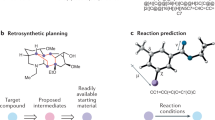
Abstract
As the enzyme toolbox for biocatalysis has expanded, so has the potential for the construction of powerful enzymatic cascades for efficient and selective synthesis of target molecules. Additionally, recent advances in computer-aided synthesis planning are revolutionizing synthesis design in both synthetic biology and organic chemistry. However, the potential for biocatalysis is not well captured by tools currently available in either field. Here we present RetroBioCat, an intuitive and accessible tool for computer-aided design of biocatalytic cascades, freely available at retrobiocat.com. Our approach uses a set of expertly encoded reaction rules encompassing the enzyme toolbox for biocatalysis, and a system for identifying literature precedent for enzymes with the correct substrate specificity where this is available. Applying these rules for automated biocatalytic retrosynthesis, we show our tool to be capable of identifying promising biocatalytic pathways to target molecules, validated using a test set of recent cascades described in the literature.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Other than literature precedent data, which can currently only be accessed at https://retrobiocat.com pending future publications, the database files for RetroBiocat at the time of publication are available at https://doi.org/10.6084/m9.figshare.12696482.v4. The 52 pathway test set is available, along with the source code at https://doi.org/10.6084/m9.figshare.12698072.v7 or https://github.com/willfinnigan/retrobiocat.
Code availability
RetroBioCat is freely available as a web application at https://retrobiocat.com. We have also made the source code freely available under the MIT license, available at https://github.com/willfinnigan/retrobiocat, or specifically for the version described here, at https://doi.org/10.6084/m9.figshare.12698072.v7.
References
Bornscheuer, U. T. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012).
Sheldon, R. A. & Brady, D. The limits to biocatalysis: pushing the envelope. Chem. Commun. 54, 6088–6104 (2018).
Hönig, M., Sondermann, P., Turner, N. J. & Carreira, E. M. Enantioselective chemo- and biocatalysis: partners in retrosynthesis. Angew. Chem. Int. Ed. Engl. 56, 8942–8973 (2017).
France, S. P., Hepworth, L. J., Turner, N. J. & Flitsch, S. L. Constructing biocatalytic cascades: in vitro and in vivo approaches to de novo multi-enzyme pathways. ACS Catal. 7, 710–724 (2017).
Huffman, M. A. et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 366, 1255–1259 (2019).
Schober, M. et al. Chiral synthesis of LSD1 inhibitor GSK2879552 enabled by directed evolution of an imine reductase. Nat. Catal. 2, 909–915 (2019).
Koch, M., Duigou, T. & Faulon, J. L. Reinforcement learning for bioretrosynthesis. ACS Synth. Biol. 9, 157–168 (2020).
Coley, C. W., Rogers, L., Green, W. H. & Jensen, K. F. Computer-assisted retrosynthesis based on molecular similarity. ACS Cent. Sci. 3, 1237–1245 (2017).
Szymkuć, S. et al. Computer-assisted synthetic planning: the end of the beginning. Angew. Chem. Int. Ed. Engl. 55, 5904–5937 (2016).
Segler, M. H. S., Preuss, M. & Waller, M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555, 604–610 (2018).
Landrum, G. RDKit: open-cource cheminformatics software (2016).
Coley, C. W., Green, W. H. & Jensen, K. F. Machine learning in computer-aided synthesis planning. Acc. Chem. Res. 51, 1281–1289 (2018).
Grzybowski, B. A. et al. Chematica: a story of computer code that started to think like a chemist. Chem 4, 390–398 (2018).
Hartenfeller, M. et al. A collection of robust organic synthesis reactions for in silico molecule design. J. Chem. Inf. Model. 51, 3093–3098 (2011).
Plehiers, P. P., Marin, G. B., Stevens, C. V. & Van Geem, K. M. Automated reaction database and reaction network analysis: extraction of reaction templates using cheminformatics. J. Cheminform. 10, 11 (2018).
Duigou, T., Du Lac, M., Carbonell, P. & Faulon, J. L. Retrorules: a database of reaction rules for engineering biology. Nucleic Acids Res. 47, D1229–D1235 (2019).
Molga, K., Gajewska, E. P., Szymkuć, S. & Grzybowski, B. A. The logic of translating chemical knowledge into machine-processable forms: a modern playground for physical-organic chemistry. React. Chem. Eng. 4, 1506–1521 (2019).
