Abstract
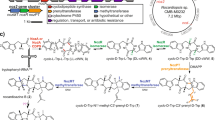
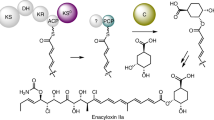
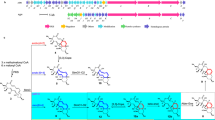
Enzymes that catalyse remarkable Diels–Alder-like [4+2] cyclizations have been previously implicated in the biosynthesis of spirotetronate and spirotetramate antibiotics. Biosynthesis of the polyether antibiotic tetronasin is not expected to require such steps, yet the tetronasin gene cluster encodes enzymes Tsn11 and Tsn15, which are homologous to authentic [4+2] cyclases. Here, we show that deletion of Tsn11 led to accumulation of a late-stage intermediate, in which the two central rings of tetronasin and four of its twelve asymmetric centres remain unformed. In vitro reconstitution showed that Tsn11 catalyses an apparent inverse-electron-demand hetero-Diels–Alder-like [4+2] cyclization of this species to form an unexpected oxadecalin compound that is then rearranged by Tsn15 to form tetronasin. To gain structural and mechanistic insight into the activity of Tsn15, the crystal structure of a Tsn15-substrate complex has been solved at 1.7 Å resolution.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The tetronasin biosynthetic gene cluster sequence is available on GenBank (accession code: FJ462704). The crystal structure data is available on the PDB (accession codes: 6NOI (Tsn15) and 6NNW (Tsn15-substrate complex)). All other data that supports the findings of this study are available from the corresponding author on reasonable request.
References
Diels, O. & Alder, K. Synthesen in der hydroaromatischen Reihe. Justus Liebigs Ann. der Chem. 460, 98–122 (1928).
Corey, E. J. Catalytic enantioselective Diels–Alder reactions: methods, mechanistic fundamentals, pathways, and applications. Angew. Chem. Int. Ed. Engl. 41, 1650–1667 (2002).
Lichman, B. R., O’Connor, S. E. & Kries, H. Biocatalytic strategies towards [4+ 2] cycloadditions. Chem. Eur. J. 25, 6864–6877 (2019).
Jeon, B., Wang, S.-A., Ruszczycky, M. W. & Liu, H. Natural [4+2]-cyclases. Chem. Rev. 117, 5367–5388 (2017).
Ma, S. M. et al. Complete reconstitution of a highly reducing iterative polyketide synthase. Science 326, 589–592 (2009).
Kasahara, K. et al. Solanapyrone synthase, a possible Diels–Alderase and iterative Type I polyketide synthase encoded in a biosynthetic gene cluster from Alternaria solani. ChemBioChem 11, 1245–1252 (2010).
Kim, H. J., Ruszczycky, M. W., Choi, S., Liu, Y. & Liu, H. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of Spinosyn A. Nature 473, 109–112 (2011).
Fage, C. D. et al. The structure of SpnF, a standalone enzyme that catalyzes [4+2] cycloaddition. Nat. Chem. Biol. 11, 256–258 (2015).
Hashimoto, T. et al. Biosynthesis of versipelostatin: identification of an enzyme-catalyzed [4+2]-cycloaddition required for macrocyclization of spirotetronate-containing polyketides. J. Am. Chem. Soc. 137, 572–575 (2015).
Byrne, M. J. et al. The catalytic mechanism of a natural Diels–Alderase revealed in molecular detail. J. Am. Chem. Soc. 138, 6095–6098 (2016).
Tian, Z. et al. An enzymatic [4+2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat. Chem. Biol. 11, 259–265 (2015).
Zhang, Z. et al. Enzyme-catalyzed inverse-electron demand Diels–Alder reaction in the biosynthesis of antifungal ilicicolin H. J. Am. Chem. Soc. 141, 5659–5663 (2019).
Cogan, D. P. et al. Structural insights into enzymatic [4+2] aza-cycloaddition in thiopeptide antibiotic biosynthesis. Proc. Natl Acad. Sci. USA 114, 12928–12933 (2017).
Ohashi, M. et al. SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis. Nature 549, 502–506 (2017).
Demetriadou, A. K. et al. Biosynthesis of the polyketide polyether Antibiotic ICI 139603 in Streptomyces longisporoflavus from 18O-labelled acetate and propionate. J. Chem. Soc., Chem. Commun. 19, 408–410 (1985).
Demydchuk, Y. et al. Analysis of the tetronomycin gene cluster: insights into the biosynthesis of a polyether tetronate antibiotic. ChemBioChem 9, 1136–1145 (2008).
Sun, Y., Hong, H., Gillies, F., Spencer, J. B. & Leadlay, P. F. Glyceryl-S-Acyl carrier protein as an intermediate in the biosynthesis of tetronate antibiotics. ChemBioChem 9, 150–156 (2008).
Hailes, H. C., Jackson, C. M., Leadlay, P. F., Ley, S. V. & Staunton, J. Biosynthesis of tetronasin: part 1 introduction and investigation of the diketide and triketide intermediates bound to the polyketide synthase. Tetrahedron Lett. 35, 307–310 (1994).
Boons, G.-J. et al. Novel polyene cyclisation routes to the acyl tetronic acid ionophore tetronasin (ICI M139603). Tetrahedron Lett. 35, 323–326 (1994).
