Abstract
Recent experimental and observational research has suggested that childhood allergic asthma and other conditions may be the result of prenatal exposure to environmental contaminants, such as di-(2-ethylhexyl) phthalate (DEHP). In a previous epidemiological study, we found that ancestral exposure (F0 generation) to endocrine disruptors or the common plasticizer DEHP promoted allergic airway inflammation via transgenerational transmission in mice from generation F1 to F4. In the current study, we employed a MethylationEPIC Beadchip microarray to examine global DNA methylation in the human placenta as a function of maternal exposure to DEHP during pregnancy. Interestingly, global DNA hypomethylation was observed in placental DNA following exposure to DEHP at high concentrations. Bioinformatic analysis confirmed that DNA methylation affected genes related to neurological disorders, such as autism and dementia. These results suggest that maternal exposure to DEHP may predispose offspring to neurological diseases. Given the small sample size in this study, the potential role of DNA methylation as a biomarker to assess the risk of these diseases deserves further investigation.
Similar content being viewed by others
Introduction
The human placenta plays a crucial role in regulating developmental programs essential to proper fetal growth1. Environmental factors, such as endocrine disruptors, can shape their functions throughout the prenatal stage and promote the development of inflammatory diseases (e.g. asthma) and developmental-related disorders, through epigenetic modifications2,3,4.
Plasticizers are commonly used in the synthesis of polyvinyl chloride to make packaging materials, such as those used for cosmetics, shampoo, and food. Some plasticizers, such as di-(2-ethylhexyl) phthalate (DEHP), are also endocrine disruptors5. Since DEHP is non-covalently bound to plastics, its probability of leaching is very high, thereby allowing entry into the human body via inhalation, ingestion, or dermal contact.
In a previous study, we reported that environmental endocrine disruptors (e.g., phthalate) increase fetal susceptibility to allergic diseases, via the neonatal immune system, including dendritic cells (DCs) and T cells. Mechanistic analysis revealed that paternal DEHP exposure can lead to the hypomethylation of the igf2r promoter with corresponding transgenerational epigenetic effects in mice from generations F1 to F44.
Epigenetic modifications, including DNA methylation, histone modification, and microRNA expression, play important roles in normal cellular differentiation and the development of various human diseases. It is widely acknowledged that aberrant epigenetic modifications play a role in the development of cancer in humans. It has also been determined that these modifications can be induced by environmental factors. One previous study reported that exposure to phthalate can induce childhood asthma by altering DNA methylation6. Recent research has also indicated that changes in DNA methylation can be induced trans-generationally4,6.
In this study, we analyzed the dose-dependent effects of DEHP on DNA methylation in placental tissue. Our results revealed DNA hypomethylation in the high DEHP exposure group. This study also sought to elucidate the mechanisms by which prenatal phthalate exposure predisposes a child to neurological diseases through the modification of DNA methylation.
Results
Genome-wide DEHP-induced DNA hypomethylation in human placenta samples
Infinium Human Methylation 850 K Beadchip was used to investigate the genome-wide DNA methylation of human placenta among 12 pregnant women exposed to DEHP (Table 1) (Fig. 1A). The distribution of beta values across all samples revealed a typical bi-modal distribution with two peaks close to 0 and 1 (Fig. 1B). Interestingly, among probes presenting at least 10% differences in methylation, hypomethylation was more pronounced in the DEHP-high group (red line) than in the DEHP-low group (blue line, Fig. 1C). This DEHP-induced hypomethylation appeared in the heatmap of differentially methylated cytosine (DMC, Fig. 1D). Taken together, we conclude that DEHP can induce DNA hypomethylation in human placenta tissue.