Segler, M. H. S. & Waller, M. P. Neural-symbolic machine learning for retrosynthesis and reaction prediction. Chem. Eur. J. 23, 5966–5971 (2017).
Fehér, T. et al. Validation of RetroPath, a computer-aided design tool for metabolic pathway engineering. Biotechnol. J. 9, 1446–1457 (2014).
Turner, N. J. & Humphreys, L. Biocatalysis in Organic Synthesis: the Retrosynthesis Approach (Royal Society of Chemistry, 2018).
Turner, N. J. & O’Reilly, E. Biocatalytic retrosynthesis. Nat. Chem. Biol. 9, 285–288 (2013).
de Souza, R. O. M. A., Miranda, L. S. M. & Bornscheuer, U. T. A retrosynthesis approach for biocatalysis in organic synthesis. Chem. Eur. J. 23, 12040–12063 (2017).
Heath, R. S. et al. An engineered alcohol oxidase for the oxidation of primary alcohols. ChemBioChem 20, 276–281 (2019).
Batista, V. F., Galman, J. L., Pinto, D. C., Silva, A. M. S. & Turner, N. J. Monoamine oxidase: tunable activity for amine resolution and functionalization. ACS Catal. 8, 11889–11907 (2018).
Devine, P. N. et al. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2, 409–421 (2018).
Arnold, F. H. Directed evolution: bringing new chemistry to life. Angew. Chem. Int. Ed. Engl. 57, 4143–4148 (2018).
Rácz, A., Bajusz, D. & Héberger, K. Life beyond the Tanimoto coefficient: similarity measures for interaction fingerprints. J. Cheminform. 10, 1–12 (2018).
Breitling, R. et al. Selenzyme: enzyme selection tool for pathway design. Bioinformatics 34, 2153–2154 (2018).
Coley, C. W., Rogers, L., Green, W. H. & Jensen, K. F. SCScore: synthetic complexity learned from a reaction corpus. J. Chem. Inf. Model. 58, 252–261 (2018).
Genheden, S. et al. AiZynthFinder: a fast, robust and flexible open-source software for retrosynthetic planning. J Cheminform. 12, 70 (2020).
Sehl, T. et al. Two steps in one pot: enzyme cascade for the synthesis of nor(pseudo)ephedrine from inexpensive starting materials. Angew. Chem. Int. Ed. Engl. 52, 6772–6775 (2013).
Wang, J. et al. Efficient production of phenylpropionic acids by an amino-group-transformation biocatalytic cascade. Biotechnol. Bioeng. 117, 614–625 (2020).
Erdmann, V. et al. Methoxamine synthesis in a biocatalytic 1-pot 2-step cascade approach. ACS Catal. 9, 7380–7388 (2019).
Lichman, B. R. et al. One-pot triangular chemoenzymatic cascades for the syntheses of chiral alkaloids from dopamine. Green Chem. 17, 852–855 (2015).
Parmeggiani, F., Lovelock, S. L., Weise, N. J., Ahmed, S. T. & Turner, N. J. Synthesis of d- and l-phenylalanine derivatives by phenylalanine ammonia lyases: a multienzymatic cascade process. Angew. Chem. Int. Ed. Engl. 54, 4608–4611 (2015).
Both, P. et al. Whole-cell biocatalysts for stereoselective C-H amination reactions. Angew. Chem. Int. Ed. Engl. 55, 1511–1513 (2016).
Oberleitner, N. et al. From waste to value—direct utilization of limonene from orange peel in a biocatalytic cascade reaction towards chiral carvolactone. Green Chem. 19, 367–371 (2017).
Wu, S. et al. Highly regio- and enantioselective multiple oxy- and amino-functionalizations of alkenes by modular cascade biocatalysis. Nat. Commun. 7, 11917 (2016).
Ramsden, J. I. et al. Biocatalytic N-alkylation of amines using either primary alcohols or carboxylic acids via reductive aminase cascades. J. Am. Chem. Soc. 141, 1201–1206 (2019).
Jakoblinnert, A. & Rother, D. A two-step biocatalytic cascade in micro-aqueous medium: using whole cells to obtain high concentrations of a vicinal diol. Green Chem. 16, 3472–3482 (2014).
Klumbys, E., Zebec, Z., Weise, N. J., Turner, N. J. & Scrutton, N. S. Bio-derived production of cinnamyl alcohol via a three step biocatalytic cascade and metabolic engineering. Green Chem. 20, 658–663 (2018).
