Abstract
The number of clinician burnouts is increasing and has been linked to a high administrative burden. Automatic speech recognition (ASR) and natural language processing (NLP) techniques may address this issue by creating the possibility of automating clinical documentation with a “digital scribe”. We reviewed the current status of the digital scribe in development towards clinical practice and present a scope for future research. We performed a literature search of four scientific databases (Medline, Web of Science, ACL, and Arxiv) and requested several companies that offer digital scribes to provide performance data. We included articles that described the use of models on clinical conversational data, either automatically or manually transcribed, to automate clinical documentation. Of 20 included articles, three described ASR models for clinical conversations. The other 17 articles presented models for entity extraction, classification, or summarization of clinical conversations. Two studies examined the system’s clinical validity and usability, while the other 18 studies only assessed their model’s technical validity on the specific NLP task. One company provided performance data. The most promising models use context-sensitive word embeddings in combination with attention-based neural networks. However, the studies on digital scribes only focus on technical validity, while companies offering digital scribes do not publish information on any of the research phases. Future research should focus on more extensive reporting, iteratively studying technical validity and clinical validity and usability, and investigating the clinical utility of digital scribes.
Similar content being viewed by others
Introduction
In the past few years, clinician burnout has become an acknowledged problem in healthcare. In a 2017 survey among 5000 US clinicians, 44% reported at least one symptom of burnout1. To investigate this problem, the National Academy of Medicine formed a committee focused on improving patient care by supporting clinician well-being. The committee’s extensive report, called Taking Action Against Clinician Burnout, describes reasons for clinician burnout. An important reason is the increasing administrative burden2. Since the introduction of the electronic health record (EHR), the time spent on administrative tasks has increased to approximately half of a clinician’s workday3,4,5. These administrative tasks decrease clinicians’ career satisfaction6 and negatively affect the clinician–patient relationship7. Other studies have assessed the relationship between EHR-use and burnout and found that more time spent on the EHR, especially after-hours, was linked to a higher risk of burnout8,9.
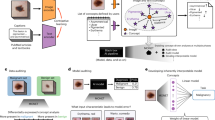
Recently, clinicians have hired medical scribes to reduce the administrative burden, i.e., persons who manage administrative tasks, such as summarizing a consultation. Studies show positive results for the use of medical scribes, with clinicians spending more face-to-face time with patients and less after-hour time on the EHR10,11. Although a medical scribe might seem like the perfect solution, it shifts the burden to other personnel. As a result, direct medical costs increase, while the administrative burden remains substantial. Two recent perspectives12,13 describe the need for a so-called digital scribe. This digital scribe uses techniques such as automatic speech recognition (ASR) and natural language processing (NLP) to automate (parts of) clinical documentation. The proposed structure for a digital scribe includes a microphone that records a conversation, an ASR system that transcribes this conversation, and a set of NLP models to extract or summarize relevant information and present it to the physician. The extracted information could, for instance, be used to create clinical notes, add billing codes, or use the extracted information for diagnosis support.
Companies like Google, Nuance, Amazon, and many startups are creating a digital scribe14,15,16. Although much needed, there are several concerns about implementing a digital scribe in healthcare. These relate to technical aspects such as the accuracy of current ASR systems for transcription of spontaneous speech13 and a digital scribe’s ability to extract all the essential information from a non-linear, fragmented conversation13,17. There are also concerns related to a digital scribe’s clinical utility, such as the effect on a physician’s workflow. Such concerns need to be addressed before digital scribes can be safely implemented in practice. More specifically, successful implementation of an artificial intelligence (AI) tool, such as a digital scribe, requires a thorough investigation of its suitability, technical validity, clinical validity and usability, and clinical utility (see Box 1). A scoping review of current evidence is needed to determine the current status of the digital scribe and to make recommendations for future research.
Objective
The purpose of the present study is to perform a scoping review of the literature and contact companies on the current status of digital scribes in healthcare. The specific research questions are:
-
Which methods are being used to develop (part of) a digital scribe? (Suitability)
-
How accurate are these methods? (Technical validity)
-
Have any of these methods been evaluated in clinical practice? (Clinical validity and usability, clinical utility)
Methods
Data search
We performed a scoping review based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews (PRISMA-ScR) statement18. We searched Medline, Web of Science, Arxiv, and ACL for all relevant articles until December 25, 2020. Furthermore, we scanned reference lists of relevant publications for additional articles. Search terms included terms describing the setting (clinical conversations) in combination with relevant methods (NLP, ASR) and usage of the output (clinical documentation). We also included “digital scribe” and “automated scribe” as search terms because these incorporate the setting, method, and goal. The full search queries can be found in Supplementary Table 1.
