Abstract
Charcoal rot disease caused by Macrophomina phaseolina (Tassi) Goid is one of the most devastating diseases in soybean in India. During 2018, 226 diverse soybean genotypes were evaluated for genetic resistance under hot-spot conditions. Out of them, a subset of 151 genotypes were selected based on Percent Disease Incidence (PDI) and better agronomic performance. Out of these 151 genotypes evaluated during 2019, 43 genotypes were selected based on PDI and superior agronomic performance for further field evaluation and molecular characterization. During 2020 and 2021, these forty-three genotypes, were evaluated for PDI, Area Under Disease Progress Curve (AUDPC), and grain yield. In 2020, genotype JS 20-20 showed least PDI (0.42) and AUDPC (9.37).Highest grain yield was recorded by the genotype JS 21-05 (515.00 g). In 2021, genotype JS 20-20 exhibited least PDI (0.00) and AUDPC (0.00).Highest grain yield was recorded in JS 20-98 (631.66 g). Across both years, JS 20-20 had the least PDI (0.21) and AUDPC (4.68), while grain yield was highest in JS 20-98 (571.67 g). Through MGIDI (multi-trait genotype-ideotype distance) analysis, JS 21-05 (G19), JS 22-01 (G43), JS 20-98 (G28) and JS 20-20 (G21) were identified as the ideotypes with respect to the traits that were evaluated. Two unique alleles, Satt588 (100 bp) on linkage group K (Chromosome no 9) and Sat_218 (200 bp) on linkage group H (Chromosome no 12), were specific for thetwo resistant genotypes JS 21-71and DS 1318, respectively. Through cluster analysis, it was observed that the genotypes bred at Jabalpur were more genetically related.
Similar content being viewed by others
Introduction
Soybean (Glycine max L. Merril) is the foremost important leguminous crop in the world, contributing 25% edible oil and two-thirds of the protein in livestock feed1. India ranks fifth in the world’s edible oil market; nevertheless, 50% of its edible oil demand is met through imports1. Soybean is the mainIndian oilseed crop with a production of 10.45 million metric tons in an area of 12.7 million hectares2. With an export share of USD 82,10,524, India has been a key player of soybean defatted oil cake (DOC) in international markets3. Soybean production in India is under rainfed conditions which results in various forms of biotic stresses such as drought. Therefore, soybean productivity has been much lower compared to the major growing countries like U.S.A and Brazil1.
Charcoal rot disease caused by Macrophomina phaseolina (Tassi) Goid is the most devastating soybean disease in India, causing as much as a 77% yield loss4. An estimate of 39.2 thousand metric tones’ yield loss is attributed to this disease in India5. Although drought/drought-like conditions aggravate this disease, significant yield losses are reported even under irrigated conditions6,7.Soybean is vulnerable to this disease across all growth stages, but disease aggravation is often observed during reproductive stages6. Greyish black appearance of the lower stem and root tissue and presence of numerous black microsclerotia are major diagnostic symptoms of soybean charcoal rot disease8,9.
M. phaseolina is a necrotrophic fungal pathogen having a broad host range including economical crop species like soybean (Glycine max, L.), sorghum (Sorghum bicolor (L.) Moench), maize (Zea mays, L.), cotton (Gossypium hirsutum L.) etc.10,11. Genetic resistance is the most effective and eco-friendly means of managing this disease12.However, breeding and genomics of charcoal rot resistance has been limited due to the quantitative nature of host-plant resistance, the pathogenic variability and multi-dimensional mode of its pathogenicity13.
Though artificial screening is less tedious, field screening should still be considered, since it is the ultimate environment where the crop is grown14. Area Under Disease Progress curve has been extensively employed in assessment of partial or quantitative disease resistance under field conditions, for temporal integration of disease progress in relation to the growth and development of host plants15. Percent Disease Index (PDI) and AUDPC have often been used for charcoal rot disease evaluation16,17.
Indian soybean mega-varieties such as JS 95-60 and JS 93-05 and many other varieties are highly susceptible to charcoal rot disease. Despite the importance of this disease in India, only a few reports on charcoal rot resistance are available16,18. Simultaneous selection for grain yield and resistance/tolerance to different forms of stresses helps in development of superior varieties. Studies on the effect of charcoal rot disease incidence on grain yield revealed that the association between disease incidence and grain yield was not always negative and the trends could vary across genotypes and environments19. Further, some soybean genotypes, which are sensitive to charcoal rot disease showed no yield penalty19. Similarly, in case of sorghum, no significant positive correlation between charcoal rot disease incidence and yield has been observed20,21. However, grain yield under high-disease pressure can be vital in soybean breeding for resistance under charcoal rot stress.
Precise estimation of genotypic values is critical for selection and recommendation of any genotype. A mixed-model based best linear unbiased prediction (BLUP) method estimates the random effects22. The shrinkage property of BLUP model tends to narrow the difference between actual and predicted genotypic values23. Superiority of BLUP method for prediction accuracy, over Best Linear Unbiased Estimation (BLUE) and other models has been demonstrated through several studies11,24,25.
Breeders often target multiple traits during the selection process, in addition to the yield performance. Multi-trait-based selections are often carried out using a linear-selection index: Smith-Hazel (SH) index26. However, the multi-collinearity effect and irrational allocation of weightage coefficients to the traits under study, renders the SH index ineffective in achieving the desired genetic gain27. A recent multivariate and BLUP based selection index, MGIDI that negates these drawbacks was found to outperform the SH index28.
