Abstract
As an alternative target to surgically resected tissue specimens, liquid biopsy has gained much attention over the past decade. Of the various circulating biomarkers, circulating tumor cells (CTCs) have particularly opened new windows into the metastatic cascade, with their functional, biochemical, and biophysical properties. Given the extreme rarity of intact CTCs and the associated technical challenges, however, analyses have been limited to bulk-cell strategies, missing out on clinically significant sources of information from cellular heterogeneity. With recent technological developments, it is now possible to probe genetic material of CTCs at the single-cell resolution to study spatial and temporal dynamics in circulation. Here, we discuss recent transcriptomic profiling efforts that enabled single-cell characterization of patient-derived CTCs spanning diverse cancer types. We further highlight how expression data of these putative biomarkers have advanced our understanding of metastatic spectrum and provided a basis for the development of CTC-based liquid biopsies to track, monitor, and predict the efficacy of therapy and any emergent resistance.
Similar content being viewed by others
Introduction
Despite the first report on breast circulating tumor cells (CTCs) in 1869,1 techniques for isolating these circulating biomarkers were only first described in 1960,2,3 and were gradually improved over the next 40 years. While much progress has been made with the albumin gradient method and FDA-approved CellSearch® system during the first generation of CTC research,4 the existence of heterogeneous CTC subpopulation highlighted the need to develop marker-independent isolation technologies.5,6 Since then, label-free techniques utilizing the principles of biophysical properties have been developing rapidly as the second generation.4,7 FDA-approved/listed platforms, such as CellSearch® (Silicon Biosystems) and ClearCell® FX (Biolidics) are exemplary technologies that have been widely used and demonstrated the clinical significance of CTCs.8,9,10,11,12
Currently, 265 clinical trials regarding CTCs are listed on clinicaltrials.gov. Despite successful CTC enumeration, achieving high yield and high purity remains challenging because of millions to billions of blood cells and a few to tens of CTCs present as background and target cells, respectively, in a milliliter of whole blood from cancer patient.13 It has been posited that the conventional EpCAM-based enrichment method would require 5 L of blood to detect at least one CTC in metastatic disease with 99% sensitivity.9 Such exceptionally low CTC frequencies could be attributed to progressively lost expression of epithelial markers during epithelial-to-mesenchymal transition (EMT) in circulation,14,15 as higher CTC counts have been reported with immunologic or physical property-based enrichment.16,17,18
In addition to the wide range of CTC detection rate reported in clinical studies, broad phenotypic plasticity and diversity have been observed at multiple molecular levels during metastatic cascade – from EMT and invasion19,20,21 to evasion of apoptosis,22 chemoresistance,23 migration,24 intravasation,25 extravasation, and organ colonization.26 While a tumor biopsy from either primary tumor or metastatic lesion alone may not always recapitulate the entire tumor harboring segregated clones,27 spatiotemporally heterogeneous CTCs collected in a sequential manner could potentially reveal comprehensive window into the metastatic disease for real-time monitoring of therapy response, which remains an unmet need in current clinical practice with tissue biopsy.
Single-cell analysis
Emerging sequencing data from spatially distinct tumors provide clear evidence of intratumoral heterogeneity.28,29,30 Owing to the technical challenges, however, CTC analyses have been limited to bulk-cell samples, missing the information on cellular heterogeneity. The inevitable leukocyte contamination in any given primarily enriched sample further complicates downstream molecular analyses. Such confounding effect is particularly pronounced in transcriptomic studies when the activated leukocytes concurrently overexpress cancer-associated biomarkers, such as MUC1 or HER2, masking the true expression of CTC-specific transcripts.31 Their mesenchymal nature and hematopoietic origin further interfere with the expression of EMT-related and stem cell markers, respectively, resulting in false-positive observations.32
The transition from bulk to single-cell analyses on patient-derived CTCs has thus been fueled in part by studies over the past five years. At the genomic level, they have identified clinically relevant alterations, ranging from small-scale (e.g., single nucleotide variation (SNV), microsatellite instability) to large-scale mutations (e.g., copy-number variation, large-scale state transition, inter/intrachromosomal rearrangement). These aberrations include time-varying SNVs during the course of chemotherapy,33 private mutations that are absent in either matched primary or metastatic tumor34 and that are not yet listed in the COSMIC database (http://cancer.sanger.ac.uk),35 and copy-number profiles that distinguish chemosensitive from chemorefractory disease.23
Although limited in sample size and number of studies, transcriptomic studies have further revealed complex and heterogeneous expression patterns within and across patients. For example, expression profiles of single CTCs have demonstrated superior diagnostic accuracy in defining lineage identity and in identifying clinically distinct subsets of tumors across multiple myeloma and prostate cancers.36,37 They have also revealed therapeutically relevant biomarkers38,39,40 (e.g., predictive of resistance and/or response to adjuvant therapies), that are involved in activated oncogenic signaling pathways41 (e.g., PI3K-AKT-mTOR) and that are potentially targetable.24,36,38,41,42,43
Integrated workflow
Despite the prevalence of EpCAM− CTCs44 and varying capture efficiency,45 epithelial marker-dependent CellSearch® technology remains as the most common enrichment method to isolate CTCs from patient-derived peripheral blood. Pre-enrichment is often required for recovery of preferably viable and intact CTCs, and can be performed with direct imaging modalities,36 density gradient centrifugation in Percoll or Ficoll,24,32 immunoaffinity,42,46,47,48 microfiltration in two43 and three41 dimensions, and microfluidic approaches.37,38,39,40,49,50,51 Table S1 summarizes cell sorting and isolation technologies, including methods, working principles, features, limitations, and the reported recovery rates of spiked cancer cells. Primarily enriched bulk CTC samples are subsequently subjected to manual cell picking or micromanipulation,24,32,36,41,42,43,46,48,49 and micro/nanoplatforms37,38,39,40,47,50,51 for single-cell isolation and downstream molecular and phenotypic characterization (Table 1).
