Abstract
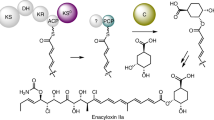
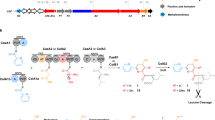
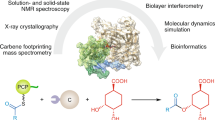
Heterochiral coupling is favoured in abiotic peptide bond formation, whereas biotic peptide bond formation is dominated by homochiral coupling. Here, we report that heterochiral coupling is a rather general paradigm in the head-to-tail macrolactamization of non-ribosomal peptide biosynthesis. The canonical cis-acting offloading cyclases, such as type I thioesterase (TE) and terminal condensation-like domains, catalyse head-to-tail macrolactamization between N- and C-terminal residues with d- and l-configurations, respectively. In contrast, the penicillin-binding protein-type TEs, a recently identified family of trans-acting cyclases, couple heterochiral residues with complementary stereoselectivity to the canonical one. Thus, a suite of cis- and trans-TE non-ribosomal peptide synthetases could overcome the stereochemical constraints present in heterochiral head-to-tail macrolactam formation in bacterial non-ribosomal peptide biosynthesis. Furthermore, we provide the structural rationale for the C-terminal stereoselectivity of non-canonical offloading cyclases. Penicillin-binding protein-type TEs with broad substrate specificity are potentially applicable as biocatalysts and genetic tools for synthetic biology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The crystallographic data that support the findings of this study are available from the Protein Data Bank (http://www.rcsb.org). The coordinates and the structure factor amplitudes for the structures of SurE apo and SurE complexed with 4 have been deposited under accession codes 6KSU and 6KSV, respectively. All other data supporting the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
References
Hill, R. R., Birch, D., Jeff, G. E. & North, M. Enantioselection in peptide bond formation. Org. Biomol. Chem. 1, 965–972 (2003).
Brady, S. F. et al. Practical synthesis of cyclic peptides, with an example of dependence of cyclization yield upon linear sequence. J. Org. Chem. 44, 3101–3105 (1979).
Rich, D. H., Bhatnagar, P., Mathiaparanam, P., Grant, J. A. & Tam, J. P. Synthesis of tentoxin and related dehydro cyclic tetrapeptides. J. Org. Chem. 43, 296–302 (1978).
Sieber, S. A. & Marahiel, M. A. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105, 715–738 (2005).
Trauger, J. W., Kohli, R. M., Mootz, H. D., Marahiel, M. A. & Walsh, C. T. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature 407, 215–218 (2000).
Gao, X. et al. Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat. Chem. Biol. 8, 823–830 (2012).
Tsomaia, N. Peptide therapeutics: targeting the undruggable space. Eur. J. Med. Chem. 94, 459–470 (2015).
Kuranaga, T. et al. Total synthesis of the nonribosomal peptide surugamide B and identification of a new offloading cyclase family. Angew. Chem. Int. Ed. 57, 9447–9451 (2018).
Takada, K. et al. Surugamides A–E, cyclic octapeptides with four d-amino acid residues, from a marine Streptomyces sp.: LC-MS-aided inspection of partial hydrolysates for the distinction of d- and l-amino acid residues in the sequence. J. Org. Chem. 78, 6746–6750 (2013).
Mohiman, H. et al. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 13, 30–37 (2016).
Xu, F., Nazari, B., Moon, K., Bushin, L. B. & Seyedsayamdost, M. R. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J. Am. Chem. Soc. 139, 9203–9212 (2017).
Ninomiya, A. et al. Biosynthetic gene cluster for surugamide A encompasses an unrelated decapeptide, surugamide F. ChemBioChem 17, 1709–1712 (2016).
Matsuda, K. et al. SurE is a trans-acting thioesterase cyclizing two distinct non-ribosomal peptides. Org. Biomol. Chem. 17, 1058–1061 (2019).
Magarvey, N. A., Haltli, B., He, M., Greenstein, M. & Hucul, J. A. Biosynthetic pathway for mannopeptimycins, lipoglycopeptide antibiotics active against drug-resistant gram-positive pathogens. Antimicrob. Agents Chemother. 50, 2167–2177 (2006).
Li, Q. et al. Identification of the biosynthetic gene cluster for the anti-infective desotamides and production of a new analogue in a heterologous host. J. Nat. Prod. 78, 944–948 (2015).
Son, S. et al. Genomics-driven discovery of chlorinated cyclic hexapeptides ulleungmycins A and B from a Streptomyces species. J. Nat. Prod. 80, 3025–3031 (2017).
Zhou, Y. et al. Investigation of penicillin binding protein (PBP)-like peptide cyclase and hydrolase in surugamide non-ribosomal peptide biosynthesis. Cell Chem. Biol. 26, 737–744 (2019).
Thankachan, D. et al. A trans-acting cyclase offloading strategy for nonribosomal peptide synthetases. ACS Chem. Biol. 14, 845–849 (2019).
White, C. J. & Yudin, A. K. Contemporary strategies for peptide macrocyclization. Nat. Chem. 3, 509–524 (2011).
Yu, X. & Sun, D. Macrocyclic drugs and synthetic methodologies toward macrocycles. Molecules 18, 6230–6268 (2013).