The genetic base of the soybean genotypes/cultivars bred in India is very narrow. An insight into their genetic base through molecular diversity analysis will be more reliable and stable as compared to the conventional, phenotype-based diversity analysis. Genetically diverse genotypes identified through molecular markers have been employed as parents in breeding programs29,30. Simple sequence repeat (SSR) markers have been extensively employed in genetic diversity assessment studies in soybean31,32,33. This current investigation was for identification and molecular characterization of high-yielding charcoal rot resistant soybean genotypes.
Materials and methods
Preliminary screening for charcoal rot resistance
During 2018, a total of 226 soybean genotypes including varieties, breeding lines and exotic accessions were evaluated for charcoal rot resistance under hot spot conditions at J.N.K.V.V, Jabalpur, India. The experimental design used was an augmented block design containing seven blocks. Out of 226 genotypes, subsets of 151 genotypes were selected based on disease reaction and better agronomic performance. This subsets was evaluated in 2019 using an augmented block design containing six blocks.
During both the years, genotypes were sown in two rows three meters long. Four checks-JS 20-29, JS 335, JS 93-05 and JS 95-60 were repeated and randomized across the blocks. Disease evaluation was done in terms of PDI at R7 (physiological maturity) growth stage34, using a disease rating scale 0 to 935 (Table 1). Disease reaction on susceptible checks ranged from susceptible to highly susceptible during both years indicating high-disease pressure at the experimental field site.
Selective screening for charcoal rot resistance
Out of 151 genotypes evaluated during 2019, 43 genotypes were selected based on disease reaction and superior agronomic performance for their further field evaluation and molecular characterization. During 2020 and 2021, these 43 genotypes, along with five checks- JS 20-29, JS 335, JS 93-05, JS 95-60 and Dsb 21 were evaluated for PDI, AUDPC, and grain yield per plot (3.0 × 0.6 m2). The experiments were conducted in a RCBD design with three replications. In order to ensure high-disease pressure and no disease escape, seeds were mixed sorghum grain infected with M. phaseolina (10 g/each genotype/each replication) before sowing. Prior to mass multiplication of the pathogen, pathogenicity of the isolate (Fig. S1) was confirmed through a cut-stem inoculation technique36. Percent Disease Index was recorded during pod development, seed filling and at physiological maturity (between R6and R7 growth stage) by recording the number of dead plants in each plot. To evaluate the genotypes based on AUDPC, progressive development of disease was recorded at reproductive stages of soybean at 45, 60, 75 and 90 days after sowing.
AUDPC was calculated as per37
where, yi = per cent incidence of charcoal rot at ith observation, ti = time (days) at ith observation, and n = number of observation
Diversity analysis of soybean genotypes
Diversity analysis of48 soybean genotypes under study were performed using SSR markers developed by38 (table). Plant genomic DNA was extracted using CTAB method (Cetyltrimethylammonium bromide)39. The purified plant genomic DNA was quantified using the nanodrop (Denovix DS-11 + spectrophotometer) and the quality of the DNA was checked on 0.8% agarose gel electrophoresis. Polymorphism among the genotypes was determined using 59 SSR markers distributed across the 20 soybean linkage groups (https://soybase.org/). For marker analysis, the purified genomic DNA was subjected to amplification using PCR in reaction mixture (10 µl) containing 1.0 µl DNA (50–70 ng/µl), 1 µl 10 × PCR master mix, 0.6 µl each forward and reverse SSR primers (100 ng/µl), and 6.8 µl molecular-grade water. Amplification using SSR markers was carried out in thermocycler (Applied Biosystems, USA) using the standard protocol conditions with initial denaturation at 94 °C for 5 min, denaturation (94 °C) for 40 s, annealing (55 °C) for 1 min, extension (72 °C) for 1 min and final extension (72 °C) for 7 min. Amplified SSR products were resolved on 3.5% Metaphor agarose (Lonza, Switzerland) and SSR sizes were estimated using 50 bp DNA ladder. All the polymorphic markers in each genotype were recorded for the number of alleles present in the particular marker. Bands were scored as 1 (presence) or 0 (absence) for each allele and missing bands were scored as 9. Polymorphic information content (PIC) and expected heterozygosity (H) values show the discriminating ability of the marker based on the number of known alleles and their frequency distribution. PIC value for each marker was analyzed using the formula given by40.
where, Pi indicates the frequency of the ith allele among the genotypes analyzed and was calculated for each SSR locus. Jaccard’s similarity coefficient was employed in estimating the genetic similarity among genotypes. The resulting similarity matrix was further analyzed using the unweighted pair-group method arithmetic average (UPGMA) clustering algorithm for construction of dendrogram.
Statistical analyses
Prior to analysis, data for Percent Disease Incidence (PDI) was transformed using arcsine transformation41 to make the residual normal. Analysis of Augmented Randomized Complete Block Design was carried out using R package “augmentedRCBD”42. The Least Significance Difference (LSD) test was carried out using the R package “agricolae”43. Analysis of variance, estimation of variance components and heritability and MGIDI analysis was done through R package “metan”27. A phylogenetic tree was constructed from genotypic data of polymorphic SSR markers using NTSYSpc version 2.244, on the basis of genetic distances.
Bioethical statement
We confirmed that all local, national or international guidelines and legislation were adhered for the use of plants in this study (https://www.nature.com/srep/journal-policies/editorial-policies#research-involving-plants).
