Abstract
The COVID-19 global pandemic has resulted in international efforts to understand, track, and mitigate the disease, yielding a significant corpus of COVID-19 and SARS-CoV-2-related publications across scientific disciplines. Throughout 2020, over 400,000 coronavirus-related publications have been collected through the COVID-19 Open Research Dataset. Here, we present CO-Search, a semantic, multi-stage, search engine designed to handle complex queries over the COVID-19 literature, potentially aiding overburdened health workers in finding scientific answers and avoiding misinformation during a time of crisis. CO-Search is built from two sequential parts: a hybrid semantic-keyword retriever, which takes an input query and returns a sorted list of the 1000 most relevant documents, and a re-ranker, which further orders them by relevance. The retriever is composed of a deep learning model (Siamese-BERT) that encodes query-level meaning, along with two keyword-based models (BM25, TF-IDF) that emphasize the most important words of a query. The re-ranker assigns a relevance score to each document, computed from the outputs of (1) a question–answering module which gauges how much each document answers the query, and (2) an abstractive summarization module which determines how well a query matches a generated summary of the document. To account for the relatively limited dataset, we develop a text augmentation technique which splits the documents into pairs of paragraphs and the citations contained in them, creating millions of (citation title, paragraph) tuples for training the retriever. We evaluate our system (http://einstein.ai/covid) on the data of the TREC-COVID information retrieval challenge, obtaining strong performance across multiple key information retrieval metrics.
Similar content being viewed by others
Introduction
The evolution of the SARS-CoV-2 virus, with its unique balance of virulence and contagiousness, has resulted in the COVID-19 pandemic. Since December 2019, the disease threatens exponential spread across our society, catalyzed by a modern air and road transportation system, along with dense urban centers where close contact amongst people yielded hubs of viral spread.
Global efforts have arisen in an attempt to quell the spread of the virus. National governments have shut down entire economic sectors, enforcing stay-at-home orders for many people. Hospitals have restructured themselves to cope with an unprecedented influx of intensive care unit patients, sometimes growing organically to increase their number of beds1. Institutions have adjusted their practices to support efforts—repurposing assembly lines to build mechanical ventilators2, delaying delivery of non-COVID-related shipments3, creating contact-tracing mobile apps4 and "digital swabs”5 to track symptoms and potential spread. Pharmaceutical enterprises and academic institutions have invested significantly in developing vaccines and therapeutics6, while deeply studying both COVID-19 and SARS-CoV-2.
The health impacts of this crisis have been matched only by the economic backlash to society. Hundreds of thousands of small businesses have shut down, entire industrial sectors have been negatively impacted7, and tens of millions of workers have been laid off or furloughed8. Even after our global society succeeds at controlling the virus’s spread, we will be faced with many challenges, including re-opening our societies, lifting stay-at-home orders, deploying better testing, developing vaccines and therapeutics, aiding the unemployed and out-of-business, etc.
The global response to COVID-19 has yielded a growing corpus of scientific publications—increasing at a rate of thousands per week—about COVID-19, SARS-CoV-2, other coronaviruses, and related topics9. The individuals on the front lines of the fight—healthcare practitioners, policy makers, medical researchers, etc.—will require specialized tools to keep up with the literature.
CO-Search is a cascaded retriever-ranker semantic search engine that takes complex search queries (e.g. natural language questions), and retrieves scientific articles strictly over the coronavirus-related literature. CO-Search indexes content from over 400,000 scientific papers made available through the COVID-19 Open Research Dataset Challenge (CORD-19)9—an initiative put forth by the US White House and other prominent institutions in early 2020. The goal of this line of work is to offer an alternative, scientific search engine, designed to limit misinformation in a time of crisis.
We evaluate CO-Search on data from the TREC-COVID challenge10—a five-round information retrieval (IR) competition for COVID-19 search engines—using several standard IR metrics: normalized discounted cumulative gain (nDCG), precision with N documents (P@N), mean average precision (MAP), and binary preference (Bpref). For full details see the “Methods” section. TREC-COVID considers IR system submissions that are either manual—in which queries and retrieved documents may be manually adjusted by a human operator—or automatic (such as CO-Search)—in which they may not. A third category is accepted in Rounds 2–5, of type feedback, in which systems are trained with supervision from the annotations of prior rounds. Submissions compete on a predefined set of topics, and are judged using a number of metrics, including those listed above. Expert human annotators provide relevance judgments on a small set of topic–document pairs, which are included, together with non-annotated pairs, in the evaluation.
The CORD-199 coronavirus-related literature corpus, primarily from PubMed, mostly published in 2020, has quickly generated a number of data science and computing works11. These cover topics from IR to natural language processing (NLP), including applications in question answering12, text summarization, and document search10.