Infinium methylation microarray analysis of Methylation changes in DEHP-exposed human placenta samples. (A) The flowchart shows how the pipeline of raw data was processed. DNA methylation was performed using the high-resolution Infinium MethylationEPIC BeadChip Kit interrogating about 850,000 methylation sites quantitatively across the genome at single-nucleotide resolution. By utilizing a cut-off p-value threshold of greater than 0.05 and an FDR of 10%, a total of 1254 probes displaying a minimum of 10% differential methylation were identified. (B) Density of DNA methylation level for 12 human placenta samples exposed to DEHP; blue lines: low DEHP exposure (n = 6), red lines: high DEHP exposure (n = 6). Individual probes with beta (β)-values (range 0–1) are approximate representations of the absolute methylation percentage of specific CpG sites within the sample population. Beta value = 1 indicates complete methylation; beta value = 0 represents no methylation. (C) Density of 10% differentially DNA methylated probes of all samples. Hypomethylation was observed in the DEHP high group (red line), as compared to DEHP low group (blue line). (D) Heatmap showing the 10% differentially DNA methylated probes; left panel: low DEHP exposure; right panel: high DEHP exposure. Heatmap showing differentially methylated cytosine (DMC) sites across the two different exposure dosages of DEHP. Most of the probes observed were hypomethylation in DEHP (red line: methylated, blue line: unmethylated).
DEHP-induced methylation changes in the intergenic region
Further analysis was performed on genomic regions showing differential methylation under the influence of DEHP. Global hypomethylation was found in the gene body and intergenic region (Fig. 2A), in which most of the hypomethylated sites occurred (Fig. 2B). Note that sites of DNA hypermethylation were also observed in the gene body, intergenic region, and promoter region (Fig. 2C). The probes with the most pronounced hypomethylation were observed at 7 genes (Fig. 3A, Table 2), while those with the most pronounced hypermethylation were observed at 5 genes (Fig. 3B, Table 3).
Spatial distribution of differentially methylated CpGs (DMCs) and gene-centric annotations. (A) Violin plot of 1261 probes showing beta values of gene body (n = 386), 5’UTR (n = 30), intergenic region (n = 677), and promoters (n = 168). The violin shape shows the quantitative distribution of DNA methylation. Pink denotes the high DEHP group, while blue denotes the low DEHP group. The distribution of (B) hypomethylated sites and (C) hypermethylated sites in the gene body, 5’UTR, intergenic region, and promoters is shown in the pie chart.
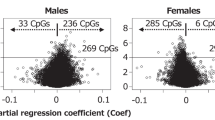
We also analyzed the correlation between concentration of DEHP metabolites and methylation level of those differential methylated sites of the patients, showing a significant correlation (positive for hypermethylation and negative for hypomethylation) in most of the methylated sites, using linear (Fig. S1) or nonlinear regression (Fig. S2). These results further suggested that DEHP can induce a methylation changes in human placenta in a dose-dependent manner.
Protein‑protein interaction (PPI) analysis
A total of 598 genes, including 117 hypermethylated and 312 hypomethylated genes, were subjected to Cytoscape analysis of protein–protein interactions (PPIs, Fig. 4A–D, and Supplementary Table S1). Note that most of the proteins associated with DEHP-induced hypermethylation were involved in the transcriptional regulation of Wnt signaling (Fig. 4A,B), while most of the proteins associated with DNA hypomethylation were linked to adhesion molecules and neural networks (Fig. 4C,D).
Cytoscape and STRING pathway analysis for genes with differential methylated CpG sites of human placenta exposed to low and high levels of DEHP. Pathways that are enriched with (A) hypermethylated genes and (C) hypomethylated genes are shown. Cytoscape analysis of protein–protein interaction (PPI) found that (B) hypermethylated genes are related to the Wnt signaling pathway, while (D) hypomethylated genes are related to neural-related adhesion molecules. The colored box was enlarged in the right panel. These figures on protein–protein interaction (B,D) were generated by Cytoscape version 3.9.1 (https://cytoscape.org/index.html)36.
Changes in methylation in the imprinted region
Shortly after fertilization (i.e. during preimplantation embryo development), the maternal genome undergoes passive demethylation, after which global remethylation corrects the embryonic methylation pattern. The differential methylation of 10 imprinted genes under exposure to DEHP was correlated with ECG abnormalities, autism, and alcohol dependence (Fig. 5, Table 4).
