferrin, T. E. A geometric approach to macromolecule–ligand interactions. J. Mol. Biol. 161, 269–288 (1982).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Liu, X. et al. Molecular dynamics simulations and novel drug discovery. Expert Opin. Drug Discovery 13, 23–37 (2018).
Wang, G. & Zhu, W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Future Med. Chem. 8, 1707–1710 (2016).
Santosh, A. K., Alpeshkumar, K. M., Evans, C. C. & Sudha, S. Pharmacophore modeling in drug discovery and development: an overview. Med. Chem. 3, 187–197 (2007).
Sanderson, K. Automation: chemistry shoots for the moon. Nature 568, 577–579 (2019).
Segler, M. H. S., Preuss, M. & Waller, M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555, 604–610 (2018).
Zheng, S., Rao, J., Zhang, Z., Xu, J. & Yang, Y. Predicting retrosynthetic reactions using self-corrected transformer neural networks. J. Chem. Inf. Model. 60, 47–55 (2020).
Marques-Lopez, E., Herrera, R. P. & Christmann, M. Asymmetric organocatalysis in total synthesis–a trial by fire. Nat. Prod. Rep. 27, 1138–1167 (2010).
Maldonado, A. G. & Rothenberg, G. Predictive modeling in homogeneous catalysis: a tutorial. Chem. Soc. Rev. 39, 1891–1902 (2010).
Brown, J. M. & Deeth, R. J. Is enantioselectivity predictable in asymmetric catalysis. Angew., Chem. Int. Ed. 48, 4476–4479 (2009).
Houk, K. N. & Cheong, P. H. Y. Computational prediction of small-molecule catalysts. Nature 455, 309–313 (2008).
Harper, K. C. & Sigman, M. S. Predicting and optimizing asymmetric catalyst performance using the principles of experimental design and steric parameters. Proc. Natl Acad. Sci. USA 108, 2179–2183 (2011).
Reid, J. P. & Sigman, M. S. Holistic prediction of enantioselectivity in asymmetric catalysis. Nature 571, 343–348 (2019).
Norrby, P. O. Holistic models of reaction selectivity. Nature 571, 332–333 (2019).
Beker, W., Gajewska, E. P., Badowski, T. & Grzybowski, B. A. Prediction of major regio-, site-, and diastereoisomers in Diels–Alder reactions by using machine-learning: the importance of physically meaningful descriptors. Angew. Chem., Int. Ed. 58, 4515–4519 (2019).
Zahrt, A. F. et al. Prediction of higher-selectivity catalysts by computer-driven workflow and machine learning. Science 363, eaau5631 (2019).
Reid, J. P. & Sigman, M. S. Comparing quantitative prediction methods for the discovery of small-molecule chiral catalysts. Nat. Rev. Chem. 2, 290–305 (2018).
Bahmanyar, S. & Houk, K. N. The origin of stereoselectivity in proline-catalyzed intramolecular aldol reactions. J. Am. Chem. Soc. 123, 12911–12912 (2001).
Gordillo, R. & Houk, K. N. Origins of stereoselectivity in Diels-Alder cycloadditions catalyzed by chiral imidazolidinones. J. Am. Chem. Soc. 128, 3543–3553 (2006).
Ford, D. D., Nielsen, L. P. C., Zuend, S. J., Musgrave, C. B. & Jacobsen, E. N. Mechanistic basis for high stereoselectivity and broad substrate scope in the (salen)Co(iii)-catalyzed hydrolytic kinetic resolution. J. Am. Chem. Soc. 135, 15595–15608 (2013).
Lin, H., Pei, W., Wang, H., Houk, K. N. & Krauss, I. J. Enantioselective homocrotylboration of aliphatic aldehydes. J. Am. Chem. Soc. 135, 82–85 (2013).
Wolf, L. M. & Denmark, S. E. A theoretical investigation on the mechanism and stereochemical course of the addition of (E)-2-butenyltrimethylsilane to acetaldehyde by electrophilic and nucleophilic activation. J. Am. Chem. Soc. 135, 4743–4756 (2013).