In 2020, more than 20 organizations have launched publicly accessible search engines using the CORD-19 corpus. For instance, Neural Covidex13 was constructed from various open source information-retrieval building blocks, as well as a deep learning transformer14 finetuned on a machine-reading comprehension dataset (MS MARCO)15 to predict query-document relevance, for ranking. SLEDGE16 extends this by using SciBERT17—the scientific text-trained version of the prominent BERT18 NLP model—also finetuned on MS MARCO, to re-rank articles retrieved with BM25.
One of the first question–answering systems built on top of the CORD-19 corpus is CovidQA (http://covidqa.ai), which includes a small number of questions from the CORD-19 tasks12. CAiRE is a multi-document summarization system19 which works by first pre-training on both a general text corpus20,21 and a biomedical review dataset, then finetuning on the CORD-19 dataset.
One of the applications of the corpus has been Named Entity Recognition (NER). Wang et al.22 introduce the COVID-NER corpus, which includes 75 fine-grained entity types, both conventional (e.g., genes, diseases, and chemicals) and corpus-specific (e.g., viral proteins, coronaviruses, substrates, and immune responses). Ahamed and Samad perform a network analysis of the corpus23, in which they use word associations to identify the phrases that co-occur with the most medically relevant keywords. This allows them to identify information about different antiviral drugs, pathogens, and pathogen hosts, as well as proteins and medical therapies, as to how they are connected to the central topic of “coronavirus”.
Broader surveys11 of the COVID-19-related literature have already arisen, covering a wider range of research perspectives including molecular, clinical, and societal factors. Roberts et al. (2020)10 offers an in-depth analysis of the TREC-COVID competition structure, including the notable differences in IR systems for pandemics, which deviate substantially from typical IR systems. They address key questions around COVID-19-specific IR systems, including: How are topics different from typical web-based search? What is the appropriate search content? How to deploy quickly? What are the appropriate IR modalities? How to customize IR systems for pandemics? Can existing data be leveraged? How to best respond to the rapidly growing literature corpus? How to evaluate systems? And so forth. COVID search engines differ from more general neural IR engines24,25 because of the relatively limited and focused, and also rapidly changing collection of documents. Another recent system paper from the challenge is ref. 26, in which the authors describe an ensemble system that combines more than 100 IR methods, including lexical rankers, embeddings, as well as relevance feedback. Our proposed method builds on these insights by selectively choosing three deep-learning methods and showing how they each enhance COVID-specific scientific search.
Results
Dataset
To quantitatively evaluate the effectiveness of our search engine, we combine the CORD-19 corpus with the TREC-COVID competition’s evaluation dataset. The evaluation dataset consists of topics, along with relevance judgments which assign topic–document pairs into one of the following groups: irrelevant, partially relevant, or relevant. See Table 1 for example topics. The relevance judgments are determined by human experts in related fields (biology, medicine, etc.).
The U.S. White House, along with the U.S. National Institutes of Health, the Allen Institute for AI, the Chan-Zuckerberg Initiative, Microsoft Research, and Georgetown University recently prepared the CORD-19 Challenge in response to the global crisis. As of February 2021, this resource consists of over 400,000 scientific publications (up from 29,000 at the challenge inception in February 2020) about COVID-19, SARS-CoV-2, and earlier coronaviruses9.
This challenge represents a call to action to the artificial intelligence (AI) and IR communities to "develop text and data mining tools that can help the medical community develop answers to high priority scientific questions”. It is currently the most extensive coronavirus literature corpus publicly available.
To build on CORD-19, the Text Retrieval Conference (TREC) recently partnered with the National Institute of Standards and Technology (NIST), to define a structured and quantitative evaluation system for coronavirus IR systems. The TREC-COVID challenge10 is composed of five successive rounds of evaluation on 30–50 topics. The first round includes 30 topics. Each subsequent round takes the prior round’s topics and adds five new ones.
Each topic is represented as a tuple consisting of a query, a question, and a narrative, with an increasing amount of detail in each. IR systems must retrieve up to 1000 ranked documents per topic from the CORD-19 publications, and are evaluated on many metrics. See the “Methods” section for further details.
System architecture
CO-Search consists of a retriever, which returns a sorted subset of documents from the general corpus, a re-ranker, which further sorts them, and an offline pre-processing step known as document indexing, which parses documents via a combination of deep learning and keyword-based techniques to make them both semantically and syntactically searchable at scale. This process converts pieces of raw text into high-dimensional vector representations, such that one vector’s proximity to another indicates similar content. The full system is shown in Fig. 2.
