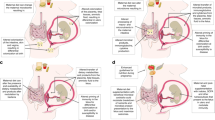
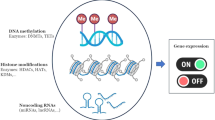
Abstract
Mounting evidence suggests that environmental stress experienced in utero (for example, maternal nutritional deficits) establishes a predisposition in the newborn to the development of chronic diseases later in life. This concept is often referred to as the “fetal origins hypothesis” or “developmental origins of health and disease”. Since its first proposal, epigenetics has emerged as an underlying mechanism explaining how environmental cues become gestationally “encoded”. Many of the enzymes that impart and maintain epigenetic modifications are highly sensitive to nutrient availability, which can be influenced by the metabolic activities of the intestinal microbiota. Therefore, the maternal microbiome has the potential to influence epigenetics in utero and modulate offspring’s long-term health trajectories. Here we summarize the current understanding of the interactions that occur between the maternal gut microbiome and the essential nutrient choline, that is not only required for fetal development and epigenetic regulation but is also a growth substrate for some microbes. Bacteria able to metabolize choline benefit from the presence of this nutrient and compete with the host for its access, which under extreme conditions may elicit signatures of choline deficiency. Another consequence of bacterial choline metabolism is the accumulation of the pro-inflammatory, pro-thrombotic metabolite trimethylamine-N-oxide (TMAO). Finally, we discuss how these different facets of microbial choline metabolism may influence infant development and health trajectories via epigenetic mechanisms and more broadly place a call to action to better understand how maternal microbial metabolism can shape their offspring’s propensity to chronic disease development later in life.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Maloney, C. A. & Rees, W. D. Gene–nutrient interactions during fetal development. Reproduction 130, 401–410 (2005).
Barker, D. J. The fetal and infant origins of adult disease. Br. Med. J. 301, 1111 (1990).
Barker, D. J. Fetal origins of coronary heart disease. Br. Med. J. 311, 171–174 (1995).
Barker, D. J. P. Mothers, Babies, and Health in Later Life (Churchill Livingstone, London, UK, 1998).
Almond, D. & Currie, J. Killing me softly: the fetal origins hypothesis. J. Econ. Perspect. 25, 153–172 (2011).
Rasmussen, K. M. The “fetal origins” hypothesis: challenges and opportunities for maternal and child nutrition. Annu. Rev. Nutr. 21, 73–95 (2001).
Danese, A., Pariante, C. M., Caspi, A., Taylor, A. & Poulton, R. Childhood maltreatment predicts adult inflammation in a life-course study. Proc. Natl. Acad. Sci. USA 104, 1319–1324 (2007).
McDade, T. W., Hoke, M., Borja, J. B., Adair, L. S. & Kuzawa, C. Do environments in infancy moderate the association between stress and inflammation in adulthood? Initial evidence from a birth cohort in the Philippines. Brain Behav. Immun. 31, 23–30 (2013).
Miller, G. E. & Chen, E. Harsh family climate in early life presages the emergence of a proinflammatory phenotype in adolescence. Psychol. Sci. 21, 848–856 (2010).
Taylor, S. E., Lehman, B. J., Kiefe, C. I. & Seeman, T. E. Relationship of early life stress and psychological functioning to adult C-reactive protein in the Coronary Artery Risk Development in Young Adults study. Biol. Psychiatry 60, 819–824 (2006).
Bale, T. L. Epigenetic and transgenerational reprogramming of brain development. Nat. Rev. Neurosci. 16, 332–344 (2015).
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(Suppl.), 245–254 (2003).
Feng, J. et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430 (2010).
Hitchings, G. H. et al. A new synthesis of cytosine and 5-methylcytosine. J. Biol. Chem. 177, 357–360 (1949).
Hsieh, C.-L. The de novo methylation activity of Dnmt3a is distinctly different than that of Dnmt1. BMC Biochem. 6, 6 (2005).