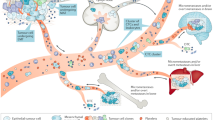
Microfluidics has particularly come to the fore in the field among the various isolating and cell sorting devices incorporating hydrodynamics,12 optics,52 dielectrophoresis,53 magnetics,54 or acoustics.55 The ability to manipulate even a small drop of whole blood and to retain cells or molecules at defined locations inside microfluidic devices enable characterization of chemical, thermal, and/or temporal variations in a multiplex manner. Having the benefits of integrated functionalities, microfluidic devices further eliminate the need for independent multiple modules required for sample preparation, purification and analysis, depending on input characteristics. Microfluidic technologies have increasingly been applied to rare cell populations, including CTCs, to explore multiple modalities of CTCs at the single-cell level (Fig. 1). These workflows coupled microfluidic systems, such as CTC-iChip,37,38,39,40,51 HBCTC-Chip,49 ClearCell® FX,12,50,56 NanoVelcroChip57,58 and Parsortix,41 with single-cell profiling technologies, such as whole genome/exome sequencing (WGS/WES),56,58 single-cell RNA sequencing (scRNA-seq) or PCR (scPCR), single-cell western blotting (scWB) and secretion profiling.59,60
Microfluidic technologies for single-cell molecular characterization of patient-derived CTCs. a ClearCell® FX-integrated workflow. Single-cell genomic analysis: High concordance rate of EGFR mutations (T790M and L858R) was found between NSCLC CTCs and matched primary tumors.12 Single-cell transcriptomic analysis: Patient classification was done for breast cancer and NSCLC through full-length mRNA transcriptomic analysis50 and targeted gene expression profiling,76 respectively. Single-cell metabolomics analysis: Supervised principal component analysis (PCA) revealed unique metabolic profiles between CTCs and lymphocytes in gastric and colorectal cancer patients.117 b Single-cell proteomic analysis: Microfluidic single-cell western blotting (scWB) enabled the rapid analysis of an eight-plex protein expression in ER+ breast cancer.60 c Single-cell secretomic analysis: The integrated microfluidic on-chip system revealed highly heterogeneous expression profiles of two secreted proteins (i.e., IL-8 and VEGF) in CTCs from lung cancer patients.118
Gene-specific targeted preamplification or whole transcriptome amplification (WTA) is required prior to sequencing or profiling to analyze less than 1 pg of mRNA from the isolated single cells. Current WTA methods include PCR-based, multiple displacement amplification (MDA) or in vitro transcription (IVT)-based amplification61 of cDNA templates transcribed from single-cell mRNA. Many, however, are limited to selective amplification of the polyadenylated RNAs, and thus may be biased to the 3’-end or the 5’-end of a transcript.62 Among a few WTA techniques developed for full-length mRNA-characterization of a single cell, modified versions of SMARTer48 (e.g., SMART-Seq2) and that of Tang’s method63 are commonly employed in single-CTC transcriptomic studies aiming to achieve improved transcript detection, coverage, accuracy, and yield (Table 1).
For quantitative transcriptomic analysis, an accurate identification of technical artifacts from intrinsic biological cellular variability is critical to prevent spurious readings from single CTCs. Ideally, quality control (QC) metrics should be performed with the amplified cDNA products after preamplification step, given the amount of genetic material minimally required. Current single-cell transcriptomic studies on patient-derived CTCs have assessed (1) the yield or concentration of amplified DNA,36,64,65,66 (2) Cq values for selected reference, or housekeeping, genes using qPCR,33,58 (3) fragment size distribution of selected DNA sequences using gel/capillary electrophoresis,36,58,64,67,68,69,70,71 and (4) genome integrity index (GII), which ranges from 0 (poor quality) to 4 (high quality) and is computed based on PCR bands of four primer pairs using gel electrophoresis.23,72,73
Cells harboring QC-passed RNAs are subsequently subjected to library construction, followed by scRNA-seq36,38,40,46,48,49,50,51 or quantitative profiling with conventional qPCR,24,32 digital droplet PCR (ddPCR)39,40 or microfluidic dynamic array.42,43 The quality of constructed libraries are further validated with (1) the proportion of reads mapping to genome, and/or (2) the number of genes detected.50 Lineage specificity of CTCs is often confirmed by high expression of cancer-specific markers and low expression of leukocyte markers with pre-specified thresholds.37 Low success rate of <60% for overall amplification and library preparation attributed to multiple processing steps has been reported in CTC studies, highlighting the need to systematically quantify QC metrics prior to the analysis.
Expression data
Single-cell transcriptome of patient-derived CTCs have been analyzed comparatively with cancer cell lines, white blood cells (WBCs), matched primary tumors and/or metastases.24,37,42,43,48 Alternatively, expression levels were assessed and compared between CTC subgroups defined by unsupervised hierarchical clustering or other classification methods.41,49 In the following sections, we focus on the most relevant gene signatures that are perceived to be critical determinants of metastasis and disease outcome and that are commonly differentially expressed in CTCs at the single-cell level: EMT, stemness, interaction with blood components, DNA repair, signaling pathways and drug targets (Table 2).