Riva, E. et al. Chemical probes for the functionalization of polyketide intermediates. Angew. Chem. Int. Ed. 53, 11944–11949 (2014).
Tosin, M., Smith, L. & Leadlay, P. F. Insights into lasalocid A ring formation by chemical chain termination in vivo. Angew. Chem. Int. Ed. Engl. 50, 11930–11933 (2011).
Zheng, Q. et al. Structural insights into a flavin-dependent [4+2] cyclase that catalyzes trans-decalin formation in pyrroindomycin biosynthesis. Cell Chem. Biol. 25, 718–727 (2018).
Bosserman, M. A., Downey, T., Noinaj, N., Buchanan, S. K. & Rohr, J. Molecular insight into substrate recognition and catalysis of Baeyer–Villiger monooxygenase MtmOIV, the key frame-modifying enzyme in the biosynthesis of anticancer agent mithramycin. ACS Chem. Biol. 8, 2466–2477 (2013).
Vieweg, L., Reichau, S., Schobert, R., Leadlay, P. F. & Süssmuth, R. D. Recent advances in the field of bioactive tetronates. Nat. Prod. Rep. 31, 1554–1584 (2014).
Jiang, X. & Wang, R. Recent developments in catalytic asymmetric inverse-electron-demand Diels–Alder reaction. Chem. Rev. 113, 5515–5546 (2013).
Pałasz, A. Recent advances in inverse-electron-demand hetero-Diels–Alder reactions of 1-oxa-1,3-butadienes. Top. Curr. Chem. 374, 24 (2016).
Zheng, Q. et al. Enzyme-dependent [4+2] cycloaddition depends on lid-like interaction of the N-terminal sequence with the catalytic core in PyrI4. Cell Chem. Biol. 23, 352–360 (2016).
Hofmann, E., Zerbe, P. & Schaller, F. The crystal structure of Arabidopsis thaliana allene oxide cyclase: insights into the oxylipin cyclization reaction. Plant Cell 18, 3201–3217 (2006).
Yoeun, S., Cho, K. & Han, O. Structural evidence for the substrate channeling of rice allene oxide cyclase in biologically analogous Nazarov reaction. Front. Chem. 6, 500 (2018).
Costa, K. C., Glasser, N. R., Conway, S. J. & Newman, D. K. Pyocyanin degradation by a tautomerizing demethylase inhibits Pseudomonas aeruginosa biofilms. Science 355, 170–173 (2017).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
MacNeil, D. J. et al. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111, 61–68 (1992).
Wilkinson, C. J. et al. Increasing the efficiency of heterologous promoters in actinomycetes. J. Mol. Microbiol. Biotechnol. 4, 417–426 (2002).
Harding S. E. & Rowe S. E. Analytical Ultracentrifugation in Biochemistry and Polymer Science (Royal Society of Chemistry, 1992).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D 65, 582–601 (2009).
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011).
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D 64, 61–69 (2008).
McCoy, A. J. Solving structures of protein complexes by molecular replacement with phaser. Acta Crystallogr. D 63, 32–41 (2007).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
DeLano, W. L. The PyMOL Molecular Graphics System (Delano Scientific, 2002).
Acknowledgements
R.L was supported by the Woolf Fisher Trust and the Cambridge Commonwealth European and International Trust. M.V.B.D was supported by the São Paulo Research Foundation under grant nos 2015/09188-8, 2017/50140-4 and 2018/00351-1. F.C.R.P was supported by a CNPq (National Council for Scientific and Technological Development) fellowship (141090/2016-2). M.T and R.J gratefully acknowledge EPSRC (DTA PhD studentship to R.J.); BBSRC (project grant no. BB/J007250/1 to M.T.) and the FAPESP–Warwick Joint Fund (for a SPRINT award to M.V.B.D. and M.T.).
Author information
Authors and Affiliations
Contributions
R.L., P.F.L., F.J.L., M.T. and M.V.B.D. developed the hypothesis and designed the study. Y.D., Y.S. and M.S. cloned, sequenced and analysed the gene clusters. R.L., Y.S. and H.H. performed gene deletions, whereas M.T. and R.J. performed and analysed the experiments with chain-terminating probes. R.L. performed protein expressions and purifications, in vitro experiments and compound isolations. R.L. and F.J.L. performed compound characterizations. F.C.R.P., R.L. and M.V.B.D. solved the crystal structures. All authors analysed and discussed the results. P.F.L., R.L. and F.C.R.P. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–35, Supplementary Tables 1–6 and Supplementary Notes 1 and 2
Rights and permissions
About this article
Cite this article
Little, R., Paiva, F.C.R., Jenkins, R. et al. Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis. Nat Catal 2, 1045–1054 (2019). https://doi.org/10.1038/s41929-019-0351-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-019-0351-2
This article is cited by
-
Catalytic asymmetric oxa-Diels–Alder reaction of acroleins with simple alkenes
Nature Communications (2023)
-
Tandem intermolecular [4 + 2] cycloadditions are catalysed by glycosylated enzymes for natural product biosynthesis
Nature Chemistry (2023)
-
A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions
Nature Chemistry (2023)
-
Discovery and characterization of a terpene biosynthetic pathway featuring a norbornene-forming Diels-Alderase
Nature Communications (2022)
-
Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels–Alderase
Nature Catalysis (2021)