Busto, E., Simon, R. C. & Kroutil, W. Vinylation of unprotected phenols using a biocatalytic system. Angew. Chem. Int. Ed. Engl. 54, 10899–10902 (2015).
Citoler, J., Derrington, S. R., Galman, J. L., Bevinakatti, H. & Turner, N. J. A biocatalytic cascade for the conversion of fatty acids to fatty amines. Green Chem. 21, 4932–4935 (2019).
Thorpe, T. W. et al. One-pot biocatalytic cascade reduction of cyclic enimines for the preparation of diastereomerically enriched N-heterocycles. J. Am. Chem. Soc. 141, 19208–19213 (2019).
Heath, R. S., Pontini, M., Hussain, S. & Turner, N. J. Combined imine reductase and amine oxidase catalyzed deracemization of nitrogen heterocycles. ChemCatChem 8, 117–120 (2016).
Tavanti, M., Mangas-Sanchez, J., Montgomery, S. L., Thompson, M. P. & Turner, N. J. A biocatalytic cascade for the amination of unfunctionalised cycloalkanes. Org. Biomol. Chem. 15, 9790–9793 (2017).
Sattler, J. H. et al. Redox self-sufficient biocatalyst network for the amination of primary alcohols. Angew. Chem. Int. Ed. Engl. 51, 9156–9159 (2012).
Mourelle-Insua, Á., Zampieri, L. A., Lavandera, I. & Gotor-Fernández, V. Conversion of γ- and δ-keto esters into optically active lactams. Transaminases in cascade processes. Adv. Synth. Catal. 360, 686–695 (2018).
Aumala, V. et al. Biocatalytic production of amino carbohydrates through oxidoreductase and transaminase cascades. ChemSusChem 12, 848–857 (2019).
Song, J.-W. et al. Multistep enzymatic synthesis of long-chain α,ω-dicarboxylic and ω-hydroxycarboxylic acids from renewable fatty acids and plant oils. Angew. Chem. Int. Ed. Engl. 52, 2534–2537 (2013).
Corrado, M. L., Knaus, T. & Mutti, F. G. Regio- and stereoselective multi-enzymatic aminohydroxylation of β-methylstyrene using dioxygen, ammonia and formate. Green Chem. 21, 6246–6251 (2019).
Fedorchuk, T. P. et al. One-pot biocatalytic transformation of adipic acid to 6-aminocaproic acid and 1,6-hexamethylenediamine using carboxylic acid reductases and transaminases. J. Am. Chem. Soc. 142, 1038–1048 (2020).
Wang, H., Zheng, Y.-C., Chen, F.-F., Xu, J.-H. & Yu, H.-L. Enantioselective bioamination of aromatic alkanes using ammonia: a multienzymatic cascade approach. ChemCatChem 12, 2077–2082 (2020).
Pickl, M., Fuchs, M., Glueck, S. M. & Faber, K. Amination of ω-functionalized aliphatic primary alcohols by a biocatalytic oxidation-transamination cascade. ChemCatChem 7, 3121–3124 (2015).
Parmeggiani, F. et al. One-pot biocatalytic synthesis of substituted d-tryptophans from indoles enabled by an engineered aminotransferase. ACS Catal. 9, 3482–3486 (2019).
Zhang, Z.-J., Cai, R.-F. & Xu, J.-H. Characterization of a new nitrilase from Hoeflea phototrophica DFL-43 for a two-step one-pot synthesis of (S)-β-amino acids. Appl. Microbiol. Biotechnol. 102, 6047–6056 (2018).
Bechi, B. et al. Catalytic bio-chemo and bio-bio tandem oxidation reactions for amide and carboxylic acid synthesis. Green Chem. 16, 4524–4529 (2014).
Jia, H.-Y., Zong, M.-H., Zheng, G.-W. & Li, N. One-pot enzyme cascade for controlled synthesis of furancarboxylic acids from 5-hydroxymethylfurfural by H2O2 internal recycling. ChemSusChem 12, 4764–4768 (2019).
Alvarenga, N. et al. Asymmetric synthesis of dihydropinidine enabled by concurrent multienzyme catalysis and a biocatalytic alternative to Krapcho dealkoxycarbonylation. ACS Catal. 10, 1607–1620 (2020).
Weise, N. J. et al. Bi-enzymatic conversion of cinnamic acids to 2-arylethylamines. ChemCatChem 12, 995–998 (2020).