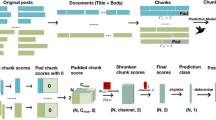
The index is created by processing documents in three ways: a deep learning model called Siamese-BERT (SBERT27) embeds single paragraphs and image captions, and two keyword-based models (TF-IDF, BM2528) vectorize entire documents (see Fig. 2a). SBERT is an extension of the widely used BERT18 language representation model which uses two BERT models with tied network parameters. It has been shown to be superior to BERT in semantic similarity search27 by being significantly more computationally efficient at learning correspondences between sentences. For instance, finding the most similar pair of sentences, using BERT, in a collection of n = 10,000 sentences would require each possible pair to be fed into the network, one sentence at a time, yielding n(n − 1)/2 = 49,995,000 inference computations, or about 65 h on an NVIDIA V100 GPU. In contrast, SBERT reduces this to 10,000 inference computations and the computation of cosine similarity distances between them, yielding about 5 s of compute time. SBERT is trained to take a short text string and a longer text document and output the correspondence between the two (i.e. their similarity) as a real-valued number between 0 and 1. In this use case, semantic embeddings from the SBERT model face the challenge of working with a relatively small number of long documents. We account for this by pre-training SBERT on a large, synthetic dataset of millions of training examples, constructed as follows. We split documents into paragraphs, extract the titles of the citations of each paragraph, and form a bipartite graph of paragraphs and citations with edges implying that a citation c came from a paragraph p. We use the graph to form tuples ((p, c) s.t.c ∈ p) for training SBERT to predict if a title was cited by a paragraph. Additionally, we generate an equivalent number of negative training samples of incorrect tuples ((p, c) s.t.c ∉ p). The full pipeline for this step is shown in Fig. 1a.
a Documents are split into paragraphs and the citations included in them to form a bipartite graph that induces training tuples (p, c). These are fed to a Siamese-BERT (SBERT) model trained to discern if a citation is contained in a given paragraph. This process makes SBERT match user search queries to scientific publication titles. b t-SNE visualization of the SBERT embeddings of entire documents, each denoted by a single point. Their color represents the topic to which they are most closely matched. Notably, queries pertaining to the same topic tend to cluster together.
The structure of the embedded space is such that proximal queries and documents share semantic meaning. Visualizing this reveals a human-understandable clustering of documents and topics. Figure 1b shows a two-dimensional t-SNE29 plot—an effective method for visualizing high-dimensional data—of the embedded space, with different colors representing topics of TREC-COVID, and points representing documents. We can observe that semantically similar documents cluster by topic.
Document retrieval (Fig. 2b, top row)—which returns a list of the top 1000 documents for a query—is accomplished by fusing the returned lists of the SBERT, TF-IDF, and BM25 models. SBERT allows for variable-length queries and documents to be embedded into the same vector space (the multi-dimensional internal representation of the data, by the model), in order to model semantic proximity and enable k-nearest-neighbor (kNN) retrieval. We use approximate kNN retrieval using the Annoy framework (https://github.com/spotify/annoy), to account for the large number of paragraphs parsed by SBERT. TF-IDF and BM25 independently return two document lists (TF-IDF uses kNN with cosine distance; BM25 uses a Lucene inverted index30, built with Anserini) that either share in the most unique keywords of the query (TF-IDF) or share many of the same keywords as the query (BM25-Anserini)28.
a Indexing: Raw documents are processed into a searchable format. Documents are split into paragraphs and image captions, embedded with an SBERT deep learning model, and stored into an index. The raw documents are also embedded with two-keyword-based models (TF-IDF and BM25). b Retrieval and re-ranking: The system computes a linear combination of TF-IDF and SBERT retrieval scores, then combines them with the retrieval scores of BM25 using reciprocal rank fusion31, to generate a sorted candidate list. k-Nearest-Neighbors are used for TF-IDF and SBERT, and the Lucene Inverted Index is used for BM25. The retrieved documents and the query are parsed using a question answering model and an abstractive summarizer prior to being re-ranked based on answer match, summarization match, and retrieval scores.
These three lists are then combined by first linearly fusing the SBERT list with the TF-IDF list, then using reciprocal rank fusion (RRF)31 to merge this with the BM25 list. This retrieval process returns the top 1000 documents as a function of their semantic and syntactic distance to the query.
Document re-ranking (Fig. 2b, bottom row) takes this set of documents, runs them through both a question–answering module (QA) and a summarizer, then ranks the documents by a weighted combination of their original retrieval scores, the QA output, and the summarizer output. Whereas standard question answering systems generate answers, our model extracts multiple answer candidates (text spans) from the paragraphs of the retrieved documents. This is accomplished by taking the query and the retrieved paragraphs, and using a sequential paragraph selector32, to filter for a set of paragraphs that, when combined, could answer the query. Specifically, the model uses multi-hop reasoning to model relationships between paragraphs, and selects sequentially ordered sets of them. It is pre-trained using a Wikipedia-derived dataset of 113k question–answer pairs and sentence-level supporting facts33, and further finetuned on a QA dataset built from PubMed34, for biomedical specificity. Once filtered, these sequential paragraph sets are fed into a reading comprehension model (trained on a standard question–answering dataset with topic structure similar to CORD-1935) to extract answer candidates.