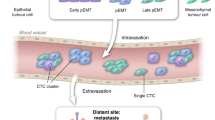
In line with molecular evidences supporting EMT-driven metastasis,24,32,41,42,43,49 bulk-cell studies have suggested the contribution of EMT to early steps of the metastatic spread (i.e., tumor invasion, intravasation, CTC generation and survival, and early seeding in secondary organs).74 Nonexclusive hypotheses of EMT’s contribution to CTC biology suggest that (1) CTCs may have been mesenchymally-shifted in primary tumors to have enhanced survival properties through activation of genes involved in survival pathways and escape from immune surveillance, or (2) undergo EMT processes within the bloodstream by means of TGFß liberated from circulating platelets.74 At the center of the research axis is to identify and characterize such premetastatic subsets of CTC population that are favored to be liberated from primary tumors and survive in the bloodstream to succeed in the early colonization phases. scRNA-seq is particularly well suited in this regard to discover distinct subsets of CTCs capable of forming metastasis.
Emerging single-cell profiling data provide clear evidence of a continuum in the development of CTC phenotypes, including epithelial (E), epithelial-mesenchymal (E/M), mesenchymal (M) and stem-like phenotype. Highly heterogeneous expression of epithelial markers (e.g., EpCAM, CK18, CK19) was observed in CTCs across colorectal,24 ovarian,32 breast,42 and prostate37,41,43 cancers. Similarly, mesenchymal or EMT-related genes (e.g., CDH2, VIM, TGFß1, ZEB1/2) were commonly enriched in single ovarian,32 breast,42 and prostate41,43 CTCs. Compared to matched primary/metastatic tumor tissue and cell lines, migration-related and cell-cell adhesion genes (e.g., TSPAN8, CD151, CD44v6, and FN1) were generally downregulated in colon24 and prostate CTCs,43 respectively, possibly suggesting a lack of their need for mobility and migratory capabilities in the bloodstream in these cancer types.
While some studies have suggested that CTCs that are ‘frozen’ in either E or M state lacking EMT plasticity are unable to form metastases,75 the repetitive observation of patient-derived CTCs expressing mesenchymal attributes directly correlated with the appearance of metastases in recent studies suggest that these mesenchymal-shifted cells, and not benign cells passively detached from a primary tumor, are precursors of metastasis.76 Clinical data linking CTC-derived EMT markers with multiple clinical parameters are discussed elsewhere.74 It remains to be elucidated whether there exist specific hybrid E/M states that are particularly prone to perturbations triggering extensive phenotypic and functional change in circulation. Computational analytical tools enabling pseudo-time reconstruction of transitioning cells77,78,79 and the topography underlying E/M plasticity80 from a static single-cell gene expression data may be applied to prospective studies to clarify the nature of hybrid E/M states and define their role in metastasis.
It has been suggested that tumor cells having an intermediate phenotype of EMT show the highest plasticity and thus represent cancer stem cells (CSCs).81 Varying expression levels of stem cell markers (e.g., CD24, CD44, ALDH1A1, NANOG, and OCT4) were found at the single-cell level in ovarian,32 breast42, and prostate CTCs.37,41 Interestingly, genes involved in oncogenic signaling pathway were found to be differentially expressed in single CTCs depending on the level of cellular plasticity or stemness. In prostate cancer, for example, expression of the key regulators in the PI3K/Akt/mTOR signaling pathway (i.e., PI3K, mTOR) were highly expressed in CD44−/CD24+ CTCs.41 This subset of CTC populations may thus be more susceptible to perifosine (Akt inhibitors) and rapamycin (mTOR inhibitors) treated with conventional chemotherapy or radiotherapy.82 Given that both CD44+/CD24− and CD44+/CD24+ tumor cells have functional significance in initiating tumor growth83 and that CTCs express these two cell surface markers across various malignancies, it remains to be investigated whether their expression has comparable functional significance in circulation.
The innate immune regulator, CD47, was the only gene that was upregulated in CTCs compared to matched tumor tissue in colorectal cancer,24 suggesting a potential immune-escape mechanism associated with CTC survival in circulation. Another form of immune evasion was suggested in melanoma, where the genes associated with the escape from immune surveillance, including HLA genes (i.e., HLA-G, HLA-H, HLA-C, and HLA-B) and TRPM1, were significantly downregulated in CTCs compared to melanoma cell lines, primary melanocytes, human embryonic stem cells, and lymphoma cell lines.48 Transcriptional repression of HLA genes has been associated with complete loss of MHC class I membrane expression, and importantly, the primary resistance to immune checkpoint inhibitor (ICI)-based immunotherapy.84 The screening of plasma-membrane proteins through whole transcriptomic analysis is thus of utmost interest to identify not only CTC-specific diagnostic biomarkers48 but also immune escape and survival mechanisms underlying resistance to immunotherapy.
Another key player is TGFß-releasing platelet, which may adhere to CTCs in the bloodstream.19,85 In line with this hypothesis, platelet markers were frequently expressed in isolated single CTCs, as well as in CTC clusters in breast cancer.49 Labelle et al. showed that TGFß liberated from platelets may induce EMT in tumor cells within the bloodstream and further promote the formation of the early metastatic niche.85,86 Based on these observations, it is speculated that TGFß expressing CTCs may represent a specific subpopulation having high metastatic potential. Importantly, platelet-dependent natural killer (NK) cell escape mechanism has been suggested by in vitro and preclinical models across diverse mouse and human cancer cell lines.74 It is thus posited that the presence of platelets may equip tumor cells with enhanced ability to escape elimination by the immune system through EMT, ultimately promoting their metastatic competency.