Medema, M. H. et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 11, 625–631 (2015).
Schultz, A. W. et al. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J. Am. Chem. Soc. 130, 4507–4516 (2008).
Ma, J. et al. Biosynthesis of ilamycins featuring unusual building blocks and engineered production of enhanced anti-tuberculosis agents. Nat. Commun. 8, 391 (2017).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Lahiri, S. D. et al. Structural insight into potent broad-spectrum inhibition with reversible recyclization mechanism: avibactam in complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-lactamases. Antimicrob. Agents Chemother. 57, 2496–2505 (2013).
Nakano, S. et al. Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline d-peptidase from Bacillus cereus DF4-B. Sci. Rep. 5, 13836–13836 (2015).
Delfosse, V. et al. Structure of the archaeal Pab87 peptidase reveals a novel self-compartmentalizing protease family. PLoS ONE 4, e4712–e4712 (2009).
Calcott, M. J. & Ackerley, D. F. Genetic manipulation of non-ribosomal peptide synthetases to generate novel bioactive peptide products. Biotechnol. Lett. 36, 2407–2416 (2014).
Bozhüyük, K. A. J. et al. De novo design and engineering of non-ribosomal peptide synthetases. Nat. Chem. 10, 275–281 (2017).
Niquille, D. L. et al. Nonribosomal biosynthesis of backbone-modified peptides. Nat. Chem. 10, 282–287 (2018).
Bozhüyük, K. A. J. et al. Modification and de novo design of non-ribosomal peptide synthetases using specific assembly points within condensation domains. Nat. Chem. 11, 653–661 (2019).
Pace, C. N., Vajdos, F., Fee, L., Grimsley, G. & Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 (1995).
Rice, L. M., Earnest, T. N. & Brunger, A. T. Single-wavelength anomalous diffraction phasing revisited. Acta Crystallogr. D 56, 1413–1420 (2000).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistic. Acta Crystallogr. D 67, 282–292 (2011).
Skubak, P. & Pannu, N. S. Automatic protein structure solution from weak X-ray data. Nat. Commun. 4, 2777 (2013).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D 64, 61–69 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Schüttelkopf, A. W. & van Aalten, D. M. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr. D 60, 1355–1363 (2004).
Bachmann, B. O. & Ravel, J. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 458, 181–217 (2009).
Kim, J. H., Komatsu, M., Shin-ya, K., Omura, S. & Ikeda, H. Distribution and functional analysis of the phosphopantetheinyl transferase superfamily in Actinomycetales microorganisms. Proc. Natl Acad. Sci. USA 115, 6828–6833 (2018).
Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E. & Ikeda, H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl Acad. Sci. USA 107, 2646–2651 (2010).
Acknowledgements
We thank H. Ikeda (Kitazato University) for providing vectors and the E. coli strain used for genetic manipulation of S. albidoflavus NBRC12854, and A. Katsuyama (Hokkaido University) for technical assistance in the conformational energy calculation. The synchrotron radiation experiments were performed at beamline BL-1A of the Photon Factory. We also thank the beamline staff of the Photon Factory for their help in collecting X-ray diffraction data. This work was partly supported by the Takeda Science Foundation, the Asahi Glass Foundation, the Naito Foundation, the Uehara Memorial Foundation, the Japan Agency for Medical Research and Development (AMED grant no. JP19ae0101045) and Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (JSPS KAKENHI grant nos. JP16703511, JP16H06443, JP18056499, JP19178402 and JP20H00490).
Author information
Authors and Affiliations
Contributions
K.M., T.M., I.A. and T.W. designed the experiments. R.Z. and T.M. performed the crystallization experiment. K.M., R.Z., T.M. and M.K. performed in vitro analysis. K.M. performed kinetic analysis. M.K. and A.S. synthesized substrate analogues. K.M., M.K. and T.W. performed structure determination of enzyme reaction products. M.K. performed genetic manipulation of Streptomyces. K.M., R.Z., T.M., M.K., A.S., I.A. and T.W. analysed the data. K.M., T.M., I.A. and T.W. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Tables 1 and 2 and Figs. 1–41.
Supplementary Data Dataset 1
Model of N-formyl-d-Leu tethered on Ser63 of SurE apo structure.
Supplementary Data Dataset 2
Model of N-formyl-l-Leu tethered on Ser63 of SurE apo structure.
Supplementary Data Dataset 3
Model of I1P tethered on Ser63 of SurE holo structure.
Rights and permissions
About this article
Cite this article
Matsuda, K., Zhai, R., Mori, T. et al. Heterochiral coupling in non-ribosomal peptide macrolactamization. Nat Catal 3, 507–515 (2020). https://doi.org/10.1038/s41929-020-0456-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-020-0456-7
This article is cited by
-
Chemoenzymatic tandem cyclization for the facile synthesis of bicyclic peptides
Communications Chemistry (2024)
-
Biocatalytic cyclization of small macrolactams by a penicillin-binding protein-type thioesterase
Nature Chemical Biology (2024)
-
Size matters
Nature Chemical Biology (2024)
-
Molecular basis for carrier protein-dependent amide bond formation in the biosynthesis of lincosamide antibiotics
Nature Catalysis (2023)
-
Total syntheses of surugamides and thioamycolamides toward understanding their biosynthesis
Journal of Natural Medicines (2023)