In a parallel fashion, the summarizer generates a single abstractive summary from the retrieved documents. It is built in an encoder–decoder fashion, in which an encoder (BERT18) first embeds an entire document, and a decoder (a modified GPT-2 model36) converts this embedding into raw text, outputting a summary. To increase the probability that a generated summary matches (and thus, helps re-rank) the contents of the retrieved paragraphs, we tuned the model to generate short summaries of fewer than 65 words37.
Finally, the system uses the generated answers and summary to compute two scores for each retrieved document. The first measures the relevance of a document, given the query, and the second measures the degree to which any single document summarizes the entire set of retrieved documents. These two scores are combined with the original relevance scores to output a final ranked list of documents.
Evaluation
We evaluate our system quantitatively using the CORD-19 document dataset and the topics and relevance judgments provided by TREC-COVID. The dataset contains five sets of topics, where each topic is represented as a (query, question, narrative) tuple. Relevance judgments—provided on a very small subset of all possible topic–document pairs—scores topic–documents as irrelevant, partially relevant, or relevant. These judgments have been iteratively gathered throughout the course of the five-round TREC-COVID competition, in which search engines submitted up to 1000 ranked documents per query, and the organizers pooled from amongst the most common topic–document pairs for judging (i.e. depth-N pooling, in which the top N documents from each response provided by the set of contributing systems are judged for relevance by human assessors38, with N ranging from 7 to 20 for the various rounds). The pool depths results in many relevant documents being missed. Though this labeling procedure is inherently sparse and somewhat biased, this is the best available method for evaluating IR systems, as obtaining relevance judgments on all possible topic–document pairs is infeasible.
In order to better evaluate our approach, we use a variety of IR metrics. Key amongst them are high-precision metrics such as nDCG, top-N precision, and MAP. The critical limitation with these is that their effectiveness relies on complete relevance judgments across all topic–document pairs. To account for this, an additional metric, Bpref, which is robust to missing relevance judgments, is considered. For full details, see the “Methods” section.
Our results on this data are shown in Table 2. We compare the performance of our system in two contexts. The first context is within the general set of submissions. This includes metric evaluations on all documents—annotated and non-annotated—and this includes ranking against the three possible system types in the competition: manual, automatic, and feedback systems. Manual submissions use human operators that can iteratively adjust the query or the retrieved documents to improve ranking. Feedback systems are trained using the relevance judgments of prior rounds. Automatic search engines may not do either. Strictly speaking, feedback systems are also automated (in that they do not use a human in the loop), though they have an inherent advantage over automatic systems and are thus considered separately. In the second context, we evaluate our system (and all others) strictly on relevance judgments, and we compare our automatic system strictly against other automatic systems. Specifically, we re-score every automatic system’s runs after removing non-judged topic–document pairs. To determine team rankings, we account for both multiple submissions per team, and for multiple submissions with the same score, assigning to each the highest one (i.e., if the top two scoring submissions for a metric have the same score, each would be ranked #1).
Each round builds on the previous rounds, adding five new topics, many documents, as well as new relevance judgments. As a result, Round 5 is the most complete round. In the first context (columns “All submissions, All pairs”), our system ranks in the top 21 (Table 2) across all rounds. In considering the rankings from Round 1 through Round 5, there is a pronounced improvement in rankings from Round 1 to Round 2, with a drop then plateau in performance from Rounds 3 to 5. The improvement from Round 1 to 2 can be explained by the judgment fraction—the percentage of relevance judgments goes up, increasing the performance across these metrics. This happens because metrics such as precision penalize search engines for retrieving relevant but non-annotated documents for a topic. Rounds 3–5 have sufficient relevance judgments from prior rounds to improve feedback systems, leading to a drop in the ranking.
In the second context, our system ranks in the top 6 across all metrics and all rounds, in the top 4 across all but four, and as the top 1 system across half of them. The stability in performance is largely due to the consistent judgment fraction (100%, implicitly), and the absence of feedback and manual systems, both of which improve with relevance judgments. This stability—evident also in the metrics—implies a system that is robust to increasing corpus size.
Of note, the availability of relevance judgments is quite sparse throughout all rounds, with Round 1 exhibiting a coverage of 0.57%, and Round 5 a coverage of 0.24%. This is precisely what motivates the use of the Bpref metric, which is robust to missing annotations, as evidenced by its consistency across contexts.
Discussion
Here we present CO-Search, a scientific search engine over the growing corpus of COVID-19 literature. We train the system using the scientific papers of the COVID-19 Open Research Dataset challenge, and evaluate its performance using the data of the TREC-COVID competition on a number of key metrics, achieving strong performance across metrics and competition rounds. The system uses a combination of semantic and keyword-based models to retrieve and score documents. It then re-ranks these documents by using a Wikipedia-trained & PubMed-trained question–answering system, together with an abstractive summarizer, to modulate retrieval scores.