Genes involved in DNA repair (e.g., RAD51, PARP1) and G2/M DNA damage checkpoint (e.g., AR, TK1, PLK1, MAGEA1, MAGEC1, MAGEC2, CTAGB1, BIRC5, TOP2A) were frequently expressed in prostate CTCs.41,46 While several transcripts (e.g., PLK1, TOP2A) have been associated with aggressiveness in localized prostate cancer,46 it is noteworthy that CTCs derived from advanced cancer patients also highly expressed these markers relative to normal prostate tissues. In contrast, compared to cancer cell lines and matched primary tumors, genes involved in cell proliferation (e.g., MYC, ATF3, TERT, RAC1, FOXA1, RRM1, CCNB1, BIRC5, Ki-67, c-Myc) were significantly downregulated in CTCs across breast42 and colorectal24 cancers, suggesting a non-proliferative, or dormant, state of CTCs in circulation.
Given the generally diminished expression of proliferation-related genes, conventional therapeutic strategies targeting proliferating cells may not be the best for eradicating “seeds” of metastasis. Promisingly genes involved in the PI3K-AKT-mTOR signaling pathway, in which many are currently in (pre)clinical trial stages or FDA-approved, and other potentially targetable genes were frequently expressed in CTCs at high levels across various malignancies including multiple myeloma,36 and breast,38,42 prostate41,43 and colorectal24 cancers. The incorporation of single-CTC analysis into clinical trials may thus be ideal from clinical perspective for the development of companion diagnostics.42
Clinical significance
scRNA-seq or scPCR technologies have been widely applied to study early mammalian development, neuronal diversity, and immune system, revealing spatial and temporal dynamics, cellular heterogeneity, clonal distribution, pathways, and crosstalk.87,88,89 Their application in the context of CTC-based liquid biopsy, however, has been limited primarily to capturing a snapshot of the cellular states at a given point in time. In clinical contexts, it is the dynamics of such cellular state (i.e., temporal heterogeneity) that is of primary interest to monitor therapeutic response during the course of treatment. Despite technical challenges, a few single-cell studies have successfully demonstrated clinically-promising use of CTC-derived transcripts particularly for serial monitoring of the disease in a prospective cohort.
Prior knowledge of genes of interest is often required in traditional gene expression analysis for clinical diagnosis of tissues or cells in circulation using immunohistochemistry, in situ hybridization, or flow cytometry, yielding semi-quantitative data. In contrast, scRNA-seq generates high-throughput expression data in an unbiased, objective manner, with superior diagnostic sensitivity over existing technologies. For example, scRNA-seq-acquired expression data of a few selected, well-established markers, which were previously used to sort multiple myeloma (MM) cells by flow cytometry, achieved near perfect accuracy in differentiating normal and malignant plasma cell.36 Similarly, the improved diagnostic performance of CTC-based multiplex assays was observed in advanced breast cancer, supporting the robust detection capability of single-CTC-derived markers.38
The benefits of scRNA-seq technologies in the CTC field are particularly pronounced in classifying sub-populations of cells that may be clinically distinct, which are overlooked by conventional diagnostics due to the insufficient resolution. Unsupervised hierarchical clustering of single circulating MM cells-derived transcripts, for example, differed considerably from one patient to another, indicating the presence of different subtypes in MM.36 The existence of key chromosomal translocations associated with clinical risk may further be inferred from scRNA-seq data; circulating MM cells overexpressed CCND1 and CCND3 indicative of chromosomal translocations of CCND1/IGH fusion from t(11;14) and CCND3/IGH fusion from t(6;14)), respectively, and the presence of these genomic aberrations were further validated in matched MM by fluorescence in situ hybridization (FISH).36 Provided that the overexpression of CCND1 has been associated with resistance to EGFR-, BRAF- and MAF-targeted therapies,90 single-CTC transcriptome may be used as a predictive indicator for diagnosis, MM classification, and therapeutic efficacy in clinical settings.
Transcriptomes of single CTCs have been analyzed comparatively with that of CTC clusters, which have been associated with enhanced metastatic competence91,92,93 and poor prognosis49,94,95 across multiple cancer types. Differential expression analysis between the two groups identified specific gene signature (e.g., cell junction component plakoglobin) required in forming CTC cluster and distant metastases, in which high expression levels were indicative of short metastasis-free survival.49 Further, single-CTC-derived transcriptomes revealed signaling pathways (i.e., non-canonical Wnt signaling) relevant to treatment response (i.e., anti-androgen resistance), which was not evident in matched primary tumors, in prostate cancer.37 Similarly, the potential role of Sonic Hedgehog, Wnt, and TGFß signaling pathways in metastatic castration-resistance and immunotherapy response was suggested by single-CTC expression profiles in prostate cancer.43
By applying label-free microfluidic approaches,12 our group recently demonstrated how previously developed prognostic index,96,97 and the resulting prognostication, can be refined with single-CTC-derived gene signatures while accounting for cellular heterogeneity.76 Expression of a subset of matrisome genes, including MMP1 and MMP12, in tumor tissues and CTCs was consistently associated with metastatic spread and early recurrence of non-small-cell lung cancer (NSCLC), respectively. In line with our earlier observations of EGFR mutations (e.g., T790M/L858R) in CTCs and matched tumor tissue,12 this study provides the molecular evidence linking a sold tumor with single-CTC-based indicator associated with clinical presentation at the transcript level, highlighting promising predictive value of circulating biomarkers.
