Abstract
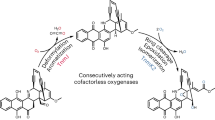
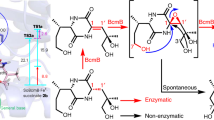
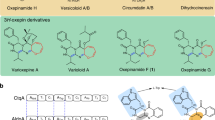
Oxygen heterocycles—in particular, tetrahydropyrans (THPs) and tetrahydrofurans—are common structural features of many biologically active polyketide natural products. Mupirocin is a clinically important antibiotic isolated from Pseudomonas fluorescens and is assembled on a THP ring, which is essential for bioactivity. However, the biosynthesis of this moiety has remained elusive. Here, we show an oxidative enzyme-catalysed cascade that generates the THP ring of mupirocin. Rieske non-haem oxygenase (MupW)-catalysed selective oxidation of the C8–C16 single bond in a complex acyclic precursor is combined with an epoxide hydrolase (MupZ) to catalyse the subsequent regioselective ring formation to give the hydroxylated THP. In the absence of MupZ, a five-membered tetrahydrofuran ring is isolated, and model studies are consistent with cyclization occurring via an epoxide intermediate. High-resolution X-ray crystallographic studies, molecular modelling and mutagenesis experiments of MupZ provide insights into THP ring formation proceeding via an anti-Baldwin 6-endo-tet cyclization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding authors upon reasonable request. X-ray crystallographic data are available in the EMBL-EBI PDB under accession number 6FXD.
References
Newman, D. J. & Cragg, G. M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016).
Carlson, J. C. et al. Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes. Nat. Chem. 3, 628–633 (2011).
Bridwell-Rabb, J., Kang, G., Zhong, A., Liu, H. W. & Drennan, C. L. An HD domain phosphohydrolase active site tailored for oxetanocin-A biosynthesis. Proc. Natl Acad. Sci. USA 113, 13750–13755 (2016).
Fuller, A. T. et al. Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature 234, 416–417 (1971).
Thomas, C. M., Hothersall, J., Willis, C. L. & Simpson, T. J. Resistance to and synthesis of the antibiotic mupirocin. Nat. Rev. Microbiol. 8, 281–289 (2010).
Martin, F. M. & Simpson, T. J. Biosynthetic studies on pseudomonic acid (mupirocin), a novel antibiotic metabolite of Pseudomonas fluorescens. J. Chem. Soc. Perkin Trans. 1, 207–209 (1989).
EI-Sayed, A. K. et al. Characterization of the mupirocin biosynthesis gene cluster from Pseudomonas fluorescens NCIMB 10586. Chem. Biol. 10, 419–430 (2003).
Hothersall, J. et al. Mutational analysis reveals that all tailoring region genes are required for production of polyketide antibiotic mupirocin by Pseudomonas fluorescens: pseudomonic acid B biosynthesis precedes pseudomonic acid A. J. Biol. Chem. 282, 15451–15461 (2007).
Gao, S.-S. et al. Biosynthesis of mupirocin by Pseudomonas fluorescens NCIMB 10586 involves parallel pathways. J. Am. Chem. Soc. 136, 5501–5507 (2014).
Gao, S.-S. et al. Selected mutations reveal new intermediates in the biosynthesis of mupirocin and the thiomarinol antibiotics. Angew. Chem. Int. Ed. 56, 3930–3934 (2017).
Silvian, L. F., Wang, J. & Steitz, T. A. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science 285, 1074–1077 (1999).
Hemmerling, F. & Hahn, F. Biosynthesis of oxygen and nitrogen-containing heterocycles in polyketides. Beilstein J. Org. Chem. 12, 1512–1550 (2016).
Taylor, R. D., MacCoss, M. & Lawson, A. D. G. Rings in drugs. J. Med. Chem. 57, 5845–5859 (2014).
Luhavaya, H. et al. Enzymology of pyran ring A formation in salinomycin biosynthesis. Angew. Chem. Int. Ed. 54, 13622–13625 (2015).
Cooper, S. M. et al. Mupirocin W, a novel pseudomonic acid produced by targeted mutation of the mupirocin biosynthetic gene cluster. Chem. Commun. 9, 1179–1181 (2005).
Kauppi, B. et al. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6, 571–586 (1998).
Sydor, P. K. et al. Regio- and stereodivergent antibiotic oxidative carbocyclizations catalysed by Rieske oxygenase-like enzymes. Nat. Chem. 3, 388–392 (2011).
Suen, W. C. & Gibson, D. T. Isolation and preliminary characterization of the subunits of the terminal component of naphthalene dioxygenase from Pseudomonas putida NCIB 9816-4. J. Bacteriol. 175, 5877–5881 (1993).
Ferraro, D. J., Gakhar, L. & Ramaswamy, S. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338, 175–190 (2005).
Li, B. et al. Whole-cell biotransformation systems for reduction of prochiral carbonyl compounds to chiral alcohol in Escherichia coli. Sci. Rep. 4, 6750 (2014).
Gally, C., Nestl, B. M. & Hauer, B. Engineering Rieske non-heme iron oxygenases for the asymmetric dihydroxylation of alkenes. Angew. Chem. Int. Ed. 54, 12952–12956 (2015).