We perform an ablation study of our system using Round 5 data (first context) in order to examine the performance effects of its components (Table 3). This is done in two steps, first for the retriever, then for the re-ranker. For each, we analyze the metric performance of various components individually, and united. The retriever’s components (TF-IDF, BM25, SBERT) each perform poorly, but benefit from substantial synergy when united into the full retrieval pipeline (top half of Table 3). This occurs because keyword-based techniques, on their own, do not perform as well on queries in natural language. Similarly, semantic techniques tend to underweight the most salient keywords of a natural language query. Combined, these two techniques work well for this unique dataset. The retrieval subsystem accounts for most of the performance of the overall system. The addition of the re-ranker, with its two other deep learning modules (Q&A, summarizer) serve to further boost this performance on the order of 1–2% across the various metrics employed.
We compare our system against three of the top-performing systems of Round 5, as shown in Table 4. As can be seen, no single system outperforms the rest across all metrics, indicating the possibility of forming hybrid systems that benefit from the strengths of each. The system covidex13 uses a transformer fine-tuned on the MedMARCO machine-reading comprehension dataset16 to predict query-document relevance. The system uogTr linearly combines a SciBert model17 trained on the medical queries of MSMarco15 and SciColBERT. The system unique_ptr leverages synthetic query generation39 for training data augmentation. RRF enables easy merging of ideas. It would be straightforward for CO-Search to be extended to benefit from these ideas: synthetic query generation could augment the SBERT training tuples shown in Fig. 1; the outputs of both a medically fine-tuned SciBert model, or a transformer fine-tuned on the MedMARCO data, could be joined with our own output via RRF.
From Round 5, the two topics on which CO-Search performs best, as ranked by Bpref, are “what kinds of complications related to COVID-19 are associated with diabetes” and “are patients taking Angiotensin-converting enzyme inhibitors (ACE) at increased risk for COVID-19?”. Conversely the system performs worst on “what are the guidelines for triaging patients infected with coronavirus?” and “what causes death from Covid-19?”. This is likely due to the hybrid semantic-syntactic nature of the system. The keyword models allow the system to focus in on important words like “diabetes” and “angiotensin”, while the semantic SBERT model would focus on broader meanings inherent in pieces of the text such as “complications..associated with...”. Note that the worst-performing topics lack the obvious keywords of the first.
The semantic search capability of CO-Search allows it to disambiguate between subtle variations in word ordering that, in biological contexts, result in critically different meanings (e.g. “What regulates expression of the ACE2 protein?” vs. “What does the ACE2 protein regulate?”), maximizing its utility to the medical and scientific communities in a time of crisis. Key to the fair evaluation of the system is considering the general use case (all IR systems, all documents), and a specific use case (automatic systems, judged documents).
This work is intended as a tool to support the fight against COVID-19. In this time of crisis, tens of thousands of documents are being published, only some of which are scientific, rigorous, and peer-reviewed. This may lead to the inclusion of misinformation and the potential rapid spread of scientifically disprovable or otherwise false research and data. People on the front lines—medical practitioners, policy makers, etc.—are time-constrained in their ability to parse this corpus, which could impede their ability to approach the returned search results with the appropriate levels of skepticism and inquiry available in less exigent circumstances. Coronavirus-specialized search capabilities are key to making this wealth of knowledge both useful and actionable. The risks are not trivial, as decisions made based on returned, incorrect, or demonstrably false results might jeopardize trust or public health and safety. The authors acknowledge these risks, but believe that the overall benefits to researchers and to the broader COVID-19 research agenda outweigh the risks.
Methods
Evaluation metrics
Below we define key metrics in evaluation. Throughout this work we adopt the standard convention that m@N refers to an evaluation using metric m, and the top N retrieved documents.
Precision (P):
nDCG: For position i ∈ {0, 1, . . . , N}, the nDCG of a retrieved set of documents over Q queries is given by
where \({\,\text{rel}\,}_{i}^{(q)}\) denotes the relevance of entry i, ranked according to query q. IDCG denotes the ideal and highest possible DCG. In the limit of perfect annotations, nDCG performs reliably in measuring search engine performance. Since it treats non-annotated documents as incorrect (reli evaluates to zero), it is less reliable for datasets with incomplete annotations.
MAP: The average precision (AP) of a retrieved document set is defined as the integral over the normalized precision-recall curve of the set’s query. MAP is defined as the mean AP over all queries:
where R is recall, Pq is precision as a function of recall, for a particular query. Note that, as in the case of nDCG, MAP penalizes search engines that yield accurate but unique (i.e. non-annotated) results, since non-annotated documents are treated as irrelevant by P.