Despite a well-established role as a biomarker prognostic of survival particularly in breast cancer,98,99 microscopy-based CTC enumeration alone may not be sufficient to predict drug resistance in the absence of robust molecular characterization. As an advanced alternative, single-CTC-derived transcripts may serve as an excellent source to develop quantitative scoring assay comprising tissue lineage-specific genes that are not present in normal blood cells. To date, very few studies have applied such metrics in a prospectively monitored patient cohort to demonstrate their practical utility in the clinical setting.38,39,40
In 2018, the Haber group presented a predictive digital CTC scoring strategy to identify patients with poor overall survival (OS) and progression-free survival (PFS) in metastatic castration-resistant prostate cancer treated with first-line abiraterone.40 Serial monitoring of CTCs further predicted early dissemination in another independent cohort of patients with localized cancer.40 The same group found that digital quantification of intracellular ER signaling in single CTCs was predictive of residual disease in localized breast cancer patients treated with neoadjuvant therapy.38 Importantly, this 17-gene CTC score predicted early progression in metastatic breast cancer treated with endocrine therapy, which was not adequate to suppress ER signaling, despite having functional ESR1.38
The greatest focus in immuno-oncology has been on tumor biopsy-derived features, such as PD-L1 expression, tumour-infiltrating lymphocyte (TIL) density, T-cell receptor (TCR) clonality, mutational burden, and immune gene signatures, for their increasingly recognized predictive values for ICI-based immunotherapy.100 Although promising, their invasive nature makes repeated sampling not clinically practical particularly for metastatic diseases over the course of treatment. The development of less-invasive CTC-based liquid biopsies as a predictive biomarkers for response to ICI treatments will therefore be particularly promising. Hong et al. recently showed that the scoring model recapitulating temporal dynamics of CTCs identified patients with better OS and PFS in ICI-treated melanoma patients, demonstrating the feasibility of quantifying transcripts derived from microfluidically enriched CTCs for predicting patients likely to benefit from ICI therapies.39 Larger studies will be required to develop and establish such generalized framework for guiding therapeutic decision-making.
Challenges and beyond
The advent of sequencing technologies has created a new era of precision medicine. The prospect of applying this concept to develop clinically applicable biomarkers for diagnosis, prognosis, and prediction of therapy response has been extensively explored on cancer patients. Particularly, liquid biopsies focusing on the analysis of CTCs and cell-free tumor DNA (ctDNA) in the bloodstream are evolving into promising clinical parameters.101 ctDNA may allow mutational analyses to monitor tumor dynamics during cancer treatment102 and offer easier handling, storage, and shipping of samples compared to CTCs.103 A small number of mutant gene fragments present in ctDNA, which are further diluted by normal circulating DNA fragments released by apoptotic cells, however, require highly sophisticated methods to accurately assess tumor-specific genomic alterations (a detailed comparison between the two types of analytes is beyond the scope of this review). Nevertheless, a recently developed CancerSEEK blood test that examines the presence of mutations in cfDNA has achieved high sensitivity ranging from 69 to 98% across five cancer types, showing great promise for early cancer detection.104
CTCs represent intact and viable tumor cells that can be analyzed at multiple biological levels, allowing sequential sampling at multiple time points from patients undergoing systemic drug treatment. It is thus possible to perform multi-dimensional molecular and phenotypic characterization of these cells, which increasingly serves as an essential tool in precision diagnosis.105 Nevertheless, challenges remain in the field as these putative metastatic precursor cells occur at extremely low frequency relative to normal leukocytes in any given clinical sample. Such rare nature of CTCs clearly raises the question of whether these cells obtained at a single time point alone would truly recapitulate spatially and temporally evolving landscape of the entire tumor and its microenvironment, or the metastatic state. Further, the recovery efficiency varies greatly across the enrichment technology, posing additional challenges in understanding their cellular heterogeneity and the functional and clinical significance of their appearance in the bloodstream. Consequently, little is known about the molecular characteristics and mechanisms, particularly in relation to drug resistance and their capacity in circulating bloodstream with metastatic potential.
The confounding effects of inherent rarity and heterogeneity of CTCs on the downstream analysis may further be exacerbated by biased positive selection during single-cell isolation (i.e., enrichment of target cells based on antibodies specific to CTC surface markers), missing out cells with low or even no surface marker expression in circulation which prove to be of clinical significance.7 CTCs in advanced disease indeed exhibited predominant epithelial-mesenchymal-mixed (E/M), or mesenchymal (M) phenotypes (i.e., expressing mesenchymal markers) across multiple cancer types, including esophageal squamous carcinoma,106 ovarian cancer,107 pancreatic cancer,108 colorectal cancer,109 triple-negative breast cancer,110 and hepatocellular carcinoma.111 These EMT-shifted CTCs would not have been detected by immunoaffinity-based enrichment solely facilitated by antibodies targeting epithelial markers (e.g., EpCAM and pan-keratins). Capture efficiency may thus be enhanced by using cocktails of antibodies,7 including both epithelial and mesenchymal biomarkers, or by utilizing tumor lineage-specific signatures38 without making a priori assumption about the type of tumor cells.
In contrast, negative depletion (i.e., removal of non-target cells) using label-free approaches which leverage unique physical properties (e.g., cell size) of CTCs may lead to relatively low purity given the size overlap with leukocytes,112 as observed across breast, colorectal and prostate cancers.113 Some may even present a similar immunofluorescence staining pattern with leukocytes expressing both leukocyte- and CTC-specific markers, adding layers of complexity. Although such “double positive” cells are often excluded from the analysis, their occurrence in healthy blood samples at a much lower frequency point towards their possible functional role and clinical impact.114 Microfluidic approaches are increasingly being applied in this regard to enable both WBC elimination and selective CTC isolation on a single platform, as demonstrated by two-stage microfluidic chips.115,116
Finally, many single-CTC studies do not state the total number of cells initially isolated during the enrichment step, and the quality and number of CTCs that have failed QC and that have been excluded from further analysis. This makes the direct comparison of transcriptional changes found in CTCs between studies extremely difficult, as such molecular findings may only be applicable to a small subpopulation of CTCs depending on the enrichment technology or the QC metrics. The development of a clearly defined and more uniform workflow is thus urgently needed to facilitate its clinical application at different stages of the antitumor therapy or cancer progression across, and within, patients. The 17-gene CTC-specific assay is an exemplary quantitative scoring metrics that has achieved high sensitivity for monitoring of therapy response in localized and metastatic breast cancer patients.38 Continuous optimization of the developed platform and prospective clinical validation of CTC-based liquid biopsy will ultimately provide clinicians with robust, yet readily understandable, test results in a shorter turnaround time compared to conventional tissue biopsy.