Discussion
The primary modulator of the intrauterine environment is the placenta, the function of which can be shaped by exposure to environmental contaminants, resulting in epigenetic alterations linked directly to fetal abnormalities7,8,9. Numerous studies have reported strong correlations between alterations in DNA methylation in the human placenta and abnormalities in the growth of the fetus10,11,12. One previous study reported that exposure to phthalate could have adverse effects on epigenetic outcomes in the human placenta13. Ravaei et al. recently reported that placental DNA methylation profiles can serve as a biomarker to predict the development of autism spectrum disorder (ASD) in fetuses14. Our current results confirmed that DEHP can indeed affect fetal development via aberrant DNA methylation.
Our previous study provided a convincing epigenetic explanation supporting a causal link between maternal exposure to DEHP and the transgenerational risk of allergic lung inflammation in offspring4. In the current study, we gauged the extent of the epigenetic effects of maternal DEHP exposure by examining global DNA methylation in 12 pregnant women exposed to DEHP. Our results were consistent with the findings in previous studies4,15,16, indicating that exposure to DEHP induced DNA hypomethylation in the placenta in a dose-dependent manner. Although the methylation levels of the CG sites seem to clusters at 0, 0.5 and 1 (Fig. 3), this phenomenon should not be caused by genetic effect, as only four probes (out of 12) were found to be overlapped with known SNP (Tables 2 and 3). On the contrary, correlation analysis using both linear and nonlinear regression demonstrated a significant correlation between concentration of DEHP metabolites and methylation level of the probes in most of the patients (Figs. S1 and S2), further indicating that DEHP affects DNA methylation in human placenta.
It is also important to note that most of the differential changes in methylation were observed in the gene body, promoter, and intergenic region. Note also that the genes associated with DEHP-induced hypermethylation were involved in transcriptional activity, particularly Wnt signaling, whereas the genes associated with DEHP-induced hypomethylation were linked to adhesion molecules and neural networks.
Wnt signaling plays an important role in cell proliferation and cell fate determination. Several developmental diseases in humans have been linked to alterations in Wnt signaling induced by environmental disruptors (e.g. phthalate)17. Zhang et al. reported that exposing pregnant rats to phthalate led to the down-regulation of Wnt/β-catenin signaling in fetal genital tubercles (GTs), indicating that phthalate may affect GT development18. Note that early exposure to phthalate has been linked to the development of neurological conditions, such as autism and dementia19,20,21,22. Those conditions have also been linked to the downregulation of Wnt signaling23,24,25; however, researchers have yet to elucidate the mechanism by which Wnt signaling is suppressed. Our findings in the current study suggest that Wnt signaling in the placenta is down-regulated by DEHP-induced hypermethylation. It also appears that these effects increase the susceptibility of the fetus to the development of these conditions.
Adhesion molecules associated with the cytoskeleton (e.g., cadherin) have also been shown to play an important role in maintaining the structural stability of neurons and preserving brain function26. Researchers have linked the dysregulation of adhesion molecules to the development of autism27,28 and dementia29,30. Maternal exposure to phthalate has been shown to upregulate cadherin levels in rodent models31,32. Our results in the current study suggest that the hypomethylation of adhesion molecules and cadherin following maternal exposure to DEHP may predispose the fetus to a wide spectrum of neurological disorders. These findings are consistent with a recent animal study in which juvenile exposure to phthalate was shown to exacerbate autism-like behavior via DNA hypomethylation16.
This study was subject to several limitations, which should be considered in the interpretation of the results. First, the sample size was small. Our findings will require confirmation using a large study cohort. Considerable effort was exercised in the selection of study subjects and age-matched controls to minimize the aging effect on DNA methylation. In addition, we excluded subjects with a history of cigarette smoking or environmental exposure to tobacco. Nonetheless, other lifestyle and environmental factors (e.g. diet and air pollutants) could not be controlled. These factors must be considered potential confounders biasing our results in either direction (i.e. under- or over-estimating the findings).
In conclusion, this study revealed a possible relationship between exposure to DEHP and global DNA methylation in the human placenta. This proof-of-concept study suggests that maternal exposure to DEHP predisposes the fetus to neurological disorders via epigenetic alteration. Future research with a larger sample size will be required to identify the mechanisms underlying the link between these effects and fetal abnormalities.