Bpref: Bpref strictly uses information from judged documents. It is a function of how frequently relevant documents are retrieved before non-relevant documents. In situations with incomplete relevance judgments (most IR datasets) it is more stable than other metrics, and it is designed to be robust to missing relevance judgments. It gives roughly the same results with incomplete judgments as MAP would give with complete judgments38. It is defined as
where R is the number of judged relevant documents, r is a relevant retrieved document, n is one of the first R irrelevant retrieved documents, and non-judged documents are ignored.
Document indexing
We train the SBERT model of the indexing step with cross-entropy loss, Adam optimization40 with a learning rate of 2e–5, a linear learning rate warm-up over 10% of the training data, and a default pooling strategy of MEAN (see Fig. 1a).
Document retrieval
At runtime, the retrieval step takes an input query, embeds it using SBERT, computes approximate nearest neighbors over the SBERT paragraph embeddings, and returns a set of paragraphs, together with each paragraph’s cosine similarity to the query. TF-IDF and BM25 take as input queries and documents, returning vectors \(t\in {{\mathbb{R}}}^{M}\) and \(b\in {{\mathbb{R}}}^{M}\) such that ti = TF-IDF(query, document i), bi = BM25(query, document i), and M is the size of the document corpus. We build a Lucene index with BM25 retrieval function with default parameters of k1 = 1.2, b = 0.75 in the Anserini IR toolkit. The formula for TF-IDF is given by
where tf(t, d) is the term frequency—the number of times term t appears in document d—and df(t) is the document frequency—the number of documents in the set that contain term t. We use the scikit-learn41 version of TF-IDF, with a vocabulary size of 13,000, a max document frequency of 0.5, a minimum document frequency of 3, and L2 normalization42 of the vectors computed from Eq. (5), above.
The SBERT and TF-IDF scores are combined linearly. For document d (containing paragraphs p), and query q, with subscript es denoting an SBERT embedding, their combination C is given by
This induces a ranking \({R}_{{\mathrm {C}}}^{q}\) on the documents, which is then combined with the BM25-induced ranking \({R}_{{\mathrm {B}}}^{q}\) using reciprocal ranked fusion31, to obtain a final retrieved ordering:
In practice, we find that the constants μ = 0.7 and k = 60 yield good results. Future work could consider using a learned layer to attend over semantic embeddings and keyword vectors, given the query.
Document re-ranking
Re-ranking combines the RRF scores of the retrieved documents with the outputs of the QA engine and the summarizer. We define Q to measure the degree to which a document answers a query:
where 1(x) is the indicator function: 1(x) = {1 if x is true, 0 otherwise}. The set A(q) contains the text span outputs of the QA model. We define S to measure the degree to which a document summarizes the set of documents retrieved for a query:
where M(q)e is the embedded abstractive summary of q, summarized across all retrieved documents. Then the final ranking score R(d, q) of a document, for a particular query, is given by
With higher scores indicating better matches. In essence, rank score R is determined by letting S and Q modulate the retrieval score of a query–document pair.
Question-Answering: We follow the HotPotQA setup32 and all model parameters contained therein. We use paragraphs with high TF-IDF scores for the given query as negative examples for the sequential paragraph selector. The original beam search is modified to include paragraph diversity and avoid extracting the same answers from different paths.
Abstractive summarization: We extend the original GPT-2 model by adding a cross-attention function alongside every existing self-attention function. We constrain the cross-attention function to attend strictly to the final layer outputs of the encoder. We use the base models and hyperparameters of Wolf et al.43, with 12 layers, 768-dimensional activations in the hidden layers, and 12 attention heads. The model is pre-trained using self-supervision with a gap-sentence generation objective44, where we select a random source sentence per document, replace it with a special mask token in the input 80% of the time, and use that sentence as a prediction target in all cases. We then finetune the model with single-document supervised training, using the first 512 tokens of CORD-19 documents after the abstract as input, and the first 300 tokens of the abstract as target output.
Abstracts are split into five groups based on the number of tokens: <65, 65–124, 125–194, 195–294, >295. During training, a special token is provided to specify the summary length in these five categories. At inference time, the model is initialized to output summaries of token lengths <65 in order to generate more concise summaries.
To adapt the model to operate on multiple retrieved paragraphs from different documents, we concatenate the first four sentences of the retrieved paragraphs until they reach an input length of 512 tokens, then feed this into the summarization model.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data used in this study was taken from the COVID-19 Open Research Dataset Challenge9, and is publicly available. The aggregated data analyzed in this study will be made available upon reasonable request.
Code availability
The code used in this study will be made available upon reasonable request.