Besides, we aimed to include real-world data on existing digital scribes to bridge the gap between research and practice. Quiroz et al.13 provided a list of active companies in the digital scribe space: Robin Healthcare, DeepScribe, Saykara, Sopris Health, Amazon, Nuance. These companies were requested to provide unpublished performance data for their digital scribe.
Inclusion and exclusion criteria
Our definition of a digital scribe is any system that uses a clinical conversation as input, either as audio or text, and automatically extracts information that can be used to generate an encounter note. We included articles that describe the performance of either ASR or NLP on clinical conversational data. A clinical conversation was defined as a conversation—in real life, over the phone, or via chat—between at least one patient and one healthcare professional. Because ASR and NLP are different fields of expertize and will often be described in separate studies, we chose to include studies that only focused on part of a digital scribe. Studies that described NLP models that were not aimed at creating an encounter note but, for example, extracted information for research purposes, were excluded. Articles written in any language other than English were excluded. Because of the rapidly evolving research field and the time lag for publications, proceedings were included.
Study selection
Two reviewers (M.M.v.B. and S.A.C.) independently screened all articles on title and abstract, using the inclusion and exclusion criteria. The selected articles were assessed for eligibility by reading the full text.
Data extraction and synthesis
The first reviewer extracted information from the included articles and the unpublished data provided by companies. The second reviewer verified the extracted information. The following aspects were extracted and assessed:
-
1.
Setting and research phase
-
2.
ASR models and performance
-
3.
NLP tasks, models, and performance
Results
Study selection
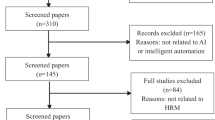
Our search resulted in 2348 articles. After screening the titles and abstracts of these articles, we assessed 144 full-text articles for eligibility. We included 20 articles19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38 for our analysis (Fig. 1 and Supplementary Table 2). Of these, ten were conference proceedings19,20,21,23,27,28,32,38, seven were workshop proceedings22,26,29,34,35,36,37, two were journal articles24,25, and three were Arxiv preprints30,31,33.
Of the six contacted companies, DeepScribe39 was the only one to provide unpublished data on their digital scribe system’s performance. We were unable to obtain performance data from other companies.
Setting and research phase
Although all 20 studies aimed to decrease the administrative burden of clinical documentation in some way, the specific approaches and the setting differed greatly among studies. Three studies focused on improving the ASR for clinical conversations as the first step towards accurately extracting information from them19,21,36. Eleven studies chose to manually transcribe the conversations and performed NLP tasks on the transcripts20,22,24,25,27,30,31,32,34,35,40. Five studies used input data representative of the input of an implemented digital scribe (ASR transcripts or chat dialogs)26,28,33,37,38.
Settings differed greatly between studies, as most did not define a specific specialty19,21,22,23,26,31,32,33,34,35,36,38, while others were focused on primary care20,25,27, home hemodialysis24, orthopedic encounters37, cardiology, family medicine, internal medicine31, and patient-clinician dialogs via a telemedicine platform28. Fifteen studies were performed by or in collaboration with a company19,20,21,23,25,26,27,28,30,33,34,35,36,37.
All included studies focused on the technical validity of the digital scribe; only two studies investigated the clinical validity and usability by performing a qualitative evaluation with end-users24,28. None of the studies investigated the clinical utility.
Automatic speech recognition (ASR)
In total, seven of 20 studies used ASR to automate clinical documentation19,21,23,26,33,36,38, and one company provided data on their ASR system. Of these, two studies and the company presented a new ASR model19,21, four used ASR to transcribe conversations as input for NLP models26,33,37,38, one presented a model to correct ASR errors36, and one compared the performance of existing ASR systems on clinical conversations23 (see Supplementary Table 3).
In all studies, the metric used to evaluate the ASR transcripts was the word error rate (WER, see Box 2). The lowest WER was 14.1%, according to the unpublished data provided by DeepScribe. This ASR system combines Google Video Model41, IBM Watson42, and a custom-made Kaldi model43. The best performing published (as opposed to the unpublished data provided by DeepScribe) ASR system had a WER of 18%19. Four studies23,26,33,36 used existing ASR systems and found WERs between 38% (IBM Watson) and 65% (Mozilla DeepSpeech44).