Results
Large-scale screening of soybean germplasm for charcoal rots resistance under sick-plot conditions
During 2018, a total of 230 soybean germplasm lines (including four checks-JS 20-29, JS 335, JS 93-05 and JS 95-60) were screened for charcoal rot resistance under hot-spot conditions. Of them,26 genotypes were highly resistant (HR), 28 genotypes were resistant (R), 36 genotypes were moderately resistant (MR), 41 were moderately susceptible (MS), 43 were susceptible (S) and 56 were highly susceptible (HS). (Table S1 and Figs. S2–S5). Out of 230 soybean genotypes, based on disease reaction and other agronomic traits, 155 genotypes (including four checks-JS 20-29, JS 335, JS 93-05 and JS 95-60)were selected for evaluation during 2019. Of them, 36 genotypes were highly resistant (HR), 21 genotypes were resistant (R), 22 genotypes were moderately resistant (MR), 41 were moderately susceptible (MS), 21 were susceptible (S) and 14 were highly susceptible (HS) (Table S2 and Fig. S1). The ANOVA, showed a significant genotypic effect (p < 0.001) for the PDI, during both the years (Table S3).The LSD test revealed that the genotypes significantly varied from each other (p < 0.05) for PDI, during both the years of experimentation. (Table S2).
Evaluation of selected soybean genotypes for PDI, AUDPC and grain yield under high-disease pressure conditions
Based on disease reaction and other agronomic traits, a total of 43 genotypes were selected from the 2019 experiment. The 43 genotypes, along with five susceptible checks (Dsb 21, JS 95-60, JS 93-05, JS 335 and JS 20-29) were evaluated for PDI, AUDPC and grain yield during 2020 and 2021. Violin plots for different traits during 2020 and 2021 are shown in Fig. 1.
During 2020, PDI ranged from 2.14 (JS 20-20) to 81.67 (JS 95-60), with an average of 30.20. AUDPC ranged from 9.37 (JS 20-20) to 2359.37 (JS 95-60) with a mean of 435.00. The range in grain yield was 15.00 g (DSb 21) to 515.00 g (JS 21-05) with a mean grain yield of 294.00 g. Genotype JS 20-20 had least PDI (0.42) followed by JS 21-05 (1.67), JS 22-01 (1.67) and JS 20-19 (2.50). Genotype JS 20-20 exhibited least AUDPC (9.37) followed by JS 21-05 (18.75), JS 22-01 (18.75) and JS 20-98 (21.87). Highest grain yield was recorded in the genotype JS 21-05 (515.00 g) followed by JS 20-98 (511.67), JS 22-01 (501.67) and JS 21-73 (451.67 g) (Table 2).
During 2021, PDI ranged from 0.00 (JS 20-20) to 75.52 (JS 95-60), with an average of 25.8.AUDPC ranged from 0.00 (JS 20-20) to 2137.50 (JS 95-60) with a mean of 327.00. Grain yield ranged from 16.66 g (JS 95-60) to 631.66 g (JS 20-98) with a mean grain yield of 373.00 g. Genotype JS 20-20 had the lowest PDI (0.00) followed by JS 21-05 (0.42), JS 22-01 (0.83), JS 20-96 (2.08) and JS 20-98 (2.06). Lowest AUDPC was observed in JS 20-20 (0.00) followed by JS 22-01 (6.25), JS 21-05 (9.37) and JS 20-98 (15.62). Highest grain yield was recorded by JS 20-98 (631.66 g) followed by JS 22-01 (580.00 g), PS 1611 (548.33 g) and DS 3106 (535.00 g) (Table 2).
Pooled means across both the years for PDI, AUDPC and grain yield were 28.0, 381.0 and 17.5 g, respectively. PDI, AUDPC and grain yield ranged from 0.20 (JS 20-20) to 95.00 (JS 95-60), 4.68 (JS 20-20) to 2248.43 (JS 95-60) and 17.5 g (JS 95-60) to 571.66 g (JS 20-98), respectively. PDI was least in JS 20-20 (0.21) followed by JS 21-05 (1.04), JS 22-01 (1.25) and JS 20-98 (2.50). The lowest AUDPC was observed in JS 20-20 (4.68) followed by JS 22-01 (12.50), JS 21-05 (14.06) and JS 20-98 (18.75). Grain yield was highest in JS 20-98 (571.67 g) followed by JS 21-05 (556.67 g), JS 22-01 (540.83 g) and JS 21-73 (492.50 g) (Table 2).
ANOVA, LRT, variance components and genetic parameters for the traits under study
The ANOVA in each year for 2020 and 2021 revealed that the genotypic effects for the three traits under study were significant at p < 0.001 (Table S4. Pooled analysis of variance (ANOVA) across years indicated that the genotypic effect, environmental effect and G × E interaction effect werehighly significant (p < 0.001) (Table 3) for the three traits.The genotypic effect for AUDPC contributed 93.6% to the total variation, followed by G × E interaction effect (4.6%) and environmental effect (1.22%). The largest portion of the total variation for PDI was explained by the genotypic effect (92.72%) followed by G × E interaction effect (3.59%) and environmental effect (1.56%). Similarly, 86.22% of the total variation for grain yield was governed by genotypic effect followed by G × E interaction effect (5.29%) and environmental effect (0.07%). Likelihood Ratio Test (LRT) revealed highly significant genotype and G × E interaction effect (p < 0.001) for all the three traits under study (data not shown).
Different variance components and genetic parameters of the traits under study across both the years are presented in Table S5. During both the years, for all three traits, genotypic variance was higher than the environmental variance. Heritability estimates were high for all the traits under study. Genotypic coefficient of variation (CVg) was high for all the three traits, in both the years. Residual coefficients of variation (CVr) was medium for PDI and AUDPC and low for grain yield, in both the years.