Lam, Y.-h & Houk, K. N. How cinchona alkaloid-derived primary amines control asymmetric electrophilic fluorination of cyclic ketones. J. Am. Chem. Soc. 136, 9556–9559 (2014).
Lam, Y.-h & Houk, K. N. Origins of stereoselectivity in intramolecular aldol reactions catalyzed by cinchona amines. J. Am. Chem. Soc. 137, 2116–2127 (2015).
Reid, J. P., Simón, L. & Goodman, J. M. A practical guide for predicting the stereochemistry of bifunctional phosphoric acid catalyzed reactions of imines. Acc. Chem. Res. 49, 1029–1041 (2016).
Rosales, A. R. et al. Application of Q2MM to predictions in stereoselective synthesis. Chem. Commun. 54, 8294–8311 (2018).
Hansen, E., Rosales, A. R., Tutkowski, B., Norrby, P.-O. & Wiest, O. Prediction of Stereochemistry using Q2MM. Acc. Chem. Res. 49, 996–1005 (2016).
Corbeil, C. R., Thielges, S., Schwartzentruber, J. A. & Moitessier, N. Toward a computational tool predicting the stereochemical outcome of asymmetric reactions: development and application of a rapid and accurate program based on organic principles. Angew. Chem., Int. Ed. 47, 2635–2638 (2008).
Weill, N., Corbeil, C. R., De Schutter, J. W. & Moitessier, N. Toward a computational tool predicting the stereochemical outcome of asymmetric reactions: development of the molecular mechanics-based program ACE and application to asymmetric epoxidation reactions. J. Comput. Chem. 32, 2878–2889 (2011).
Schneebeli, S. T., Hall, M. L., Breslow, R. & Friesner, R. A. Quantitative DFT modeling of the enantiomeric excess for dioxirane-catalyzed epoxidations. J. Am. Chem. Soc. 131, 3965–3973 (2009).
Bootsma, A. N. & Wheeler, S. Popular integration grids can result in large errors in DFT-computed free energies. Preprint at https://doi.org/10.26434/chemrxiv.8864204.v5 (2019).
Rosales, A. R. et al. Rapid virtual screening of enantioselective catalysts using CatVS. Nat. Catal. 2, 41–45 (2019).
Therrien, E. et al. Integrating medicinal chemistry, organic/combinatorial chemistry, and computational chemistry for the discovery of selective estrogen receptor modulators with FORECASTER, a novel platform for drug discovery. J. Chem. Inf. Model. 52, 210–224 (2012).
Pottel, J. & Moitessier, N. Customizable generation of synthetically accessible, local chemical subspaces. J. Chem. Inf. Model. 57, 454–467 (2017).
van Hilten, N., Chevillard, F. & Kolb, P. Virtual compound libraries in computer-assisted drug discovery. J. Chem. Inf. Model. 59, 644–651 (2019).
Rasmussen, T. & Norrby, P. O. Modeling the stereoselectivity of the beta-amino alcohol-promoted addition of dialkylzinc to aldehydes. J. Am. Chem. Soc. 125, 5130–5138 (2003).
Seminario, J. M. Calculation of intramolecular force fields from second-derivative tensors. Int. J. Quantum Chem. 60, 1271–1277 (1996).
Allen, A. E. A., Payne, M. C. & Cole, D. J. Harmonic force constants for molecular mechanics force fields via hessian matrix projection. J. Chem. Theory Comput. 14, 274–281 (2018).
Norrby, P. O., Rasmussen, T., Haller, J., Strassner, T. & Houk, K. N. Rationalizing the stereoselectivity of osmium tetroxide asymmetric dihydroxylations with transition state modeling using quantum mechanics-guided molecular mechanics. J. Am. Chem. Soc. 121, 10186–10192 (1999).
Donoghue, P. J., Helquist, P., Norrby, P.-O. & Wiest, O. Prediction of enantioselectivity in rhodium catalyzed hydrogenations. J. Am. Chem. Soc. 131, 410–411 (2009).
Harvey, J. N., Himo, F., Maseras, F. & Perrin, L. Scope and challenge of computational methods for studying mechanism and reactivity in homogeneous catalysis. ACS Catal. 9, 6803–6813 (2019).