One study36 presented a postprocessing model to correct ASR errors. By using an attention-based neural network, WERs were improved from 41 to 35% (Google Speech-to-Text45) and 36 to 35% (off-the-shelf open-source model46).
Natural language processing (NLP) tasks and models
The NLP tasks that were performed could be split into three categories: entity extraction20,25,26,27,30,32,35,38, classification22,24,30,31,32,33,34,35, and summarization22,24,28,29,31,37 (see Fig. 2 and Supplementary Table 4). All except one study used word embeddings (see Box 3) as input to their model. This study did not use word embeddings as input but used a clustering model to create 2000 clusters24. The model’s input consisted of the current words’ clusters, the number of words, and the previous words’ clusters.
Entity extraction
The eight studies using entity extraction focused on extracting symptoms20,25,27,32,38, medication regimen20,26,27,32,35, and conditions27. However, the studies differed in the combination of entities and properties they extracted. Several studies examined the possibility of extracting symptoms and identifying whether a symptom was present or not20,27,38, while only one study focused on all the other combinations (i.e., medication dosage, frequency, symptom properties). Almost all studies reported their results as F1 scores (see Box 2). The tasks of extracting the medication, medication dosage, and symptom resulted in the highest F1 scores and thus showed the best performance (see Fig. 3).
All studies used neural networks, although the type of neural network differed. Some studies used general neural networks22,30,35, but most used neural network-based sequence models with attention (see Box 3). In the studies that compared different types of models, the neural networks with attention layer achieved higher F1 scores than the neural networks without attention layer (see Fig. 3).
Three studies27,32,38 performed an error analysis of which one investigated the symptoms that were incorrectly labeled as “absent”. The authors reported that these symptoms were often discussed in multiple talk-turns. In the other study27, ten human annotators categorized the cause of all labeling errors and the impact on the clinical note. They concluded that 16 to 32% of the errors did not affect the clinical note’s content. Furthermore, most errors were caused by a failure of the model to take context into account or the lack of knowledge about a patient’s medical background. In 29 to 42% of the errors, the human annotators agreed with the model, showing the difficulty of annotating the data. One study reported that most errors originated from informal language use and describing symptoms in physical manifestations (“I only sleep for 4 h”)32.
Two studies26,38 made a comparison between manually transcribed and automatically transcribed data regarding the performance of their entity extraction model. Both found that models trained on manually transcribed data outperform the model trained on the automatically transcribed data. The difference in F1 for extracting symptoms was 0.79 versus 0.72, whereas the difference in ROUGE-1 (see Box 2) for extracting medication dosage was 85 versus 7926.
Classification
Six studies performed a type of classification22,24,30,33,34,35, which varied greatly: in which summary section it belonged22,24,33,34; if a sentence was said by the patient or the phycisian33; relevant diagnoses of the patient22,30; if any abnormalities were found in the medical history30 (see Supplementary Table 4). A greater variety of models was used for classification than for entity extraction, although neural networks were used most often. The classification tasks resulting in the highest F1 scores were the classification of primary diagnosis, utterance type, and entity status (see Supplementary Table 4). In two of these tasks, support vector machines were used.
One study33 tested their classification model on manually transcribed data and automatically transcribed data. The model performed better on the manually transcribed data, with a difference in F1 score ranging from 0.03 to 0.06, although they did not mention if the difference was significant.
One study assessed possible disparities of their classification model towards disadvantaged groups34. They formed 18 disadvantaged and advantaged groups based on gender, ethnicity, socioeconomic status, age, obesity, mental health, and location. In 7 of 90 cases, there was a statistically significant difference in favor of the advantaged group. The main reason for the disparity is a difference in the type of medical visit. For example, “blood” is a strong lexical cue to classify a sentence as important for the “Plan” section of the summary, but this word is said less often in conversations with Asian patients.
Summarization
Six studies22,24,28,29,31,37 used NLP to summarize the conversation between patient and healthcare professional automatically. Four studies used pointer generator networks to create a hybrid extractive and abstractive summary28,29,31,37. One of these studies approached the summarization problem as a machine translation problem, where the transcript has to be “translated” to a summary37. This study compared the pointer generator network to three other attention-based models (see Supplementary Table 4).
The other two studies used extractive methods, where the output of the classification or entity extraction models was used to extract the most important utterances from the conversation22,24. The combination of these utterances formed the summary. One of these studies did not compare their summaries to a gold standard24; the other study asked physicians to extract the most important utterances as gold standard22. The F1 score for the latter study was 0.61.