Kang, E. S., Park, C. W. & Chung, J. H. Dnmt3b, de novo DNA methyltransferase, interacts with SUMO-1 and Ubc9 through its N-terminal region and is subject to modification by SUMO-1. Biochem. Biophys. Res. Commun. 289, 862–868 (2001).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Song, J., Rechkoblit, O., Bestor, T. H. & Patel, D. J. Structure of DNMT1–DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331, 1036–1040 (2011).
Wyatt, G. R. Occurrence of 5-methylcytosine in nucleic acids. Nature 166, 237–238 (1950).
Boyes, J. & Bird, A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 11, 327–333 (1992).
Yang, X. et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26, 577–590 (2014).
Reik, W., Dean, W. & Walter, J. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 (2001).
Dolinoy, D. C. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutr. Rev. 66(Suppl. 1), S7–S11 (2008).
Hall, E. et al. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol. 15, 522 (2014).
McCarthy, M. M., Arnold, A. P., Ball, G. F., Blaustein, J. D. & De Vries, G. J. Sex differences in the brain: the not so inconvenient truth. J. Neurosci. 32, 2241–2247 (2012).
Paul, B. et al. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 7, 112 (2015).
Pogribny, I. P. & Beland, F. A. DNA hypomethylation in the origin and pathogenesis of human diseases. Cell. Mol. Life Sci. 66, 2249–2261 (2009).
Pogribny, I. P. et al. Epigenetic alterations in the brains of Fisher 344 rats induced by long-term administration of folate/methyl-deficient diet. Brain Res. 1237, 25–34 (2008).
Blumberg, J. B., Frei, B., Fulgoni, V. L. III, Weaver, C. M. & Zeisel, S. H. Vitamin and mineral intake is inadequate for most Americans: what should we advise patients about supplements? J. Fam. Pract. 65(Suppl.), S1–S8 (2016).
LeBlanc, J. G. et al. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 24, 160–168 (2013).
Müller, O. & Krawinkel, M. Malnutrition and health in developing countries. CMAJ 173, 279–286 (2005).
Blanton, L. V., Barratt, M. J., Charbonneau, M. R., Ahmed, T. & Gordon, J. I. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science 352, 1533 (2016).
Martorell, R. & Zongrone, A. Intergenerational influences on child growth and undernutrition. Paediatr. Perinat. Epidemiol. 26(Suppl. 1), 302–314 (2012).
Prendergast, A. J. & Humphrey, J. H. The stunting syndrome in developing countries. Paediatr. Int. Child Health 34, 250–265 (2014).
Romano, K. A., Vivas, E. I., Amador-Noguez, D. & Rey, F. E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio 6, e02481–e14 (2015).
Romano, K. A. et al. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe 22, 279–290 (2017).
Smith, M. I. et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 (2013).
den Besten, G. et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340 (2013).
Biesalski, H. K. Nutrition meets the microbiome: micronutrients and the microbiota. Ann. NY Acad. Sci. 1372, 53–64 (2016).
Culligan, E. P., Sleator, R. D., Marchesi, J. R. & Hill, C. Metagenomic identification of a novel salt tolerance gene from the human gut microbiome which encodes a membrane protein with homology to a brp/blh-family β-carotene 15,15′-monooxygenase. PLoS One 9, e103318 (2014).
Degnan, P. H., Barry, N. A., Mok, K. C., Taga, M. E. & Goodman, A. L. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe 15, 47–57 (2014).
Degnan, P. H., Taga, M. E. & Goodman, A. L. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20, 769–778 (2014).
Gustafsson, B. E. Vitamin K deficiency in germfree rats. Ann. NY Acad. Sci. 78, 166–174 (1959).
Karl, J. P. et al. Fecal menaquinone profiles of overweight adults are associated with gut microbiota composition during a gut microbiota-targeted dietary intervention. Am. J. Clin. Nutr. 102, 84–93 (2015).
