Abstract
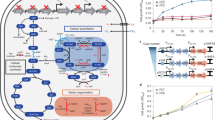
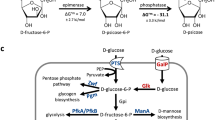
Haem has widespread applications in healthcare and food supplement industries. Escherichia coli has previously been engineered to produce a small amount of haem intracellularly through the C4 pathway, requiring extraction for applications. Here we report secretory production of free haem by engineered E. coli strains, using the C5 pathway and the optimized downstream pathway for haem biosynthesis. Furthermore, knocking out ldhA, pta and also yfeX—encoding a putative haem-degrading enzyme—results in 7.88 mg l−1 of total haem with 1.26 mg l−1 of extracellular haem in flask cultivation. Fed-batch fermentations of the engineered strain overexpressing a haem exporter CcmABC from glucose only and glucose supplemented with l-glutamate secrete 73.4 and 151.4 mg l−1 of haem, respectively, which are 63.5% of 115.5 mg l−1 and 63.3% of 239.2 mg l−1 of total haem produced. The engineered E. coli strain reported here will be useful for microbial production of free haem.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ponka, P. Cell biology of heme. Am. J. Med. Sci. 318, 241–256 (1999).
Poulos, T. L. Heme enzyme structure and function. Chem. Rev. 114, 3919–3962 (2014).
Mayfield, J. A., Dehner, C. A. & DuBois, J. L. Recent advances in bacterial heme protein biochemistry. Curr. Opin. Chem. Biol. 15, 260–266 (2011).
Hoppe, M., Brun, B., Larsson, M. P., Moraeus, L. & Hulthen, L. Heme iron-based dietary intervention for improvement of iron status in young women. Nutrition 29, 89–95 (2013).
Seligman, P. A., Moore, G. M. & Schleicher, R. B. Clinical studies of HIP: an oral heme-iron product. Nutr. Res. 20, 1279–1286 (2000).
Anderson, K. E. & Collins, S. D. Hemin treatment for acute porphyria: implications for clinical practice of an open-label study of 130 patients. J. Investig. Med. 54, S290 (2006).
Fischer, H. & Zeile, K. Synthese des haematoporphyrins, protoporphyrins und haemins. Eur. J. Org. Chem. 468, 98–116 (1929).
Espinas, N. A., Kobayashi, K., Takahashi, S., Mochizuki, N. & Masuda, T. Evaluation of unbound free heme in plant cells by differential acetone extraction. Plant Cell Physiol. 53, 1344–1354 (2012).
In, M. J., Kim, D. C., Chae, H. J. & Oh, N. S. Effects of degree of hydrolysis and pH on the solubility of heme-iron enriched peptide in hemoglobin hydrolysate. Biosci., Biotechnol., Biochem. 67, 365–367 (2003).
Kwon, S. J., de Boer, A. L., Petri, R. & Schmidt-Dannert, C. High-level production of porphyrins in metabolically engineered Escherichia coli: systematic extension of a pathway assembled from overexpressed genes involved in heme biosynthesis. Appl. Environ. Microbiol. 69, 4875–4883 (2003).
Kwon, O. H., Kim, S., Hahm, D. H., Lee, S. Y. & Kim, P. Potential application of the recombinant Escherichia coli-synthesized heme as a bioavailable iron source. J. Microbiol. Biotechnol. 19, 604–609 (2009).
Lee, M. J. et al. Effect of gene amplifications in porphyrin pathway on heme biosynthesis in a recombinant Escherichia coli. J. Microbiol. Biotechnol. 23, 668–673 (2013).
Pranawidjaja, S., Choi, S. I., Lay, B. W. & Kim, P. Analysis of heme biosynthetic pathways in a recombinant Escherichia coli. J. Microbiol. Biotechnol. 25, 880–886 (2015).
Fraser, R., Davis, S.C. & Brown, P.O. Secretion of heme-containing polypeptides. US Patent Application No. 15/021,447 (2016).
Andersen, H.D., Jensen, E.B. & Welinder, K.G. A process for producing heme proteins. European Patent Application No. 0505311A2 PCT/DK1993/000094 (1997).
Anzaldi, L. L. & Skaar, E. P. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78, 4977–4989 (2010).
Feissner, R. E., Richard-Fogal, C. L., Frawley, E. R. & Kranz, R. G. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol. Microbiol. 61, 219–231 (2006).
Layer, G., Reichelt, J., Jahn, D. & Heinz, D. W. Structure and function of enzymes in heme biosynthesis. Protein Sci. 19, 1137–1161 (2010).
Dailey, H. A. et al. Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol. Mol. Biol. Rev. 81, e00048–00016 (2017).
Kang, Z. et al. Recent advances in production of 5-aminolevulinic acid using biological strategies. World J. Microbiol. Biotechnol. 33, 200 (2017).
Ferreira, G. C. & Gong, J. 5-Aminolevulinate synthase and the first step of heme-biosynthesis. J. Bioenerg. Biomembr. 27, 151–159 (1995).