All studies using pointer generator networks reported their results as ROUGE-scores. However, one study only reported their results as ROUGE-L relative error rate reduction37, limiting the comparability with the other studies.
The ROUGE-L scores in the other three studies were 0.4231, 0.5528, and 0.5529. One study also presented a model that returned summaries with a ROUGE-L of 0.58, but this was based on manually extracting noteworthy utterances31. When using the same model with automatically extracted noteworthy utterances, the performance dropped to 0.42.
The best performing model used a pretrained pointer generator network (see Box 3) fine-tuned on medical dialog summarization, with an added penalty for the generator distribution to force the model to favor copying text from the transcript over generating new text28. The other models were: a topic-aware pointer-generator network using embeddings (see Box 3)29, which takes the topic of the current segment into account when copying or generating the next word; an LSTM architecture with BERT embeddings to extract noteworthy utterances (see Box 3)31; a combination of a transformer and pointer generator network that creates a summary per summary section (see Box 3)37.
Two studies included physicians to evaluate their summaries24,28. One study examined physicians’ ability to answer questions about patient care based on the automatic summary24. They did not find any significant difference in physicians’ answers using the human-made summaries compared to the automatic summaries. Another study asked physicians to rate the amount of relevant information in the summaries28. Physicians found that 80% of the summaries included “all” or “most” relevant facts. The study did not specify which parts were deemed relevant or not or if the model missed specific information.
DeepScribe did not provide information on the models used for summarization but included how often a summary needed to be adjusted in practice. They report that 77% of their summaries do not need modification by a medical scribe before being sent to the physician. Furthermore, 74% of their summaries do not need modification from a medical scribe or a physician before being accepted as part of the patient’s record, saving time on administrative tasks.
Discussion
This scoping review provides an overview of the current state of the development, validation, and implementation of digital scribes. Although the digital scribe is still in an early research phase, there appears to be a substantial research body testing various techniques in different settings. The first results are promising: state-of-the-art models are trained on vast corpora of annotated clinical conversations. Although the performance of these models varies per task, the results give a clear view of which tasks and which models yield high performance. Reports of clinical validity and usability, and especially clinical utility are, however, mostly lacking.
All studies focusing on ASR used physician–patient dialogs without further specification of the setting. In general, existing ASR systems not explicitly trained on clinical conversations did not perform well, with WERs up to 65%. The speech recognition systems trained on thousands of clinical conversations had WERs as low as 18%. This WER is still high compared to the claimed WERs of general, state-of-the-art, available ASR systems that attain WERs as low as 5%47. The difference in performance can be explained by the uncontrolled setting of clinical conversations with background noise, multiple speakers, and the spontaneity of the speech13. However, these aspects were not reported by any of the studies, complicating the comparison of WERs. Two new approaches decreased the WER by postprocessing the automatic transcript36 and combining multiple ASR systems (DeepScribe). These approaches are promising new ways to decrease the WER. However, what is most important is whether the WER is good enough to extract all the relevant information. Currently, the NLP models trained on manually transcribed data outperform those trained on automatically transcribed data, which means there is room for improvement of the WER.
When comparing the different NLP tasks, the diverseness in both tasks and underlying models was large. The classification models focused mainly on extracting metadata, such as relevance or structure induction of an utterance, and used various models ranging from logistic regression to neural networks. The entity extraction models were more homogeneous in models but extracted many different entities, complicating the comparison, whereas the summarization task was mostly uniform, both in models and in metrics. One notable aspect of the NLP tasks overall is the use of word embeddings. Only one study did not use word embeddings, but this was a study from 2006 when context-sensitive word embeddings were not yet available. All the other studies were published after 2019 and used various word embeddings as input. The introduction of context-sensitive word embeddings has been essential for extracting entities and summarizing clinical conversations.
In the entity extraction task, the specific tasks, such as extracting symptoms, led to better performance than more general tasks, such as extracting symptoms and their properties. An explanation for this is the heterogeneity in, for example, symptom properties, which entail the location, severity, duration, and other characteristics of a symptom. These properties can be phrased in various ways, in contrast to medication or frequency, which will be much more homogeneous in phrasing. Therefore, this homogeneity leads to many more annotations per entity, increasing performance.
The same pattern was observed in the models, where the addition of an attention layer increased performance. This finding is in line with previous studies on neural attention48,49, which describe the decrease in neural networks’ performance with increased input length. By adding weights to the input text, the model knows which parts of the text are important for its task. Adding attention not only improves performance; it also decreases the amount of training data needed, which is useful in a field such as healthcare, where gathering large datasets can be challenging.