Russell, W. R. et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 93, 1062–1072 (2011).
Semova, I. et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288 (2012).
Xu, J. et al. A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003).
Martens, E. C. et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 9, e1001221 (2011).
Donohoe, D. R. et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13, 517–526 (2011).
Bäckhed, F., Manchester, J. K., Semenkovich, C. F. & Gordon, J. I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 104, 979–984 (2007).
Vitaglione, P. et al. Protocatechuic acid is the major human metabolite of cyanidin-glucosides. J. Nutr. 137, 2043–2048 (2007).
Craciun, S. & Balskus, E. P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. USA 109, 21307–21312 (2012).
Baker, J. R. & Chaykin, S. The biosynthesis of trimethylamine-N-oxide. J. Biol. Chem. 237, 1309–1313 (1962).
Velasquez, M. T., Ramezani, A., Manal, A. & Raj, D. S. Trimethylamine N-oxide: the food, the bad and the unknown. Toxins 8, E326 (2016).
Tang, W. H. W. & Hazen, S. L. Microbiome, trimethylamine N-oxide, and cardiometabolic disease. Transl. Res. 179, 108–115 (2017).
Fennema, D., Phillips, I. R. & Shephard, E. A. Trimethylamine and trimethylamine N-oxide, a flavin-containing monooxygenase 3 (FMO3)-mediated host–microbiome metabolic axis implicated in health and disease. Drug Metab. Dispos. 44, 1839–1850 (2016).
Nam, J. et al. Choline prevents fetal overgrowth and normalizes placental fatty acid and glucose metabolism in a mouse model of maternal obesity. J. Nutr. Biochem. 49, 80–88 (2017).
King, J. H. et al. Maternal choline supplementation alters fetal growth patterns in a mouse model of placental insufficiency. Nutrients 9, 765 (2017).
Craig, S. A. S. Betaine in human nutrition. Am. J. Clin. Nutr. 80, 539–549 (2004).
Jack-Roberts, C. et al. Choline supplementation normalizes fetal adiposity and reduces lipogenic gene expression in a mouse model of maternal obesity. Nutrients 9, 899 (2017).
Kennedy, E. P. & Weiss, S. B. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 222, 193–214 (1956).
Fungwe, T. V., Cagen, L., Wilcox, H. G. & Heimberg, M. Regulation of hepatic secretion of very low density lipoprotein by dietary cholesterol. J. Lipid Res. 33, 179–191 (1992).
Randrianarisoa, E. et al. Relationship of serum trimethylamine N-oxide (TMAO) levels with early atherosclerosis in humans. Sci. Rep. 6, 26745 (2016).
Ozarda Ilcol, Y., Uncu, G. & Ulus, I. H. Free and phospholipid-bound choline concentrations in serum during pregnancy, after delivery and in newborns. Arch. Physiol. Biochem. 110, 393–399 (2002).
McMahon, K. E. & Farrell, P. M. Measurement of free choline concentrations in maternal and neonatal blood by micropyrolysis gas chromatography. Clin. Chim. Acta 149, 1–12 (1985).
Brunst, K. J. et al. Racial/ethnic and sociodemographic factors associated with micronutrient intakes and inadequacies among pregnant women in an urban US population. Public Health Nutr. 17, 1960–1970 (2014).
Resseguie, M. et al. Phosphatidylethanolamine N-methyltransferase (PEMT) gene expression is induced by estrogen in human and mouse primary hepatocytes. FASEB J. 21, 2622–2632 (2007).
Kovacheva, V. P. et al. Gestational choline deficiency causes global and Igf2 gene DNA hypermethylation by up-regulation of Dnmt1 expression. J. Biol. Chem. 282, 31777–31788 (2007).
Zeisel, S. H. Dietary choline deficiency causes DNA strand breaks and alters epigenetic marks on DNA and histones. Mutat. Res. 733, 34–38 (2012).