Genotypic BLUP values for PDI, AUDPC and Grain yield
Genotypic BLUP values for the traits PDI, AUDPC and Grain yield across two years are shown in Table S5. During 2020, BLUP value of AUDPC was least in JS 20-20 (10.3) followed by JS 21-05 (19.7), JS 22-01 (19.7) and JS 20-98 (22.8). The least BLUP value for PDI was recorded in JS 20-20 (2.4) followed by JS 21-05 (6.3), JS 22-01 (6.3) and JS 20-98 (8.3). The top four genotypes for grain yield were JS 21-05 (513.7 g), JS 20-98 (510.4 g), JS 22-01 (500.5 g) and JS 20-96 (460.7 g). Likewise, during 2021, the least BLUP value of AUDPC was recorded in JS 20-20 (0.70) followed by JS 22-01 (6.9), JS 21-05 (10.00) and JS 20-98 (16.3). The least BLUP value for PDI was observed in JS 20-20 (0.30) followed by JS 21-05 (2.40), JS 22-01 (3.30) and JS 20-96 (7.00). The grain yield BLUP was highest in the genotype JS 20-98 (630.7 g) followed by JS 21-05 (597.5 g), JS 22-01 (579.20 g) and PS 1611 (547.7 g) (Table S6).
Identification of ideotypes using MGIDI index
Using MGIDI index at a 10% selection intensity, genotypic selection was carried out based on multiple traits simultaneously (Table 3). A lower value was desirable for AUDPC and PDI and a higher value was desirable for grain yield. Across both the years, percentage selection differential was highest for AUDPC (94.9% during 2020 and 95.8% in 2021), followed by PDI (76.8% in 2020 and 80.5% in 2021) and grain yield (61.4% in 2020 and 49.4% in 2021). In 2020, JS 21-05 (G19), JS 22-01 (G43), JS 20-98 (G28), JS 20-20 (G21) and JS 20-96 (G6) were identified as ideotypes. Likewise, in 2021, JS 21-05 (G19), JS 20-98 (G28), JS 22-01 (G43), JS 20-20 (G21) and PS 1611 (G48) were ideotypes (Table 4 and Fig. 2). Across both the years, JS 21-05 (G19), JS 22-01 (G43), JS 20-98 (G28) and JS 20-20 (G21) were identified to be common ideotypes with respect to the traits that were evaluated.
Molecular diversity analysis
Fifty-four primers were polymorphic (91.5%) while five primers (Satt155, Satt252, Satt484, Satt575 and Satt724) were monomorphic across genotypes. A total of 142 alleles were amplified with an average of 2.63 alleles per locus. The number of alleles per polymorphic primer pair (locus) ranged from 2 to 6 (Satt373). One locus amplified 5 alleles, 6 loci amplified 4 alleles, 15 loci had 3 alleles and 31 loci had 2 alleles. Out of 142 alleles, 54 alleles had a frequency of 0.25 or less, 22 alleles exhibited a frequency of 0.75 or higher and the remaining 64 alleles had a frequency between 0.25 and 0.75. The size of the allele fragments ranged from 50 to 280 bp. The PIC value, for the 59 SSR markers ranged from 0.00 to 0.737 with 0.357 being the average PIC/ locus (Table 5). Out of 142 alleles identified, 5 alleles (3.5%) were unique and were amplified in a single genotype, they were; Satt 459 (LG D1b & Chromosome no 2) in Dsb1, Satt557 (LG C2 & Chromosome 6) in JS 95-60, Satt588 (LG K & Chromosome 9) in JS 21-71, Sat_218 (LG H & Chromosome 12) in DS 1318 and Satt701 (LG D1b & Chromosome 2) in SL 955. Size of these unique alleles 130, 155, 100, 200 and 150 bp respectively (Table 6). Some of the charcoal rot resistant genotypes viz. JS 21-71and DS 1318 can be identified by the unique alleles based on size of 100 and 200 bp in Satt588 and Sat_218, respectively. All 142 SSR alleles were used for the genetic diversity analysis. Jaccard’s similarity coefficient was calculated to assess the genetic proximity among the genotypes and the similarity coefficient matrix was used for UPGMA cluster analysis. Forty-eight (48) genotypes were grouped into four major clusters. Cluster I contained only two genotypes—JS 20-20 and JS 20-39. JS 20-20 was the most resistant genotype. SSR based diversity analysis separated it in a distinct cluster along with only JS20-39. Therefore, JS 20-20 may be useful for molecular mapping and resistance gene identification studies. Cluster II includes EC 393228, Harder and CAT 492A. Cluster III consists of a single genotype—AMS-MB-5-18. Cluster IV was sub-divided into IVa and IVb. Cluster IVa consists of PS 1641, JS 20-19 and SL 955. The remaining 39 genotypes were included in cluster IVb (Fig. 3).