In the studies performing the entity extraction task, the error analyses showed that often, symptoms, medications, or properties are hard to interpret even by human annotators. This result is in line with the concerns discussed in the introduction, questioning if a model would accurately extract all relevant information from a non-linear, fragmented conversation. However, this takes the concern one step further, namely how the “gold standard” will be determined if there is ambiguity between human annotators. More research is needed to define methods for developing gold standards. Shafran et al.27 have taken an exciting first step towards such a method by publishing an article about the development of their corpus, including how they dealt with ambiguity and labeling errors.
The studies investigating summarization of the clinical conversation used both extractive and abstractive summarization techniques. However, the extractive techniques resulted in a list of the most important utterances instead of a new, full summary. Therefore, the studies performing abstractive summarization are more interesting to discuss. All four studies used the same model, the pointer generator network28,29,31,37. This network’s advantage, especially with the studies’ additions, makes sure it copies more words than it generates, keeping the summary as close to the conversation as possible. Two studies also included a quality check by physicians, which gives more insight into the possibility of implementation24,28. However, it would have been interesting to include error analyses to investigate the models’ blind spots.
Future work
First of all, we believe it is vital to improve the ASR for clinical conversations further and use them as input for NLP models. A remarkable finding was that most studies used manually transcribed conversations as input to their NLP model. These manual transcripts may outperform automatically transcribed conversations regarding data quality, leading to an overestimation of the results. NLP models that require manual transcription may increase administrative burden when implemented in clinical practice.
Secondly, the current body of research is mostly focused on improving the performance of different models. Although some studies performed error analyses and qualitative analyses of the model’s output, most did not. Moreover, most studies did not fully cover the technical validity phase because of insufficient reporting on the setting, data, and situations in which the model succeeded and failed. This information is essential to describe for a model that could potentially be implemented in clinical practice. The proposed models might contain bias or lead to unintended results, as Ferracane and Konam34,37 show. This study is an inspirational example of how researchers can investigate the strengths and weaknesses of their model. A recent paper by Hernandez-Boussard et al.50 proposes reporting standards for AI in healthcare, which should be the basis for reporting on digital scribes as well.
Although most studies are in an early development phase, including qualitative analyses of the model’s output is necessary to know if the solution researchers or developers are working on is applicable in practice. The lack of implementation following the development of an AI model is common in healthcare51, which can be limited by investigating clinical validity and usability while working on technical validity. A good example is the study by Joshi et al.28, where physicians qualitatively analyze the model’s output. These results lead to new insights for improving technical validity. Studying these two research phases iteratively leads to a solution that is well-suited for clinical practice.
Most of the presented models need to be technically and clinically validated before moving on to the clinical utility phase. However, the companies already offering digital scribes seem to have skipped all four research phases, including clinical utility. We urge these companies to publish data on their digital scribes’ technical validity, clinical validity and usability, and clinical utility. Not only is transparency in the model and its performance crucial for clinical practice, but it also helps the community better understand the models and enables researchers to build on past work52.
The suitability phase falls outside the scope of this review but is nevertheless vital for developing and implementing the digital scribe. One research group has published several studies investigating which parts of a clinical conversation are relevant for creating a summary and how physicians see the potential role of a digital scribe53,54. These studies should be the starting point for researchers and developers working on a digital scribe.
Strengths and limitations
The current work is the first effort to review all available literature on developing a digital scribe. We believe our search strategy was complete, leading to a comprehensive and focused scope of the digital scribe’s current research body. By adding the company’s data, we create a broader overview than just the digital scribe’s scientific status. However, this data is unpublished, which means we have to trust the company in providing us with legitimate data. We hope this review is an encouragement for other companies to study their digital scribes scientifically.
One limitation is the small amount of journal papers included in this review, as opposed to the amount of Arxiv preprints and workshop proceedings. These types of papers are often refereed very loosely. However, only including journal papers would not lead to a complete scope of this quickly evolving field.
Contacting various digital scribe companies was a first step towards gaining insight into implemented digital scribes and their performance on the different ASR and NLP tasks. Although only one company replied, we believe it is a valuable addition to this review. It indicates that their implemented digital scribe does not differ significantly in techniques or performance from the included studies’ models while already saving physicians’ time. Nevertheless, it highlights the gap between research and practice. The studies published by companies all describe techniques that are not part of a fully functional digital scribe (yet). However, none of the companies offering digital scribes have published about the technical validity, clinical validity and usability, or clinical utility of their systems.