Lin, J. P., Fu, W. Q. & Cen, P. L. Characterization of 5-aminolevulinate synthase from Agrobacterium radiobacter, screening new inhibitors for 5-aminolevulinate dehydratase from Escherichia coli and their potential use for high 5-aminolevulinate production. Bioresour. Technol. 100, 2293–2297 (2009).
Chung, S. Y., Seo, K. H. & Rhee, J. I. Influence of culture conditions on the production of extra-cellular 5-aminolevulinic acid (ALA) by recombinant E. coli. Process Biochem. 40, 385–394 (2005).
Ding, W., Weng, H., Du, G., Chen, J. & Kang, Z. 5-Aminolevulinic acid production from inexpensive glucose by engineering the C4 pathway in Escherichia coli. J. Ind. Microbiol. Biotechnol. 44, 1127–1135 (2017).
Kang, Z., Wang, Y., Gu, P., Wang, Q. & Qi, Q. Engineering Escherichia coli for efficient production of 5-aminolevulinic acid from glucose. Metab. Eng. 13, 492–498 (2011).
Wang, L. Y., Wilson, S. & Elliott, T. A mutant HemA protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J. Bacteriol. 181, 6033–6041 (1999).
Zhang, J., Kang, Z., Chen, J. & Du, G. Optimization of the heme biosynthesis pathway for the production of 5-aminolevulinic acid in Escherichia coli. Sci. Rep. 5, 8584 (2015).
Li, F. et al. Constitutive expression of RyhB regulates the heme biosynthesis pathway and increases the 5-aminolevulinic acid accumulation in Escherichia coli. FEMS Microbiol. Lett. 350, 209–215 (2014).
Vuoristo, K. S. et al. Metabolic engineering of the mixed-acid fermentation pathway of Escherichia coli for anaerobic production of glutamate and itaconate. AMB Express 5, 61 (2015).
Letoffe, S., Heuck, G., Delepelaire, P., Lange, N. & Wandersman, C. Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc. Natl Acad. Sci. USA 106, 11719–11724 (2009).
Dailey, H. A. et al. The Escherichia coli protein YfeX functions as a porphyrinogen oxidase, not a heme dechelatase. MBio 2, e00248–00211 (2011).
Turlin, E. et al. Protoporphyrin (PPIX) efflux by the MacAB-TolC pump in Escherichia coli. MicrobiologyOpen 3, 849–859 (2014).
Suzuki, T., Yamane, T. & Shimizu, S. Phenomenological background and some preliminary trials of automated substrate supply in pH-stat modal fed-batch culture using a setpoint of high limit. J. Ferment. Bioeng. 69, 292–297 (1990).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–U341 (2009).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Kim, J. M., Lee, K. H. & Lee, S. Y. Development of a markerless gene knock-out system for Mannheimia succiniciproducens using a temperature-sensitive plasmid. FEMS Microbiol. Lett. 278, 78–85 (2008).
Palmeros, B. et al. A family of removable cassettes designed to obtain antibiotic-resistance-free genomic modifications of Escherichia coli and other bacteria. Gene 247, 255–264 (2000).
Burnham, B. F. δ-Aminolevulinic acid synthase (Rhodopseudomonas spheroides). Methods Enzymol. 17, 195–200 (1970).
Orth, J. D. et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism—2011. Mol. Syst. Biol. 7, 535 (2011).
Ebrahim, A., Lerman, J. A., Palsson, B. O. & Hyduke, D. R. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst. Biol. 7, 74 (2013).
Acknowledgements
We thank Won Jun Kim for discussion. This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT, through the National Research Foundation (NRF) of Korea.
Author information
Authors and Affiliations
Contributions
X.R.Z. and S.Y.L. designed the project. X.R.Z. and K.R.C. performed the experiments and analysed the data. X.R.Z., K.R.C. and S.Y.L. wrote the manuscript together.
Corresponding author
Ethics declarations
Competing interests
The authors declare competing financial interests as a patent (Korean Patent Application No. 10-2017-0170185) of commercial interest entitled “Method for Producing Exracellular Haem Using Metabolically Engineered Microorganism” is filed on this technology by Korea Advanced Institute of Science and Technology. S.Y.L., X.R.Z. and K.R.C. are the inventors of this patent that covers the construction and fermentation of mircroorganisms capable of secretory production of haem reported in this manuscript.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–6, Supplementary Figures 1–17, Supplementary Tables 1–3, Supplementary References
Rights and permissions
About this article
Cite this article
Zhao, X.R., Choi, K.R. & Lee, S.Y. Metabolic engineering of Escherichia coli for secretory production of free haem. Nat Catal 1, 720–728 (2018). https://doi.org/10.1038/s41929-018-0126-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-018-0126-1
This article is cited by
-
A microbial process for the production of benzyl acetate
Nature Chemical Engineering (2024)
-
Recent advances in microbial synthesis of free heme
Applied Microbiology and Biotechnology (2024)
-
Heme biosensor-guided in vivo pathway optimization and directed evolution for efficient biosynthesis of heme
Biotechnology for Biofuels and Bioproducts (2023)
-
Improved biosynthesis of heme in Bacillus subtilis through metabolic engineering assisted fed-batch fermentation
Microbial Cell Factories (2023)
-
Engineering Escherichia coli for efficient assembly of heme proteins
Microbial Cell Factories (2023)