Structure analysis
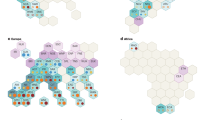
The 59 SSR markers werealso used to study the population structure of the 48 genotypes. A sharp peak in DK at K = 2 suggested the presence of two major populations (Fig. 4a). Population 1 (POP1)contained seventeen genotypes viz., NRC 2755, JS 20-73, JS 71-73, JS 21-05, AMS MB 5-18, PS 1613, JS 22-01, JSM 228, PS 1225, JS 20-96, NRC 86, JS 21-71, CAT 1847, NRC 138, AMS 264, CAT 1957 and PS 1611. The remaining 31 genotypes were grouped into Population 2 (POP2). Overall proportion of membership of the sample in POP1 and POP2 was 0.398 and 0.602, respectively. Average distance (expected heterozygosity) between individuals in same cluster was 0.260 in POP1 and 0.307 in POP2. Mean value of Wrights fixation index (Fst) was 0.273 in POP1 and 0.081 in POP2. Allele-frequency divergence among the two populations was 0.039 (Table 7). Population 1 included the majority of the genotypes bred at Jabalpur (06) followed by genotypes developed at Indore (03), germplasm lines (03), developed at Amravati (02) and Pantnagar (02). Population 2 consisted of the majority of genotypes developed at Jabalpur (13), followed by germplasm lines (05), genotypes developed at New Delhi (03), genotypes developed at Pune (02), Pantnagar (02), Ludhiana (01), KasbeDigraj (01), Amravati (01), Indore (01) and Pantnagar (01) (Fig. 4b).
Discussion
Though charcoal rot is a major fungal disease in India, to date, no systematic study on identification of high-yielding and charcoal rot resistant genotypes has been carried out. Based on grain yield and resistance reaction, the purpose of current study was identification of high-yielding charcoal rot resistant genotypes and molecular characterization of these genotypes using SSR markers under high-disease pressure.
Above ground charcoal rot symptoms start to appear from R4stage (2 cm longpod at one of the four upper most nodes with a completely unrolled leaf). It was observed that the increase in colonization of soybean by M.phaseolina was low during the vegetative and early reproductive stages, and reached its peak during R5 (beginning of pod development)to R7 growth stages6.Therefore, in this study, AUDPC was recorded during reproductive stages and PDI was recorded at R7 stage, the ideal growth stage to evaluate charcoal rot plant resistance7.
Lower levels of residual coefficient of variation (CVr) indicate the quality of experimentation. In this study, CVr was lower for grain yield and intermediate for AUDPC and PDI indicating relative uniformity of the disease pressure across the experimental site.Higher genotypic variance and heritability estimates indicate higher response to selection for the traits under this study. The pooled ANOVA revealed that the genotypic variance contributed predominantly to the total variation. The significant G × E interaction effect was observed, indicating that genotypes did not respond the same to the disease across the environments.
Improvement in any economic trait depends on understanding its mode of inheritance and heritability. No extensive studies have been carried out related to soybean charcoal rot resistance12. A polygenic mode of inheritance for resistance to M.phaseolina in soybean was reported in a few studies [18 and 12]. One QTL on chromosome 15 and two QTL on chromosome 16, governing charcoal rot resistance in soybean was mapped using F2:3 derived lines from the cross of PI 567562A (R) × PI 567,437 (S)12. In other crops like Sorghum45 and common bean46 epistatic interactions were observed. Inadequate information on the genetic mechanisms underpinning resistance and significant effects of environment has hindered the progress in breeding for resistance14. Nevertheless, transgressive segregation of progeny derived from resistant parents can be useful to identify novel and durable resistant sources47. In the current study, based on the pooled data, JS 20-20 was identified as the best genotype for mapping of genes/QTLs governing charcoal rot resistance. JS 20-98 was identified as an appropriate genotype for use as a parent in breeding for higher yield and resistance under high-disease pressure.The use of MGIDI index in selecting ideal genotypeswith resistance based on the multiple traits evaluated in this study, was effective in selecting for yield48 and quality traits49. In the current study, JS 21-05, JS 22-01, JS 20-98 and JS 20-20 were identified as the ideotypes based on the traits- PDI, AUDPC and grain yield. These genotypes were determined to be potentially high yielding sources with charcoal rot resistance. Hybridization among these ideotypes can result in selection of superior segregants having higher yield and CR resistance.
In addition, traits such as 100-seed weight, plant height, number of nodes, number of branches, biomass and harvest index should be considered in future studies to identify traits associated with yieldand disease indices under high-disease pressure similar to grain mold resistance in sorghum 50,51, and fall armyworm resistance in maize52.
To assess variation, SSR markers have been widely used for the screening of soybean germplasm53,54. In our study, 59 markers distributed uniformly across 20 linkage groups were used for molecular characterization. The high percentage of polymorph ism and high mean PIC value detected in this study is consistent with the previous studies31,55,56. However, lower number of alleles per locus indicates a relatively narrow genetic base among the genotypes used in this study.
In the current study, Satt373 had a PIC value of 0.619 with 6 alleles and Sat_244 had 5 alleles with a high PIC value of 0.737. However, satt440 marker with PIC value > 0.6 with the highest number of alleles (4) denotes a strong correlation between PIC value and allele richness. Two unique alleles that can identify different resistant genotypes were identified in this study. Cluster analysis, indicated that majority of genotypes developed at Jabalpur were grouped under a single cluster, IIb2. This indicates the genetic relatedness and narrow genetic base of the genotypes bred at Jabalpur. Except for the AMS-MB-5-18,the remaining genotypes included in POP 1 in structure analysis were included in the cluster IVb, indicating the consistency between cluster analysis and structure analysis for determining genetic relatedness among genotypes evaluated in this study.
In India, apart from charcoal rot disease, Rhizoctonia aerial blight (RAB), YMV and anthracnose are the predominant diseases that can cause significant yield losses. Mega-varieties such as JS 95-60 and JS 93-05 are highly susceptible to all these diseases. Genotypes such as JS 21-71, JS 21-72, JS 21-05, JS 21-17, PS 1611, JS 20-98 and JS20-20 were identified to be resistant to charcoal rot in the current study, were also reported as RAB resistant57,58,59. Genotypes JS 20-98, JS 21-05, JS 21-17 and PS 1611 were reported to be YMV resistant58,60, while JS 20-98, PS 1611were reported to be anthracnose resistant59,61. These genotypes can be utilized as parents to develop multiple disease resistant varieties that can play a crucial role in enhancing soybean productivity in India.