Conclusion
Although the digital scribe field has only recently started to accelerate, the presented techniques achieve promising results. The most promising models use context-sensitive word embeddings in combination with attention-based neural networks. However, the studies on digital scribes only focus on technical validity, while companies offering digital scribes do not publish on any of the research phases. Future research should focus on more extensive reporting, iteratively studying technical validity and clinical validity and usability, and investigating the clinical utility of digital scribes.
Data availability
Any data generated or analyzed are included in this article and the Supplementary Information files. Aggregate data analyzed in this study are available from the corresponding author on reasonable request.
References
Shanafelt, T. D. et al. Changes in burnout and satisfaction with work-life integration in physicians and the general US working population between 2011 and 2017. Mayo Clin. Proc. 94, 1681–1694 (2019).
National Academies of Sciences, Engineering, and Medicine. Taking Action Against Clinician Burnout: A Systems Approach to Professional Well-Being (The National Academies Press, 2019).
Arndt, B. G. et al. Tethered to the EHR: primary care physician workload assessment using EHR event log data and time-motion observations. Ann. Fam. Med. 15, 419–426 (2017).
Sinsky, C. et al. Allocation of physician time in ambulatory practice: a time and motion study in 4 specialties. Ann. Intern. Med. 165, 753–760 (2016).
Tai-Seale, M. et al. Electronic health record logs indicate that physicians split time evenly between seeing patients and desktop medicine. Health Aff. 36, 655–662 (2017).
Rao, S. K. et al. The impact of administrative burden on academic physicians. Acad. Med. 92, 237–243 (2017).
Pelland, K. D., Baier, R. R. & Gardner, R. L. “It’s like texting at the dinner table”: a qualitative analysis of the impact of electronic health records on patient-physician interaction in hospitals. J. Innov. Health Inform. 24, 216–223 (2017).
Robertson, S. L., Robinson, M. D. & Reid, A. Electronic health record effects on work-life balance and burnout within the I3 population collaborative. J. Grad. Med. Educ. 9, 479–484 (2017).
Gardner, R. L. et al. Physician stress and burnout: the impact of health information technology. J. Am. Med. Inform. Assoc. 26, 106–114 (2019).
Gidwani, R. et al. Impact of scribes on physician satisfaction, patient satisfaction, and charting efficiency: a randomized controlled trial. Ann. Fam. Med. 15, 427–433 (2017).
Mishra, P., Kiang, J. C. & Grant, R. W. Association of medical scribes in primary care with physician workflow and patient experience. JAMA Intern. Med. 178, 1467 (2018).
Coiera, E., Kocaballi, B., Halamka, J. & Laranjo, L. The digital scribe. Npj Digital Med. 1, 1–5 (2018).
Quiroz, J. C. et al. Challenges of developing a digital scribe to reduce clinical documentation burden. Npj Digital Med. 2, 1–6 (2019).
Ambient clinical intelligence: the exam of the future has arrived. Nuance Communications (2019). Available at: https://www.nuance.com/healthcare/ambient-clinical-intelligence.html. (Accessed: 18th February 2021).
Amazon comprehend medical. Amazon Web Services, Inc (2018). Available at: https://aws.amazon.com/comprehend/medical/. (Accessed: 18th February 2021).
Robin Healthcare | automated clinic notes, coding and more. Robin Healthcare (2019). Available at: https://www.robinhealthcare.com. (Accessed: 18th February 2021).
Lin, S. Y., Shanafelt, T. D. & Asch, S. M. Reimagining clinical documentation with artificial intelligence. Mayo Clin. Proc. 93, 563–565 (2018).
Tricco, A. C. et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann. Intern. Med. 169, 467–473 (2018).
Chiu, C.-C. et al. Speech recognition for medical conversations. Proc. Interspeech 2018, 2972–2976 (2018).
Du, N., Wang, M., Tran, L., Li, G. & Shafran, I. Learning to infer entities, properties and their relations from clinical conversations. In Proc. of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) 4979–4990 (Association for Computational Linguistics, 2019).
Shafey, L. E., Soltau, H. & Shafran, I. Joint speech recognition and speaker diarization via sequence transduction. Proc. Interspeech 2019, 396–400 (2019).
Jeblee, S., Khattak, F. K., Crampton, N., Mamdani, M. & Rudzicz, F. Extracting relevant information from physician-patient dialogues for automated clinical note taking. In Proc. of the Tenth International Workshop on Health Text Mining and Information Analysis (LOUHI), 65–74 (Association for Computational Linguistics, 2019).