Sun, X. et al. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS–TXNIP–NLRP3 inflammasome. Biochem. Biophys. Res. Commun. 481, 63–70 (2016).
Ávila, J. G., Echeverri, I., de Plata, C. A. & Castillo, A. Impact of oxidative stress during pregnancy on fetal epigenetic patterns and early origin of vascular diseases. Nutr. Rev. 73, 12–21 (2015).
Wang, Y., Surzenko, N., Friday, W. B. & Zeisel, S. H. Maternal dietary intake of choline in mice regulates development of the cerebral cortex in the offspring. FASEB J. 30, 1566–1578 (2016).
Niculescu, M. D., Craciunescu, C. N. & Zeisel, S. H. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 20, 43–49 (2006).
Ross, R. G. et al. Perinatal phosphatidylcholine supplementation and early childhood behavior problems: evidence for CHRNA7 moderation. Am. J. Psychiatry 173, 509–516 (2016).
Semba, R. D. et al. The association of serum choline with linear growth failure in young children from rural Malawi. Am. J. Clin. Nutr. 104, 191–197 (2016).
da Costa, K.-A. et al. Common genetic polymorphisms affect the human requirement for the nutrient choline. FASEB J. 20, 1336–1344 (2006).
Kohlmeier, M., da Costa, K.-A., Fischer, L. M. & Zeisel, S. H. Genetic variation of folate-mediated one-carbon transfer pathway predicts susceptibility to choline deficiency in humans. Proc. Natl. Acad. Sci. USA 102, 16025–16030 (2005).
Resseguie, M. E. et al. Aberrant estrogen regulation of PEMT results in choline deficiency–associated liver dysfunction. J. Biol. Chem. 286, 1649–1658 (2011).
Sha, W. et al. Metabolomic profiling can predict which humans will develop liver dysfunction when deprived of dietary choline. FASEB J. 24, 2962–2975 (2010).
Kumar, H. et al. Gut microbiota as an epigenetic regulator: pilot study based on whole-genome methylation analysis. MBio 5, e02113–e02114 (2014).
Schulfer, A. F. et al. Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat. Microbiol. 3, 234–242 (2018).
Leclercq, S. et al. Low-dose penicillin in early life induces long-term changes in murine gut microbiota, brain cytokines and behavior. Nat. Commun. 8, 15062 (2017).
McDade, T. W. et al. Social and physical environments early in development predict DNA methylation of inflammatory genes in young adulthood. Proc. Natl. Acad. Sci. USA 114, 7611–7616 (2017).
Lemas, D. J. et al. Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. Am. J. Clin. Nutr. 103, 1291–1300 (2016).
Canfora, E. E., Jocken, J. W. & Blaak, E. E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 11, 577–591 (2015).
Morrison, D. J. & Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200 (2016).
Ríos-Covián, D. et al. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. https://doi.org/10.3389/fmicb.2016.00185 (2016).
Perry, R. J. et al. Acetate mediates a microbiome–brain–β-cell axis to promote metabolic syndrome. Nature 534, 213–217 (2016).
Krautkramer, K. A. et al. Diet–microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol. Cell 64, 982–992 (2016).
Trenteseaux, C. et al. Perinatal hypercholesterolemia exacerbates atherosclerosis lesions in offspring by altering metabolism of trimethylamine-N-oxide and bile acids. Arterioscler. Thromb. Vasc. Biol. 37, 2053–2063 (2017).
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Romano, K.A., Rey, F.E. Is maternal microbial metabolism an early-life determinant of health?. Lab Anim 47, 239–243 (2018). https://doi.org/10.1038/s41684-018-0129-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41684-018-0129-1
This article is cited by
-
Maternal vitamin B1 is a determinant for the fate of primordial follicle formation in offspring
Nature Communications (2023)
-
Genetic and epigenetic perspective of microbiota
Applied Microbiology and Biotechnology (2020)