Conclusion
In the current study, JS 20-20 was identified as the best genotype for resistance and JS 20-98 was superior in grain yield under high disease pressure. Genotypes JS 21-05, JS 22-01, JS 20-98 and JS 20-20 were identified as ideal ideotypes with respect to AUDPC, PDI and grain yield for charcoal rot resistance. These genotypes will be used as parents to develop high-yielding charcoal rot resistant varieties. Two unique alleles Satt588 (100 bp) and Sat_218 (200 bp) were specific in two resistant genotypes JS 21-71and DS 1318, respectively. In the molecular diversity study, JS20-20 formed a distinct cluster and therefore may be useful in resistance gene mapping and characterization studies. Clustering pattern, showed that the genotypes bred at Jabalpur were genetically more closely related compared to other genotypes.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Agarwal, D., Billore, S. D., Sharma, A., & Dupare, B., Srivastava, S. Soybean: Introduction, improvement, and utilization in India—problems and prospects. Agric. Res. 2, https://doi.org/10.1007/s40003-013-0088-0 (2013).
USDA. World Agricultural Production. Foreign Agricultural Service/ USDA. (2021). https://apps.fas.usda.gov/psdonline/circulars/production.pdf.
Directorate General of Foreign Trade. https://www.dgft.gov.in/CP/ (2021).
Sharma, A. N., Gupta, G. K., Verma, R. K., Sharma, O. P., Bhagat, S., Amaresan, N., Saini, M. R., Chattopadhyay, C., Sushil, S. N., Asre, R., Kapoor, K. S., Satyagopal, K., Jeyakumar, P. Integrated pest management for Soybean. New Delhi, India: ICAR – National Center for Integrated Pest Management. p. 2 (2014).
Wrather, A. et al. Effect of diseases on soybean yield in the top eight producing countries in 2006. Plant Health Prog 11, 1. https://doi.org/10.1094/PHP-2010-0102-01-RS (2010).
Mengistu, A., Smith, J. R. & Ray, J. D. Seasonal progress of charcoal rot and its impact on soybean productivity. Plant Dis. 95, 1159–1166 (2011).
Mengistu, A. et al. Effect of charcoal rot on selected putative drought tolerant soybean genotypes and yield. Crop Prot. 105, 90–10 (2018).
Luna, M. P. R. et al. Advancing our understanding of charcoal rot in soybeans. J Integr Pest Manag. 8(1), 1–8 (2017).
Amrate, P. K., Shrivastava, M. K., Pancheshwar, D. K. & Stuti, S. Charcoal rot and yellow mosaic virus disease of soybean under hot spot condition: symptoms, incidence and resistance characterization. Int. J. Bio-Resour. Stress Manag. 11(3), 268–273 (2020).
Almeida, A.M.R., Seixa, C.D.S., Farias, J.R.B., Oliveira, M.C.N., Franchini, J.C., Debiase, H., Costa, J.M., Gaudencio, C. A. Macrophomina phaseolina em soja. Embrapa Soja, Londrina. http://www.infoteca.cnptia.embrapa.br/infoteca/handle/doc/989352 (2014).
Nataraj, V. Charcoal rot resistance in Soybean: current understanding and future perspectives. In eds Wani, S. (ed) Disease resistance in crop plants. Springer, Cham. https://doi.org/10.1007/978-3-030-20728-1_10 (2019).
Silva, M. P. et al. QTL mapping of charcoal rot resistance in PI 567562A soybean accession. Crop Sci. 59, 1–6 (2019).
Gupta, G. K., Sharma, S. K., Ramteke, R. Biology, epidemiology and management of the pathogenic fungus Macrophominaphaseolina (Tassi) Goid with special reference to Charcoal Rot of soybean (Glycine max (L.) Merrill). J. Phytopathol. 160, 167–180. https://doi.org/10.1111/j.1439-0434.2012.01884.x (2012).
Coser, S. M. et al. Genetic architecture of charcoal rot (Macrophomina phaseolina) resistance in soybean revealed using a diverse panel. Front Plant Sci. 8, 1626. https://doi.org/10.3389/fpls.2017.01626 (2017).
Jeger, M. J. & Viljanen-Rollinson, S. L. H. The use of the area under the disease-progress curve (AUDPC) to assess quantitative disease resistance in crop cultivars. Theor Appl Genet. 102, 32–40 (2001).
Amrate, P. K., Shrivastava, M. K., Bhale, M. S. Resistance in soybean varieties against charcoal rot disease caused by Macrophomina phaseolina. Pl. Dis. Res. 34 (2), 124–128. https://doi.org/10.5958/2249-8788.2019.00021.0 (2019).
Vinholes, P. et al. Single nucleotide polymorphism-based haplotypes associated with charcoal rot resistance in Brazilian Soybean Germplasm. Agron. J. 111, 182–192 (2019).
Talukdar, A., Verma, K., Gowda, D. S. S., et al., Molecular breeding for charcoal rot resistance in soybean I. Screening and mapping population development. Indian J. Genet. 69, 367–370 (2009).
Smith, J. R., Ray, J. D. & Mengistu, A. Genotypic differences in yield loss of irrigated soybean attributable to charcoal rot. J. Crop Improv. https://doi.org/10.1080/15427528.2018.1516262 (2018).