Kodish-Wachs, J., Agassi, E., Kenny, P. & Overhage, J. M. A systematic comparison of contemporary automatic speech recognition engines for conversational clinical speech. In Proc. of the Annual AMIA Symposium, 683–689 (American Medical Informatics Association, 2018).
Lacson, R. C., Barzilay, R. & Long, W. J. Automatic analysis of medical dialogue in the home hemodialysis domain: structure induction and summarization. J. Biomed. Inform. 39, 541–555 (2006).
Rajkomar, A. et al. Automatically charting symptoms from patient-physician conversations using machine learning. JAMA Intern. Med. 179, 836 (2019).
Selvaraj, S. P. & Konam, S. Medication regimen extraction from medical conversations. In Proc. of International Workshop on Health Intelligence of the 34th AAAI Conference on Artificial Intelligence (Association for Computational Linguistics, 2020).
Shafran, I. et al. The medical scribe: corpus development and model performance analyses. In Proc. of the 12th Language Resources and Evaluation Conference (European Language Resources Association, 2020).
Joshi, A., Katariya, N., Amatriain, X. & Kannan, A. Dr. summarize: global summarization of medical dialogue by exploiting local structures. In Findings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), 3755–3763 (Association for Computational Linguistics, 2020).
Liu, Z., Ng, A., Lee, S., Aw, A. T. & Chen, N. F. Topic-aware pointer-generator networks for summarizing spoken conversations. In Proc. IEEE Automatic Speech Recognition Understanding Workshop 2019, 814–821 (IEEE, 2019).
Krishna, K., Pavel, A., Schloss, B., Bigham, J. P. & Lipton, Z. C. Extracting Structured Data from Physician-Patient Conversations by Predicting Noteworthy Utterances. in Shaban-Nejad A., Michalowski M., Buckeridge D.L. (eds) Explainable AI in Healthcare and Medicine. Studies in Computational Intelligence, vol 914 (Springer International Publishing, 2021).
Krishna, K., Khosla, S., Bigham, J. P. & Lipton, Z. C. Generating SOAP notes from doctor-patient conversations. Preprint at arXiv (2020).
Khosla, S., Vashishth, S., Lehman, J. F. & Rose, C. MedFilter: improving extraction of task-relevant utterances through integration of discourse structure and ontological knowledge. In Proc. of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), 7781–7797 (Association for Computational Linguistics, 2020).
Schloss, B. & Konam, S. Towards an automated SOAP note: classifying utterances from medical conversations. Preprint at arXiv (2020).
Ferracane, E. & Konam, S. Towards fairness in classifying medical conversations into SOAP sections. In To be presented at AAAI 2021 Workshop: Trustworthy AI for Healthcare (AAAI Press, 2020).
Patel, D., Konam, S. & Selvaraj, S. P. Weakly supervised medication regimen extraction from medical conversations. In Proc. of the 3rd Clinical Natural Language Processing Workshop, 178–193 (Association for Computational Linguistics, 2020).
Mani, A., Palaskar, S. & Konam, S. Towards understanding ASR error correction for medical conversations. In Proc. of the First Workshop on Natural Language Processing for Medical Conversations, 7–11 (Association for Computational Linguistics, 2020).
Enarvi, S. et al. Generating medical reports from patient-doctor conversations using sequence-to-sequence models. In Proc. of the First Workshop on Natural Language Processing for Medical Conversations, 22–30 (Association for Computational Linguistics, 2020).
Du, N. et al. Extracting symptoms and their status from clinical conversations. In Proc. of the 57th Annual Meeting of the Association for Computational Linguistics, 915–925 (Association for Computational Linguistics, 2019).
DeepScribe - AI-Powered Medical Scribe. DeepScribe (2020). Available at: https://www.deepscribe.ai. (Accessed 18th February 2021).
Liu, X. et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med. 26, 1364–1374 (2020).
Transcribing videos | Cloud speech-to-text documentation. Google Cloud (2016). Available at: https://cloud.google.com/speech-to-text/docs/video-model. (Accessed 18th February 2021).
Watson speech to text - Overview. IBM (2021). Available at: https://www.ibm.com/cloud/watson-speech-to-text. (Accessed 18th February 2021).