References
Ashworth, T. R. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Australas. Med. J. 14, 146–149 (1869).
Salgado, I. et al. Tumour cells in the blood. Can. Med. Assoc. J. 81, 619–622 (1959).
Alexander, R. F. & Spriggs, A. I. The differential diagnosis of tumour cells in circulating blood. J. Clin. Pathol. 13, 414–424 (1960).
Gwak, H. et al. Progress in circulating tumor cell research using microfluidic devices. Micromachines 9, 353 (2018).
Pecot, C. V. et al. A novel platform for detection of CK+ and CK- CTCs. Cancer Discov. 1, 580–586 (2011).
Mikolajczyk, S. D. et al. Detection of EpCAM-Negative and cytokeratin-negative circulating tumor cells in peripheral blood. J. Oncol. 2011, 252361 (2011).
Poudineh, M., Sargent, E. H., Pantel, K. & Kelley, S. O. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat. Biomed. Eng. 2, 72–84 (2018).
Allard, W. J. et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res 10, 6897–6904 (2004).
Coumans, F. A., Ligthart, S. T., Uhr, J. W. & Terstappen, L. W. Challenges in the enumeration and phenotyping of CTC. Clin. Cancer Res 18, 5711–5718 (2012).
Stoecklein, N. H., Fischer, J. C., Niederacher, D. & Terstappen, L. W. Challenges for CTC-based liquid biopsies: low CTC frequency and diagnostic leukapheresis as a potential solution. Expert Rev. Mol. Diagn. 16, 147–164 (2016).
Tan, C. L. et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 7, 23251–23262 (2016).
Yeo, T. et al. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci. Rep. 6, 22076 (2016).
Ferreira, M. M., Ramani, V. C. & Jeffrey, S. S. Circulating tumor cell technologies. Mol. Oncol. 10, 374–394 (2016).
Paterlini-Brechot, P. & Benali, N. L. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 253, 180–204 (2007).
Konigsberg, R. et al. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 50, 700–710 (2011).
Tan, S. J. et al. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens. Bioelectron. 26, 1701–1705 (2010).
Warkiani, M. E. et al. Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 11, 134–148 (2016).
Warkiani, M. E. et al. Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab a chip 14, 128–137 (2014).
Yu, M. et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013).
Lecharpentier, A. et al. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br. J. Cancer 105, 1338–1341 (2011).
Satelli, A. et al. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin. Cancer Res 21, 899–906 (2015).
Yu, M. et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature 487, 510–513 (2012).
Carter, L. et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat. Med 23, 114–119 (2017).
Steinert, G. et al. Immune Escape and Survival Mechanisms in Circulating Tumor Cells of Colorectal Cancer. Cancer Res. 74, 1694–1704 (2014).
Wang, Y. et al. Single nucleotide variant profiles of viable single circulating tumour cells reveal CTC behaviours in breast cancer. Oncol. Rep. 39, 2147–2159 (2018).
Barbazan, J. et al. Molecular characterization of circulating tumor cells in human metastatic colorectal cancer. PloS ONE 7, e40476 (2012).
Alizadeh, A. A. et al. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 21, 846–853 (2015).
Li, H. et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 49, 708–718 (2017).
Brooks, M. D., Burness, M. L. & Wicha, M. S. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. cell stem cell 17, 260–271 (2015).
Janiszewska, M. et al. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat. Genet. 47, 1212–1219 (2015).
Aktas, B. et al. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res 11, R46 (2009).
Blassl, C. et al. Gene expression profiling of single circulating tumor cells in ovarian cancer—establishment of a multi-marker gene panel. Mol. Oncol. 10, 1030–1042 (2016).
Ni, X. et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl Acad. Sci. USA 110, 21083–21088 (2013).
Heitzer, E. et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 73, 2965–2975 (2013).
Forbes, S. A. et al. COSMIC: mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic acids Res. 39, D945–D950 (2011).
Lohr, J. G. et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci. Transl. Med. 8, 363ra147 (2016).
Miyamoto, D. T. et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356 (2015).
Kwan, T. T. et al. A digital RNA Signature of circulating tumor cells predicting early therapeutic response in localized and metastatic breast cancer. Cancer Discov. 8, 1286–1299 (2018).
Hong, X. et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc. Natl Acad. Sci. USA 115, 2467–2472 (2018).
Miyamoto, D. T. et al. An RNA-based digital circulating tumor cell signature is predictive of drug response and early dissemination in prostate cancer. Cancer Discov. 8, 288–303 (2018).
Gorges, T. M. et al. Accession of tumor heterogeneity by multiplex transcriptome profiling of single circulating tumor cells. Clin. Chem. 62, 1504–1515 (2016).
Powell, A. A. et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PloS ONE 7, e33788 (2012).
Chen, C. L. et al. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate 73, 813–826 (2013).
Mego, M. et al. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. Int. J. cancer 129, 417–423 (2011).
Farace, F. et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br. J. Cancer 105, 847–853 (2011).