Das, I. K. & Prabhakar Indira, S. Role of stalk-anatomy and yield parameters in development of charcoal rot caused by Macrophomina phaseolina in winter sorghum. Phytoparasitica 36, 199–208 (2008).
Williams, A., Hector, P. Q., Victor, M. G. Grain sorghum varieties reaction [Sorghum bicolor (L.) Moench] to Macrophomina phaseolina (Tassi) Goid. Revista Mexicana de Fitopatología. 27, 148–155 (2009).
Piepho, H. P., Möhring, J., Melchinger, A. E. & Büchse, A. BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 161, 209–228. https://doi.org/10.1007/s10681-007-9449-8 (2007).
Searle, S. R., Casella, G., McCulloch, C. E. Variance Components. Wiley (1992).
Piepho, H. P. Best linear unbiased prediction (BLUP) for regional yield trials: A comparison to additive main effects multiplicative interaction (AMMI) analysis. Theor Appl Genet. 89, 647–654 (1994).
Olivoto, T. et al. Mean performance and stability in multienvironment trials I: Combining Features of AMMI and BLUP techniques. Agron. J. 111, 2949–2960. https://doi.org/10.2134/agronj2019.03.0220 (2019).
Smith, H. F. Discriminant function for plant selection. Ann. Eugenics. 7, 240–250 (1936).
Olivoto, T. & Lúcio, A. D. C. Metan: An R package for multi environment trial analysis. Methods Ecol. Evol. 11, 783–789. https://doi.org/10.1111/2041-210X.13384 (2020).
Olivoto, T., and Nardino, M. MGIDI: A novel multi-trait index for genotype selection in plant breeding. Bioinformatics. https://doi.org/10.1101/2020.07.23.217778 (2020b).
Maughan, P. J. et al. Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance, and near-isogenic line analysis. Theoret. Appl. Genet. 93, 392–401. https://doi.org/10.1007/BF00223181 (1996).
Thompson, J. A., Nelson, R. L. & Vodkin, L. O. Identification of diverse soybean germplasm using RAPD markers. Crop Sci. 38(5), 1348–1355 (1998).
Kumawat, G., Singh, G., Gireesh, C., Shivakumar, M., Arya, M., Agarwal, D.K., Husain, S.M. Molecular characterization and genetic diversity analysis of soybean (Glycine max (L.) Merr.) germplasm accessions in India. Physiol. Mol. Biol. Plants. 21(1), 101–107. https://doi.org/10.1007/s12298-014-0266-y (2015).
Kumawat, G., Yadav, A., Satpute, G. K., Gireesh, C., Patel, R., Shivakumar, M., Gupta, S., Chand, S., Bhatia, V. S. Genetic relationship, population structure analysis and allelic characterization of flowering and maturity genes E1, E2, E3 and E4 among 90 Indian soybean landraces. Physiol. Mol. Biol. Plants 25(2), 387–398. https://doi.org/10.1007/s12298-018-0615-3 (2019).
Surbhi, K., Singh, K. P., Singh, N. K., Aravind, T. Assessment of genetic diversity among soybean genotypes differing in response to aerial blight (Rhizoctonia solani Kuhn) using SSR markers. J. Phytopathol. 1–8. https://doi.org/10.1111/jph.12956 (2020).
Fehr, W. R., Caviness, C. E., Burmood, D. T., Pennington, J. S. Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci. 11, 929–931. https://doi.org/10.2135/cropsci1971.0011183X001100060051x (1971).
Anonymous: Annual Report 2016–2017. Directorate of Soybean Research Indore.
Twizeyimana, M., Hill, C. B., Pawlowski, M., Paul, C., Hartman G. L. A cut-stem inoculation technique to evaluate soybean for resistance to Macrophomina phaseolina. Am. Phytopathol. Soc. (APS). 96(8), (2012).
Shaner, G., and Finney, R. The effect of nitrogen fertilization on the expression of slowmildewing resistance in Knox wheat. Phytopathology 67, 1051–1056. https://doi.org/10.1094/Phyto-67-1051 (1977).
Cregan, P. B. et al. An integrated genetic linkage map of the soybean genome. Crop Sci. 39(5), 1464–1490. https://doi.org/10.2135/cropsci1999.3951464x (1999).
Doyle, J. J. & Doyle, J. L. Isolation of plant DNA from fresh tissue. Focus. 12, 13–15 (1990)
Botstein, D., White, R. L., Skalnick, M. H. & Davies, R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet. 32, 314–331 (1980).
Gomez, K. A. & Gomez, A. A. Statistical Procedure for Agricultural Research. 2ndEdn.,NewYork, NY:Wiley (1984).
Aravind, J., Sankar, M. S., Dhammaprakash, P. W., Kaur, V. Augmented RCBD: Analysis of Augmented Randomised Complete Block Designs. R package version 0.1.0, https:// aravind-j.github.io/augmentedRCBD/ (2018).
Mendiburu, F. & Yaseen, M. Agricolae: Statistical Procedures for Agricultural Research. Available at: https://cran.r-project.org/package=agricolae (2020).
Rohlf, F. J. NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2.2. Exeter Software, Setauket, New York, USA (2000).
Rao, D. N. V. & Shinde, V. K. Inheritance of charcoal rot resistance in sorghum. J Maharashtra Agric Univ 10, 54–56 (1985).
Olaya, G., Abawi, G. S. & Weeden, N. F. Inheritance of resistance to Macrophomina phaseolina and identification of RAPD markers linked to the resistance genes in beans. Phytopathology 86, 674–679 (1996).