Kaldi ASR. Kaldi (2015). Available at: https://kaldi-asr.org. (Accessed 18th February 2021).
mozilla/DeepSpeech. GitHub (2020). Available at: https://github.com/mozilla/DeepSpeech. (Accessed 18th February 2021).
Speech-to-text: automatic speech recognition | Google Cloud. Google Cloud (2016). Available at: https://cloud.google.com/speech-to-text. (Accessed 18th February 2021).
Peddinti, V. et al. Jhu aspire system: robust LVCSR with TDNNs, Ivector adaptation and RNN-LMs. In IEEE Workshop on Automatic Speech Recognition and Understanding (ASRU), 539–546 (IEEE, 2015).
Hu, K., Sainath, T. N., Pang, R. & Prabhavalkar, R. Deliberation model based two-pass end-to-end speech recognition. In IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 7799–7803 (IEEE 2020).
Bahdanau, D., Cho, K. & Bengio, Y. Neural machine translation by jointly learning to align and translate. Preprint at arXiv (2014).
Cho, K. et al. Learning phrase representations using RNN encoder–decoder for statistical machine translation. In Proc. of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 1724–1734 (Association for Computational Linguistics, 2014).
Hernandez-Boussard, T., Bozkurt, S., Ioannidis, J. P. A. & Shah, N. H. MINIMAR (MINimum Information for Medical AI Reporting): developing reporting standards for artificial intelligence in health care. J. Am. Med. Inform. Assoc. 27, 2011–2015 (2020).
He, J. et al. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 25, 30–36 (2019).
Wiens, J. et al. Do no harm: a roadmap for responsible machine learning for health care. Nat. Med. 25, 1337–1340 (2019).
Kocaballi, A. B. et al. Envisioning an artificial intelligence documentation assistant for future primary care consultations: a co-design study with general practitioners. J. Am. Med. Inform. Assoc. 27, 1695–1704 (2020).
Quiroz, J. C. et al. Identifying relevant information in medical conversations to summarize a clinician-patient encounter. Health Inform. J. 26, 2906–2914 (2020).
Larson, D. B. et al. Regulatory frameworks for development and evaluation of artificial intelligence–based diagnostic imaging models: summary and recommendations. J. Am. Coll. Radiol. 18, 413–424 (2020).
Tatman, R. Gender and dialect bias in YouTube’s automatic captions. In Proc. of the First ACL Workshop on Ethics in Natural Language Processing, 53–59 (Association for Computational Linguistics, 2017).
Vasey, B. et al. DECIDE-AI: new reporting guidelines to bridge the development-to-implementation gap in clinical artificial intelligence. Nat. Med. 27, 186–187 (2021).
Sutskever, I., Vinyals, O. & Le, Q. V. Sequence to sequence learning with neural networks. In Proc. of the 27th International Conference on Neural Information Processing Systems (NIPS) 2, 3104–3112 (MIT Press, 2014).
See, A., Liu, P. J. & Manning, C. D. Get to the point: summarization with pointer-generator networks. In Proc. of the 55th Annual Meeting of the Association for Computational Linguistics, 1, 1073–1083 (Association for Computational Linguistics, 2017).
Mikolov, T., Sutskever, I., Chen, K., Corrado, G. & Dean, J. Distributed representations of words and phrases and their compositionality. In Proc. of the 26th International Conference on Neural Information Processing Systems (NIPS) 2, 3111–3119 (Curran Associates Inc., 2013).
Peters, M. E. et al. Deep contextualized word representations. In Proc. of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (NAACL) 1, 2227–2237 (Association for Computational Linguistics, 2018).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (NAACL), 1, 4171–4186 (Association for Computational Linguistics, 2019).
Author information
Authors and Affiliations
Contributions
M.M.v.B. contributed to conception, design, literature search, data synthesis, drafting, and critical revision of the manuscript. H.B., M.P.B., and E.W.S. contributed to conception, design, and critical revision of the manuscript. S.A.C. contributed to conception, literature search, and data synthesis. I.M.J.K. contributed to design and critical revision of the manuscript. All authors gave their final approval and accepted accountability for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van Buchem, M.M., Boosman, H., Bauer, M.P. et al. The digital scribe in clinical practice: a scoping review and research agenda. npj Digit. Med. 4, 57 (2021). https://doi.org/10.1038/s41746-021-00432-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-021-00432-5
This article is cited by
-
Prioritizing educational initiatives on emerging technologies for Italian pediatricians: bibliometric review and a survey
Italian Journal of Pediatrics (2023)
-
Harnessing oncology real-world data with AI
Nature Cancer (2023)