Yoon, S. et al. Deracemization of racemic amines to enantiopure (R)- and (S)-amines by biocatalytic cascade employing ω-transaminase and amine dehydrogenase. ChemCatChem 11, 1898–1902 (2019).
Steinreiber, J. et al. Overcoming thermodynamic and kinetic limitations of aldolase-catalyzed reactions by applying multienzymatic dynamic kinetic asymmetric transformations. Angew. Chem. Int. Ed. Engl. 46, 1624–1626 (2007).
Shanmuganathan, S., Natalia, D., Greiner, L. & Domínguez de María, P. Oxidation-hydroxymethylation-reduction: a one-pot three-step biocatalytic synthesis of optically active α-aryl vicinal diols. Green Chem. 14, 94–97 (2012).
Montgomery, S. L. et al. Direct alkylation of amines with primary and secondary alcohols through biocatalytic hydrogen borrowing. Angew. Chem. Int. Ed. Engl. 129, 10627–10630 (2017).
Guérard-Hélaine, C. et al. Stereoselective synthesis of γ-hydroxy-α-amino acids through aldolase-transaminase recycling cascades. Chem. Commun. 53, 5465–5468 (2017).
Siirola, E. et al. Asymmetric synthesis of 3-substituted cyclohexylamine derivatives from prochiral diketones via three biocatalytic steps. Adv. Synth. Catal. 355, 1703–1708 (2013).
Zhang, J.-D. et al. Asymmetric ring opening of racemic epoxides for enantioselective synthesis of (S)-β-amino alcohols by a cofactor self-sufficient cascade biocatalysis system. Catal. Sci. Technol. 9, 70–74 (2019).
France, S. P. et al. One-pot cascade synthesis of mono- and disubstituted piperidines and pyrrolidines using carboxylic acid reductase (CAR), ω-transaminase (ω-TA), and imine reductase (IRED) biocatalysts. ACS Catal. 6, 3753–3759 (2016).
Hernandez, K. et al. Combining aldolases and transaminases for the synthesis of 2-amino-4-hydroxybutanoic acid. ACS Catal. 7, 1707–1711 (2017).
Monti, D. et al. Cascade coupling of ene-reductases and ω-transaminases for the stereoselective synthesis of diastereomerically enriched amines. ChemCatChem 7, 3106–3109 (2015).
Liao, C. & Seebeck, F. P. Asymmetric β-methylation of l- and d-α-amino acids by a self-contained enzyme cascade. Angew. Chem. Int. Ed. Engl. 59, 7184–7187 (2020).
Schmidt, S. et al. Biocatalytic access to chiral polyesters by an artificial enzyme cascade synthesis. ChemCatChem 7, 3951–3955 (2015).
Li, X. et al. DeepChemStable: chemical stability prediction with an attention-based graph convolution network. J. Chem. Inf. Model. 59, 1044–1049 (2019).
Finnigan, W. et al. Engineering a seven enzyme biotransformation using mathematical modelling and characterized enzyme parts. ChemCatChem 11, 3474–3489 (2019).
Chen, B., Li, C., Dai, H. & Song, L. Retro*: learning retrosynthetic planning with neural guided A* search. Preprint at https://arxiv.org/abs/2006.15820 (2020).
Kishimoto, A., Buesser, B., Chen, B. & Botea, A. in Advances in Neural Information Processing Systems (eds Wallach, H. et al.) 7226–7236 (Curran Associates, 2019).
Coley, C. W., Green, W. H. & Jensen, K. F. RDChiral: an RDKit wrapper for handling stereochemistry in retrosynthetic template extraction and application. J. Chem. Inf. Model. 59, 2529–2537 (2019).
Acknowledgements
We acknowledge financial support from the European Research Council (788231-ProgrES-ERC-2017-ADG to S.L.F.; BIO-HBORROW: grant no. 742987 to N.J.T.). We also thank all the beta-testers of RetroBioCat, particularly S. Cosgrove and R. Speight.
Author information
Authors and Affiliations
Contributions
W.F., L.J.H., S.L.F. and N.J.T. planned the work. Code for RetroBioCat written by W.F. Pathway test set generated by L.J.H. Initial draft of the manuscript written by W.F., with subsequent contributions from L.J.H., S.L.F. and N.J.T. All authors have given approval to the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Catalysis thanks Connor Coley, Nicolas Moitessier, Dörte Rother and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 An example generated using Network Explorer to illustrate changes in molecular complexity.