Muchero, W., Ehlers J. D., Close, T. J., et al. Genic SNP markers and legume synteny reveal candidate genes underlying QTL for Macrophomina phaseolina resistance and maturity in cowpea[Vigna unguiculata (L) Walp.]. BMC Genomics. 128 (2011).
Maranna, S., Nataraj, V., Kumawat, G., Chandra, S., Rajesh, V., Ramteke, R., Patel, R. M., Ratnaparkhe, M. B., Husain, S. M., Gupta, S., Khandekar, N. Breeding for higher yield, early maturity, wider adaptability and waterlogging tolerance in soybean (Glycine max L.): A case study. Sci. Rep. (2021).
Benakanahalli, N. K., Sridhara, S., Ramesh, N., Olivoto, T., Sreekantappa, G., Tamam, N., Abdelbacki, A. M. M., Elansary, H. O., Abdelmohsen, S. A. M. A framework for identification of stable genotypes based on MTSI and MGDII indexes: An example in Guar (Cymopsis tetragonoloba L.). Agronomy. 11, 1221. https://doi.org/10.3390/agronomy11061221 (2021).
Diatta, C. et al. Development of sorghum hybrids for stable yield and resistance to grain mold for the Center and South-East of Senegal. Crop Prot. 119, 197–207 (2019).
Aruna, C., Das, I. K., Reddy, P. S., Ghorade, R. B., Gulhane, A. R., Kalpande, V. V., Kajjidoni, S. T., Hanamaratti, N. G., Chattannavar, S. N., Mehtre, S., Gholve, V., Kamble, K. R., Deepika, C., Kannababu, N., Bahadure, D. M., Govindaraj, M., Tonapi, V. A. Development of sorghum genotypes for improved yield and resistance to grain mold using population breeding approach. Front. Plant Sci. 12, 687332. doi: https://doi.org/10.3389/fpls.2021.687332 (2021).
Kasoma, C., Shimelis, H., Laing, M., Shayanowako, A. I. T. & Mathew, I. Screening of inbred lines of tropical maize for resistance to fall armyworm, and for yield and yield-related traits. Crop Prot. https://doi.org/10.1016/j.cropro.2020.105218 (2020).
Li, Y., Guan, R., Liu, Z., Ma, Y., Wang, L., Li, L., Lin, F., Luan, W., Chen, P., Yan, Z., Guan, Y., Zhu, L., Ning, X., Smulders, M. J. M., Li, W., Piao, R., Cui, Y., Yu. Z., Guan, M., Chang, R., Hou, A., Shi, A., Zhang, B., Zhu, S., Qiu, L. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theor. Appl. Genet. 117, 857–871 (2008).
Zhang, G., Xu, S., Mao, W., Hu, Q., Gong, Y. Determinationof the genetic diversity of vegetable soybean [Glycine max (L.)Merr.] using EST-SSR markers. J. Zhejiang Sci. B. 14(4), 279–28(2014)
Diwan, N. & Cregan, P. B. Automated sizing of fluorescent labeled simple sequence repeat (SSR) markers to assay genetic variation in soybean. Theor Appl Genet. 95, 723–733 (1997).
Tantasawat, P., Trongchuen, J., Prajongjai, T., Seehalak, W., Jittayasothorn,Y. Variety identification and comparative analysisof genetic diversity in yard long bean (Vigna unguiculata spp. sesquipedalis) using morphological characters, SSR and ISSR analysis. Sci Hort. 124, 204–216. (2011).
Amrate, P. K., Shrivastava, M. K., Singh, G. Screening of genotypes to identify the resistance source against major diseases of Soybean under high disease pressure conditions. Int. J. Curr. Microbiol. Appl. Sci. 9(5), 1739–1745. https://doi.org/10.20546/ijcmas.2020.905.195 (2020).
Amrate, P. K. & Shrivastava, M. K. Yield response and pathological characterization of promising genotypes of soybean against major diseases in Madhya Pradesh. J. Oilseeds Res. 38(4), 380–384 (2021).
Amrate, P. K., Shrivastava, M. K., Singh, G. Identification of sources of resistance and yield loss assessment for aerial blight and anthracnose/pod blight diseases in Soybean. Legume Research. https://doi.org/10.18805/LR-4452 (2021b).
Annual Report. Director’s Report of AICRP on Soybean 2020–2021, Ed.: Nita Khandekar. ICAR-Indian Institute of Soybean Research, Indore, Madhya Pradesh, India. p 280
Rajput, L. et al. WAASB index revealed stable resistance sources for soybean anthracnose in India. J. Agric. Sci. 159(9–10), 710–720. https://doi.org/10.1017/S0021859622000016 (2021).
Acknowledgements
Authors sincerely express our gratitude to the Director, ICAR-Indian Institute of Soybean Research (IISR), Indore for providing valuable materials for field screening; for providing lab facilities and for providing consumables for carrying out molecular studies through the institute project (IISR 3.11b/18). Authors are grateful to DST-SERB for providing manpower to carry out the molecular studies through the project-CRG/2020/002890.
Author information
Authors and Affiliations
Contributions
V.N. planned all experiments and coordinated the work; P.K.A., M.K.S. and M.S.B. performed Phenotyping in Jabalpur and identified the resistant genotypes. N.A. conducted molecular work and genotyping; G.K. and S.M. analyzed molecular data and contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amrate, P.K., Shrivastava, M.K., Bhale, M.S. et al. Identification and genetic diversity analysis of high-yielding charcoal rot resistant soybean genotypes. Sci Rep 13, 8905 (2023). https://doi.org/10.1038/s41598-023-35688-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-35688-2
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.