Cann, G. M. et al. mRNA-Seq of single prostate cancer circulating tumor cells reveals recapitulation of gene expression and pathways found in prostate cancer. PloS one 7, e49144 (2012).
Park, S. M. et al. Molecular profiling of single circulating tumor cells from lung cancer patients. Proc. Natl Acad. Sci. USA 113, E8379–E8386 (2016).
Ramskold, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Aceto, N. et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).
Ramalingam, N. et al. Abstract 2923: Label-free enrichment and integrated full-length mRNA transcriptome analysis of single live circulating tumor cells from breast cancer patients. Cancer Res. 77, 2923 (2017).
Ting, D. T. et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 8, 1905–1918 (2014).
Chiu, T. K. et al. Application of optically-induced-dielectrophoresis in microfluidic system for purification of circulating tumour cells for gene expression analysis- Cancer cell line model. Sci. Rep. 6, 32851 (2016).
Chan, J. Y. et al. Dielectrophoresis-based microfluidic platforms for cancer diagnostics. Biomicrofluidics 12, 011503 (2018).
Poudineh, M. et al. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 12, 274–281 (2017).
Gale, D. et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS ONE 13, e0194630 (2018).
Yin, J. et al. Characterization of circulating tumor cells in breast cancer patients by spiral microfluidics. Cell Biol. Toxicol. 35, 59–66 (2019).
Court, C. M. et al. Reality of single circulating tumor cell sequencing for molecular diagnostics in pancreatic cancer. J. Mol. Diagn. 18, 688–696 (2016).
Jiang, R. et al. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget 6, 44781–44793 (2015).
Deng, G. et al. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC cancer 14, 456 (2014).
Sinkala, E. et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat. Commun. 8, 14622 (2017).
Hashimshony, T., Wagner, F., Sher, N. & Yanai, I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012).
Van Loo, P. & Voet, T. Single cell analysis of cancer genomes. Curr. Opin. Genet. Dev. 24, 82–91 (2014).
Tang, F. et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6, 377–382 (2009).
Babayan, A. et al. Heterogeneity of estrogen receptor expression in circulating tumor cells from metastatic breast cancer patients. PLoS ONE 8, e75038 (2013).
Dago, A. E. et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PloS ONE 9, e101777 (2014).
Lohr, J. G. et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 32, 479–484 (2014).
Fernandez, S. V. et al. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast Cancer Res. 16, 445 (2014).
Gasch, C. et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin. Chem. 59, 252–260 (2013).
Gasch, C. et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Mol. Oncol. 10, 1330–1343 (2016).
Neumann, M. H. D. et al. Isolation and characterization of circulating tumor cells using a novel workflow combining the CellSearch® system and the CellCelector™. Biotechnol. Prog. 33, 125–132 (2017).
Neves, R. P. et al. Genomic high-resolution profiling of single CKpos/CD45neg flow-sorting purified circulating tumor cells from patients with metastatic breast cancer. Clin. Chem. 60, 1290–1297 (2014).
Polzer, B. et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 6, 1371–1386 (2014).
Shaw, J. A. et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin. Cancer Res. 23, 88–96 (2017).
Francart, M.-E. et al. Epithelial–mesenchymal plasticity and circulating tumor cells: travel companions to metastases. Developmental Dyn. 247, 432–450 (2018).
Kang, Y. & Pantel, K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell 23, 573–581 (2013).
Lim, S. B. et al. Addressing cellular heterogeneity in tumor and circulation for refined prognostication. Proc. Natl Acad. Sci. USA, 201907904, https://doi.org/10.1073/pnas.1907904116 (2019).
Ji, Z. & Ji, H. TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic acids Res. 44, e117 (2016).
Marco, E. et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc. Natl Acad. Sci. USA 111, E5643–E5650 (2014).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Font-Clos, F., Zapperi, S. & La Porta, C. A. M. Topography of epithelial-mesenchymal plasticity. Proc. Natl. Acad. Sci. USA 115, 5902–5907 (2018).
Tam, W. L. & Weinberg, R. A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med 19, 1438–1449 (2013).
LoPiccolo, J., Blumenthal, G. M., Bernstein, W. B. & Dennis, P. A. Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist Updat 11, 32–50 (2008).
Yeung, T. M., Gandhi, S. C., Wilding, J. L., Muschel, R. & Bodmer, W. F. Cancer stem cells from colorectal cancer-derived cell lines. Proc. Natl. Acad. Sci. 107, 3722–3727 (2010).
Rodig, S. J. et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 10, https://doi.org/10.1126/scitranslmed.aar3342 (2018).
Labelle, M., Begum, S. & Hynes, R. O. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer cell 20, 576–590 (2011).
Labelle, M., Begum, S. & Hynes, R. O. Platelets guide the formation of early metastatic niches. Proc. Natl. Acad. Sci. USA 111, E3053–E3061 (2014).
Cohen, M. et al. Lung single-cell signaling interaction Map reveals basophil role in macrophage imprinting. Cell 175, 1031–1044 (2018). e1018.
Johnson, M. B. & Walsh, C. A. Cerebral cortical neuron diversity and development at single-cell resolution. Curr. Opin. Neurobiol. 42, 9–16 (2017).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. https://doi.org/10.1038/s41586-019-0933-9 (2019).
Musgrove, E. A., Caldon, C. E., Barraclough, J., Stone, A. & Sutherland, R. L. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 11, 558–572 (2011).
Aceto, N., Toner, M., Maheswaran, S. & Haber, D. A. En route to metastasis: circulating tumor cell clusters and epithelial-to-mesenchymal transition. Trends Cancer 1, 44–52 (2015).
Cheung, K. J. et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl Acad. Sci. USA 113, E854–E863 (2016).