References
Thomala, L. L. Number of new hospital beds to be added in the designated hospitals after the coronavirus covid-19 outbreak in Wuhan, China as of February 2, 2020. Statista. https://www.statista.com/statistics/1095434/china-changes-in-the-number-of-hospital-beds-in-designated-hospitals-after-coronavirus-outbreak-in-wuhan/.
Bogage, J. Tesla unveils ventilator prototype made with car parts on youtube. Wash. Post. https://www.washingtonpost.com/business/2020/04/06/tesla-coronavirus-ventilators-musk/ (2020).
Day, M. & Soper, S. Amazon is prioritizing essential products as online orders spike. Bloomberg. https://www.bloomberg.com/news/articles/2020-03-17/amazon-prioritizing-essentials-medical-goods-in-virus-response.
Nicas, J. & Wakabayashi, D. Apple and google team up to ‘contact trace’ the coronavirus. N. Y. Times. https://www.nytimes.com/2020/04/10/technology/apple-google-coronavirus-contact-tracing.html (2020).
Armitage, H. Stanford medicine launches national daily health survey to predict covid-19 surges, inform response efforts. Stanford Medicine News Center. http://med.stanford.edu/news/all-news/2020/04/daily-health-survey-for-covid-19-launched0.html.
Liu, C. et al. Research and Development on Therapeutic Agents and Vaccines for Covid-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 6, 315–331 (2020).
Warwick, M., and F. Roshen. The global macroeconomic impacts of COVID-19: Seven scenarios. Centre for Applied Macroeconomic Analysis (CAMA) Working Paper 19/2020, Australia National University (2020).
Thomas, P., Chaney, S. & Cutter, C. New covid-19 layoffs make job reductions permanent. Wall Street J. https://www.wsj.com/articles/new-covid-19-layoffs-make-job-reductions-permanent-11598654257 (2020).
Wang, L. L. et al. Cord-19: The covid-19 open research dataset. arXiv preprint arXiv:2004.10706 (2020).
Roberts, K. et al. TREC-Covid: rationale and structure of an information retrieval shared task for covid-19. J. Am. Med. Inform. Assoc. 27, 1431–1436 (2020).
Bullock, J., Luccioni, A., Pham, K. H., Lam, C. S. N. & Luengo-Oroz, M. Mapping the landscape of artificial intelligence applications against COVID-19. J. Artif. Intell. Res. 69, 807–845 (2020).
Tang, R. et al. Rapidly bootstrapping a question answering dataset for COVID-19. Preprint at arXiv:2004.11339 (2020).
Zhang, E., Gupta, N., Nogueira, R., Cho, K. & Lin, J. Rapidly deploying a neural search engine for the COVID-19 open research dataset: Preliminary thoughts and lessons learned. Preprint at arXiv:2004.05125 (2020).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21, 1–67 (2020)
Bajaj, P. et al. MS Marco: a human generated machine reading comprehension dataset. arXiv preprint arXiv:1611.09268 (2016).
MacAvaney, S., Cohan, A. & Goharian, N. SLEDGE: a simple yet effective baseline for coronavirus scientific knowledge search. Preprint at https://arxiv.org/abs/2005.02365 (2020).
Beltagy, I., Lo, K. & Cohan, A. Scibert: A pretrained language model for scientific text. In (eds. Inui, K., Jiang, J., Ng, V. & Wan, X.), EMNLP-IJCNLP, 3613–3618 (Association for Computational Linguistics, 2019).
Devlin, J., Chang, M. W., Lee, K. & Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805 (2018).
Su, D. et al. Caire-covid: a question answering and multi-document summarization system for COVID-19 research. Preprint at arXiv:2005.03975 (2020).
Dong, L. et al. Unified language model pre-training for natural language understanding andgeneration. In (eds Wallach, H. M. et al.) NeurIPS, 13042–13054 (Curran Associates, Inc., 2019).
Lewis, M. et al. BART: Denoising sequence-to-sequence pre-training for natural language generation, translation, and comprehension. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, 7871–7880 (Association for Computational Linguistics, Online, 2020).
Wang, X., Song, X., Guan, Y., Li, B. & Han, J. Comprehensive named entity recognition on CORD-19 with distant or weak supervision. Preprint at arXiv:2003.12218 (2020).
Ahamed, S. & Samad, M. Information mining for COVID-19 research from a large volume of scientific literature. Preprint at arXiv:2004.02085 (2020).
Mitra, B. & Craswell, N. Neural models for information retrieval. Preprint at arxiv: 1705.01509 (2017).
Guo, J. et al. A deep look into neural ranking models for information retrieval. Inf. Process. Manag. 57, 102067 (2020).
Bendersky, M. et al. Rrf102: meeting the trec-covid challenge with a 100+ runs ensemble. Preprint at arxiv: 2010.00200 (2020).
Reimers, N. & Gurevych, I. Sentence-BERT: Sentence embeddings using Siamese BERT-Networks. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processingand the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), 3973–3983 (2019).