Arrows and reactions are coloured by the change in molecular complexity, determined using the SC-Score. Green indicates a negative change in molecule complexity, which in most cases corresponds to a synthetically useful transformation. Red indicates a positive change in molecular complexity. Colours are determined relative to the other transformations leading to a specific molecule. Some reactions have been removed for clarity. Pathway published in reference68.
Extended Data Fig. 2 Rankings for pathways 12 to 20 of the test-set for Pathway explorer.
A continuation of Fig. 4, showing rankings for pathways 12 to 20 by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. TPL: tyrosine phenol lyase, AAD: amino acid deaminase, AADH: amino acid dehydrogenase, TDL: thiamine-dependent lyase, TA: transaminase, PSase: Pictet-Spenglerase, PAL: phenylalanine ammonia lyase, P450: cytochrome P450, ADH: alcohol dehydrogenase, CumDO: cumene dioxygenase, ERED: ene reductase, BVMO: Baeyer-Villiger monooxygenase, SMO: styrene monooxygenase, AlDH: aldehyde dehydrogenase, AlOx: alcohol oxidase.
Extended Data Fig. 3 Rankings for pathways 21 to 30 of the test-set for Pathway explorer.
A continuation of Fig. 4, showing rankings for pathways 21 to 30 by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. TDL: thiamine-dependent lyase, ADH: alcohol dehydrogenase, PAL: phenylalanine ammonia lyase, CAR: carboxylic acid reductase, ERED: ene reductase, IRED: imine reductase, AmOx: amine oxidase, P450: cytochrome P450, ATP: adenosine triphosphate, NADP: nicotinamide adenine dinucleotide phosphate.
Extended Data Fig. 4 Rankings for pathways 31 to 40 of the test-set for Pathway explorer.
A continuation of Fig. 4, showing rankings for pathways 31 to 40 by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. ADH: alcohol dehydrogenase, BVMO: Baeyer-Villiger monooxygenase, SMO: styrene monooxygenase, EH: epoxide hydrolase, AmDH: amine dehydrogenase, CAR: carboxylic acid reductase, TA: transaminase, AlOx: alcohol oxidase, ERED: ene reductase, TrpS: tryptophan synthase, XOR: xanthine oxidoreductase, AAD: amino acid deaminase, ATP: adenosine triphosphate, NADP: nicotinamide adenine dinucleotide phosphate, BNA: 1-benzyl-1,4-dihydropyridine-3-carboxamide.
Extended Data Fig. 5 Rankings for pathways 41 to 52 of the test-set for Pathway explorer.
A continuation of Fig. 4, showing rankings for pathways 41 to 52 by RetroBioCat using a maximum of 4 steps and either the default scoring weights, or default weights but with the weight for number of steps with literature precedent set to zero, are shown. Pathways are marked as identified even where RetroBioCat suggests additional steps. * indicates pathways where the data from the relevant paper has not been added to the database of literature precedent reactions in RetroBioCat. TA: transaminase, IRED: imine reductase, PAL: phenylalanine ammonia lyase, DC: decarboxylase, AmDH: amine dehydrogenase, TPL: tyrosine phenol lyase, AAD: amino acid deaminase, TAM: tyrosine aminomutase, TDL: thiamine-dependent lyase, ADH: alcohol dehydrogenase, AlOx: alcohol oxidase, EH: epoxide hydrolase, CAR: carboxylic acid reductase, ATP: adenosine triphosphate, NADP: nicotinamide adenine dinucleotide phosphate.
Supplementary information
Rights and permissions
About this article
Cite this article
Finnigan, W., Hepworth, L.J., Flitsch, S.L. et al. RetroBioCat as a computer-aided synthesis planning tool for biocatalytic reactions and cascades. Nat Catal 4, 98–104 (2021). https://doi.org/10.1038/s41929-020-00556-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-020-00556-z
This article is cited by
-
LinChemIn: SynGraph—a data model and a toolkit to analyze and compare synthetic routes
Journal of Cheminformatics (2023)
-
Pickaxe: a Python library for the prediction of novel metabolic reactions
BMC Bioinformatics (2023)
-
A platform for distributed production of synthetic nitrated proteins in live bacteria
Nature Chemical Biology (2023)
-
Microbial engineering strategies to utilize waste feedstock for sustainable bioproduction
Nature Reviews Bioengineering (2023)
-
A selective and atom-economic rearrangement of uridine by cascade biocatalysis for production of pseudouridine
Nature Communications (2023)