Duda, D. G. et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proc. Natl Acad. Sci. USA 107, 21677–21682 (2010).
Hou, J. M. et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 30, 525–532 (2012).
Paoletti, C. et al. Significance of circulating tumor cells in metastatic triple-negative breast cancer patients within a randomized, phase II trial: TBCRC 019. Clin. Cancer Res. 21, 2771–2779 (2015).
Lim, S. B. et al. Pan-cancer analysis connects tumor matrisome to immune response. NPJ Precis. Oncol. 3, 15 (2019).
Lim, S. B., Tan, S. J., Lim, W. T. & Lim, C. T. An extracellular matrix-related prognostic and predictive indicator for early-stage non-small cell lung cancer. Nat. Commun. 8, 1734 (2017).
Budd, G. T. et al. Circulating Tumor Cells versus Imaging—Predicting Overall Survival in Metastatic Breast Cancer. Clin. Cancer Res. 12, 6403 (2006).
Hayes, D. F. et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 12, 4218 (2006).
Gibney, G. T., Weiner, L. M. & Atkins, M. B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 17, e542–e551 (2016).
Heidary, M. et al. The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Res. 16, 421 (2014).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med 14, 985–990 (2008).
Neumann, M. H. D., Bender, S., Krahn, T. & Schlange, T. ctDNA and CTCs in liquid biopsy – Current Status And Where We Need To Progress. Computational Struct. Biotechnol. J. 16, 190–195 (2018).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Shin, S. H., Bode, A. M. & Dong, Z. Addressing the challenges of applying precision oncology. NPJ Precis. Oncol. 1, 28 (2017).
Han, D., Chen, K., Che, J., Hang, J. & Li, H. Detection of epithelial-mesenchymal transition status of circulating tumor cells in patients with esophageal squamous carcinoma. BioMed. Res. Int. 2018, 7610154 (2018).
Po, J. W. et al. Improved ovarian cancer EMT-CTC isolation by immunomagnetic targeting of epithelial EpCAM and mesenchymal N-cadherin. J. circulating Biomark. 7, 1849454418782617 (2018).
Zhao, X. H. et al. Molecular detection of epithelial-mesenchymal transition markers in circulating tumor cells from pancreatic cancer patients: Potential role in clinical practice. World J. Gastroenterol. 25, 138–150 (2019).
Nicolazzo, C. et al. Circulating tumor cells in right-and left-sided colorectal cancer. Cancers 11, 1042 (2019).
Horimoto, Y. et al. Analysis of circulating tumour cell and the epithelial mesenchymal transition (EMT) status during eribulin-based treatment in 22 patients with metastatic breast cancer: a pilot study. J. Transl. Med. 16, 287 (2018).
Wang, Z. et al. Correlation between postoperative early recurrence of hepatocellular carcinoma and mesenchymal circulating tumor cells in peripheral blood. J. Gastrointest. Surg. 22, 633–639 (2018).
Tang, W. et al. Recent advances in microfluidic cell sorting techniques based on both physical and biochemical principles. Electrophoresis 40, 930–954 (2019).
Ligthart, S. T. et al. Circulating tumor cells count and morphological features in breast, colorectal and prostate cancer. PloS one 8, e67148 (2013).
Stott, S. L. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl Acad. Sci. USA 107, 18392 (2010).
Au, S. H. et al. Microfluidic isolation of circulating tumor cell clusters by size and asymmetry. Sci. Rep. 7, 2433 (2017).
Hyun, K. A., Lee, T. Y., Lee, S. H. & Jung, H. I. Two-stage microfluidic chip for selective isolation of circulating tumor cells (CTCs). Biosens. Bioelectron. 67, 86–92 (2015).
Abouleila, Y. et al. Live single cell mass spectrometry reveals cancer-specific metabolic profiles of circulating tumor cells. Cancer Sci. 110, 697–706 (2019).
Deng, Y. et al. An integrated microfluidic chip system for single-cell secretion profiling of rare circulating tumor cells. Sci. Rep. 4, 7499 (2014).
Acknowledgements
This work was carried out at the MechanoBioengineering Laboratory at the Department of Biomedical Engineering, National University of Singapore (NUS). We thank support provided by the Institute for Health Innovation and Technology (iHealthtech) at NUS. W.-T.L. is supported by the National Medical Research Council (NMRC/CSA/040/2012 and NMRC/CSA-INV/0025/2017). S.B.L. is supported by NUS Graduate School for Integrative Sciences and Engineering (NGS), Mogam Science Scholarship Foundation, and Daewoong Foundation. We thank Zac Goh for the illustration of single-cell microfluidic device.
Author information
Authors and Affiliations
Contributions
C.T.L. supervised the conception of the work. S.B.L., W.D.L., J.V., W.-T.L., and C.T.L. wrote and revised the paper.
Corresponding author
Ethics declarations
Competing interests
C.T.L. is a co-founder of Biolidics Ltd. W.-T.L. and C.T.L. are shareholders of Biolidics Ltd. The remaining authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lim, S.B., Di Lee, W., Vasudevan, J. et al. Liquid biopsy: one cell at a time. npj Precis. Onc. 3, 23 (2019). https://doi.org/10.1038/s41698-019-0095-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-019-0095-0
This article is cited by
-
Nano-omics: nanotechnology-based multidimensional harvesting of the blood-circulating cancerome
Nature Reviews Clinical Oncology (2022)
-
Hydrophoresis — A Microfluidic Principle for Directed Particle Migration in Flow
BioChip Journal (2020)
-
Progress toward liquid biopsies in pediatric solid tumors
Cancer and Metastasis Reviews (2019)