Yang, P., Fang, H. & Lin, J. Anserini: Enabling the use of lucene for information retrieval research. In Proc. 40th International ACM SIGIR Conference on Research and Development in Information Retrieval, 1253–1256. https://doi.org/10.1145/3077136.3080721 (2017).
Maaten, L. v. d. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Białecki, A., Muir, R., Ingersoll, G. & Imagination, L. Apache lucene 4. In SIGIR 2012 Workshop on Open Source Information Retrieval, Vol. 17. https://www.semanticscholar.org/paper/Apache-Lucene-4-Bialecki-Muir/2795d9d165607b5ad6d8b9718373b82e55f41606 (2012).
Cormack, G. V., Clarke, C. L. & Buettcher, S. Reciprocal rank fusion outperforms condorcet and individual rank learning methods. In SIGIR 2009, 758–759. https://www.semanticscholar.org/paper/Reciprocal-rank-fusion-outperforms-condorcet-and-Cormack-Clarke/9e698010f9d8fa374e7f49f776af301dd200c548 (2009).
Asai, A., Hashimoto, K., Hajishirzi, H., Socher, R. & Xiong, C. Learning to retrieve reasoning paths over wikipedia graph for question answering. In ICLR 2020. https://openreview.net/forum?id=SJgVHkrYDH (2020).
Yang, Z. et al. HotpotQA: A dataset for diverse, explainable multi-hop question answering. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, 2369–2380 (Association for Computational Linguistics, Brussels, Belgium, 2018).
Jin, Q., Dhingra, B., Liu, Z., Cohen, W. W. & Lu, X. PubMedQA: a dataset for biomedical research question answering. arXiv preprint arXiv:1909.06146 (2019).
Rajpurkar, P., Zhang, J., Lopyrev, K. & Liang, P. Squad: 100,000+ questions for machine comprehension of text. arXiv preprint arXiv:1606.05250 (2016).
Radford, A. et al. Language models are unsupervised multitask learners. OpenAI Blog 1, 9 (2019).
Fan, A., Grangier, D. & Auli, M. Controllable Abstractive Summarization. In Proceedings of the 2nd Workshop on Neural Machine Translation and Generation, 45–54 (Association forComputational Linguistics, Melbourne, Australia, 2018).
Lu, X., Moffat, A. & Culpepper, J. S. The effect of pooling and evaluation depth on ir metrics. Inf. Retr. J. 19, 416–445 (2016).
Ma, J., Korotkov, I., Yang, Y., Hall, K. & McDonald, R. Zero-shot neural retrieval via domain-targeted synthetic query generation. arXiv preprint arXiv:2004.14503 (2020).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. In (eds Bengio, Y. & LeCun, Y.) 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings (2015).
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Yang, P., Fang, H. & Lin, J. Anserini: reproducible ranking baselines using lucene. J. Data Inf. Qual. 10, 1–20 (2018).
Wolf, T. et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, 38–45 (Association for Computational Linguistics, Online, 2020).
Zhang, J., Zhao, Y., Saleh, M. & Liu, P. PEGASUS: Pre-training with extracted gap-sentences forabstractive summarization. In (eds III, H. D. & Singh, A.) Proceedings of the 37th International Conference on Machine Learning, vol. 119 of Proceedings of Machine Learning Research, 11328–11339 (PMLR, 2020).
Acknowledgements
Funding for this study provided by Salesforce.com, Inc.
Author information
Authors and Affiliations
Contributions
A.E. led the work, managed the team, designed the experiments, and created the infrastructure for the system. A.K. built the retriever and the ranker. K.H. trained the QA module. R.P. trained the abstractive summarization module. W.Y. worked on various exploratory components. D.R. advised on general IR systems and contributed substantially to the writing. R.S. supervised the work.
Corresponding author
Ethics declarations
Competing interests
All authors were employees of Salesforce.com, Inc., at the time of manuscript preparation.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Esteva, A., Kale, A., Paulus, R. et al. COVID-19 information retrieval with deep-learning based semantic search, question answering, and abstractive summarization. npj Digit. Med. 4, 68 (2021). https://doi.org/10.1038/s41746-021-00437-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-021-00437-0
This article is cited by
-
Semantic matching based legal information retrieval system for COVID-19 pandemic
Artificial Intelligence and Law (2023)
-
Cross-lingual extreme summarization of scholarly documents
International Journal on Digital Libraries (2023)
-
COBERT: COVID-19 Question Answering System Using BERT
Arabian Journal for Science and Engineering (2023)
-
Multi-probe attention neural network for COVID-19 semantic indexing
BMC Bioinformatics (2022)
-
Exploration of biomedical knowledge for recurrent glioblastoma using natural language processing deep learning models
BMC Medical Informatics and Decision Making (2022)