Yang, X. et al. Chiral pyrrolidine derivatives as catalysts in the enantioselective addition of diethylzinc to aldehydes. Tetrahedron: Asym. 10, 133–138 (1999).
Liang, G., Bays, J. P. & Bowen, J. P. Ab initio calculations and molecular mechanics (MM3) force field development for sulfonamide and its alkyl derivatives. J. Mol. Struct. THEOCHEM 401, 165–179 (1997).
Immirzi, A. & Musco, A. A method to measure the size of phosphorus ligands in coordination complexes. Inorg. Chim. Acta 25, L41–L42 (1977).
Liu, Z. et al. Elucidating hyperconjugation from electronegativity to predict drug conformational energy in a high throughput manner. J. Chem. Inf. Model. 56, 788–801 (2016).
Liu, Z., Barigye, S. J., Shahamat, M., Labute, P. & Moitessier, N. Atom Types Independent Molecular Mechanics Method for Predicting the Conformational Energy of Small Molecules. J. Chem. Inf. Model. 58, 194–205 (2018).
Champion, C. et al. Atom type independent modeling of the conformational energy of benzylic, allylic, and other bonds adjacent to conjugated systems. J. Chem. Inf. Model. 59, 4750–4763 (2019).
Wei, W. et al. Torsional energy barriers of biaryls could be predicted by electron-richness/deficiency of aromatic rings; advancement of molecular mechanics towards atom-type independence. J. Chem. Inf. Model. 59, 4764–4777 (2019).
Sterling, T. & Irwin, J. J. ZINC 15—Ligand discovery for everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015).
Hall, H. K. Correlation of the base strengths of amines. J. Am. Chem. Soc. 79, 5441–5444 (1957).
Gerosa, G. G., Spanevello, R. A., Suárez, A. G. & Sarotti, A. M. Joint experimental, in silico, and NMR studies toward the rational design of iminium-based organocatalyst derived from renewable sources. J. Org. Chem. 80, 7626–7634 (2015).
DelMonte, A. J. et al. Experimental and theoretical kinetic isotope effects for asymmetric dihydroxylation. evidence supporting a rate-limiting ‘(3 + 2)’ cycloaddition. J. Am. Chem. Soc. 119, 9907–9908 (1997).
Acknowledgements
We thank NSERC (Discovery programme) for financial support. Calcul Québec and Compute Canada are acknowledged for generous CPU allocations.
Author information
Authors and Affiliations
Contributions
N.M., M.B.P. and J.P. designed and wrote the programs REACT2D, FINDERS and CONTRUCTS (J.P., N.M.), QUEMIST (M.B.P.), UI, REDUCE, SELECT and ACE (N.M.). S.P. and M.B.P. have tested the usability and contributed to the design of the platform. P.O.N. contributed to the design of the platform. The testing (four scenarios) and data analysis were performed by S.P., M.B.P. and N.M. All the authors contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
Virtual Chemist is distributed by Molecular Forecaster (free of charge for academic research) co-founded by N.M. (CEO: J.P.).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Figs. 1,2, Tables 1–18, and References.
Supplementary Data 1
Supplementary Data 1.
Rights and permissions
About this article
Cite this article
Burai Patrascu, M., Pottel, J., Pinus, S. et al. From desktop to benchtop with automated computational workflows for computer-aided design in asymmetric catalysis. Nat Catal 3, 574–584 (2020). https://doi.org/10.1038/s41929-020-0468-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-020-0468-3
This article is cited by
-
Valence-isomer selective cycloaddition reaction of cycloheptatrienes-norcaradienes
Nature Communications (2024)
-
Reaction performance prediction with an extrapolative and interpretable graph model based on chemical knowledge
Nature Communications (2023)
-
Late-stage C–H functionalization offers new opportunities in drug discovery
Nature Reviews Chemistry (2021)
-
Organic reactivity from mechanism to machine learning
Nature Reviews Chemistry (2021)
-
Proofreading experimentally assigned stereochemistry through Q2MM predictions in Pd-catalyzed allylic aminations
Nature Communications (2021)