Jouanneau, Y., Meyer, C. & Duraffourg, N. Dihydroxylation of four- and five-ring aromatic hydrocarbons by the naphthalene dioxygenase from Sphingomonas CHY-1. Appl. Microbiol. Biotechnol. 100, 1253–1263 (2016).
Kan, S. B. J., Huang, X., Gumulya, Y., Chen, K. & Arnold, F. H. Genetically programmed chiral organoborane synthesis. Nature 552, 132–136 (2017).
Baldwin, J. E. Rules for ring closure. J. Chem. Soc. Chem. Comm. 734–736 (1976).
Gilmore, K. & Alabugin, I. V. Cyclizations of alkynes: revisiting Baldwin’s rules for ring closure. Chem. Rev. 111, 6513–6556 (2011).
Gilmore, K., Mohamed, R. K. & Alabugin, I. V. The Baldwin rules: revised and extended. WIREs Comput. Mol. Sci. 6, 487–514 (2016).
Buchan, D. W., Minneci, F., Nugent, T. C., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–W357 (2013).
Hmelo, L. R. et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 10, 1820–1841 (2015).
Holm, L. & Laakso, L. M. Dali server update. Nucleic Acids Res. 44, W351–W355 (2016).
Hotta, K. et al. Enzymatic catalysis of anti-Baldwin ring closure in polyether biosynthesis. Nature 483, 355–358 (2012).
Minami, A. et al. Allosteric regulation of epoxide opening cascades by a pair of epoxide hydrolases in monensin biosynthesis. ACS Chem. Biol. 9, 562–569 (2014).
Capyk, J. K., D’Angelo, I., Strynadka, N. C. & Eltis, L. D. Characterization of 3-ketosteroid 9α-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J. Biol. Chem. 284, 9937–9946 (2009).
Yoshiyama-Yanagawa, T. et al. The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J. Biol. Chem. 286, 25756–25762 (2011).
Li., J., van Belkum, M. J. & Vederas, J. C. Functional characterization of recombinant hyoscyamine 6β-hydroxylase from Atropa belladonna. Bioorg. Med. Chem. 20, 4356–4363 (2012).
Mao, X.-M. et al. Efficient biosynthesis of fungal polyketides containing the dioxabicyclo-octane ring system. J. Am. Chem. Soc. 137, 11904–11907 (2015).
Minami, A. et al. Sequential enzymatic epoxidation involved in polyether lasalocid biosynthesis. J. Am. Chem. Soc. 134, 7246–7249 (2012).
Shichijo, Y. et al. Epoxide hydrolase Lsd19 for polyether formation in the biosynthesis of lasalocid A: direct experimental evidence on polyene-polyepoxide hypothesis in polyether biosynthesis. J. Am. Chem. Soc. 130, 12230–12231 (2008).
Barry, S. M. & Challis, G. L. Mechanism and catalytic diversity of Rieske non-heme iron-dependent oxygenases. ACS Catal. 3, 2362–2370 (2013).
Parkhurst, J. M. et al. Dials. J. Appl. Crystallogr. 49, 1912–1921 (2016).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Vagin, A. & Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010).
Potterton, L. et al. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr. D Struct. Biol. 74, 68–84 (2018).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Struct. Biol. 62, 1002–1011 (2006).
Emsley, P., Lohkamp, B., Scottc, W. G. & Cowtand, K. Features and development of Coot. Acta Crystallogr. D Struct. Biol. 66, 486–501 (2010).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53, 240–255 (1997).
Frisch, M. J. et al. Gaussian (Wallingford, 2009).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Case, D. A. et al. AMBER 2016 (Univ. California, 2016).
Sondergaard, C. R., Olsson, M. H. M., Rostkowski, M. & Jensen, J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 (2011).
Olsson, M. H. M., Sondergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Acknowledgements
We are grateful to the BBSRC and EPSRC for funding through the Bristol Centre for Synthetic Biology (BB/L01386X/1), and the BBSRC for funding through BB/M012107/1 and BB/R007853/1, as well as a David Phillips Fellowship (BB/M026280/1) to M.W.v.d.K. We thank M. Malaysia for a scholarship to N.A.B., and H. P. Schweizer for kindly providing plasmids for gene inactivation and complementation.
Author information
Authors and Affiliations
Contributions
L.W. and C.L.W. designed the experiments, analysed the data and, together with M.P.C. and T.J.S., drafted the manuscript. L.W. conducted the heterologous expression, protein purification, biotransformations, isolation and characterization of metabolites, and site-directed mutagenesis. A.P. and C.W. assisted with protein crystallization and structure determination. M.R.C. assisted with cloning and protein purification. N.A.B. conducted substrate synthesis and model cyclization. M.W.v.d.K. performed the molecular modelling. P.R.R. and M.P.C. led the protein structural studies and contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Figures 1–48, Supplementary Tables 1–2
Rights and permissions
About this article
Cite this article
Wang, L., Parnell, A., Williams, C. et al. A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis. Nat Catal 1, 968–976 (2018). https://doi.org/10.1038/s41929-018-0183-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-018-0183-5
This article is cited by
-
Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nature Catalysis (2023)
-
rpeA, a global regulator involved in mupirocin biosynthesis in Pseudomonas fluorescens NCIMB 10586
Applied Microbiology and Biotechnology (2021)
-
Heterocycle biosynthesis via C–H functionalization
Nature Catalysis (2018)