Methods
Human birth cohort and study design
This study was a part of the Taiwan Maternal and Infant Cohort Study (TMICS), a nationwide prospective birth cohort established by epidemiologists between October 2012 and May 2015. This study recruited pregnant women who visited one of nine hospitals (three in northern, three in central, two in southern, and one in eastern Taiwan) for routine pre-birth examinations during their third trimester (weeks 29 to 40). After providing written consent, the subjects were interviewed by nursing staff using a standardized questionnaire. Non-invasive biological specimens (blood, urine, and hair) were collected at the same time. Following the birth of the babies, samples of cord blood and placental tissue were aliquoted and stored at − 70 °C for future research. Details pertaining to the study design can be found in previous articles33,34.
In the current study, 146 potential subjects were recruited from E-Da Hospital. Note that phthalate metabolite data were available for all candidate subjects, based on one-spot urine samples collected during the third trimester. We excluded all individuals with a history of cigarette smoking and/or environmental exposure to tobacco during pregnancy. The sum of five DEHP metabolites (corrected via urinary creatinine) was ranked from highest to lowest and divided into four quartiles. The five DEHP metabolites included the following: MEHP (mono-(2-ethylhexyl) phthalate), MEOHP (mono-(2-ethyl-5-hydroxylhexyl) phthalate), MEHHP (mono-(2-ethyl-5-oxohexyl) phthalate), MECPP (mono(2-ethyl-5-carboxypentyl)phthalate), and MCMHP (mono(2-carboxymethylhexyl)phthalate). Note that all analysis was performed at an internationally certified laboratory (G-EQUAS 59) under the auspices of the National Health Research Institute (NHRI), as previously described33. We selected six subjects from the highest quartile and six subjects from the lowest quartile age-matched within two years during the third trimester of pregnancy. Table 1 lists the age distribution (26–41 years). Note that the mean sum of the 5 DEHP metabolites (μg/g creatinine) in the DEHP-high group (1694.08) was significantly higher than in the DEHP exposure group (59.92, P < 0.01, Table 1). This study was further approved by the IRB of E-Da Hospital, Taiwan (EMRP41101N (RI), EMRP31102N). Written informed consent was obtained from all study subjects, in accordance with the Declaration of Helsinki. All methods were performed in accordance with the relevant guidelines and regulations.
DNA extraction
DNA was extracted using a Genomic DNA Mini Kit (Geneaid, Taiwan) in accordance with the manufacturer's instructions. DNA was then eluted in 50 µl of distilled water and stored at 0 °C until use.
MethylationEPIC Beadchip analysis
Genome-wide methylation analysis was performed using the high-resolution Infinium MethylationEPIC BeadChip Kit, quantitatively interrogating roughly 850,000 methylation sites across the genome at single-nucleotide resolution. The GenomeStudio Methylation Module was used to facilitate the analysis of MethylationEPIC data. GenomeStudio Software was used to reveal valuable information, such as chromosomal coordinates, percentage of GC, location of the CpG island, and methylation \(\beta \)-values. Individual probe \(\beta \)-values (range 0–1) are approximate representations of the absolute methylation percentage of specific CpG sites within the sample population. Beta (\(\beta \)) = 1 indicates 100% methylation, whereas \(\beta \)=0 indicates 0% methylation. The values were derived by comparing the ratio of intensities between the methylated and unmethylated alleles, using the following formula35:
where signal M and signal U respectively indicate the array intensity values for the methylated and non-methylated alleles. Samples were processed using the Bioconductor package designed explicitly for Illumina data. The DNA methylation data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE153475. To facilitate comparisons among DEHP groups, we focused exclusively on probes with a mean change in methylation level of 10% or more (0.1 in β-value). P-values of < 0.05 or FDR values of < 0.1 were deemed indicative of significant changes.
Statistical analysis
All statistical analysis was performed using GraphPad Prism version 8.0 for Windows (GraphPad Software, La Jolla, CA, USA) or R statistical software (version 4.2.1; R Foundation for Statistical Computing, Vienna, Austria). The ggplot2 package was used to perform Student’s T test and FDR for comparison of methylation differences. A P-value of < 0.05 was considered significant. Protein–protein interaction networks were integrated with DNA methylation differential data using Cytoscape version 3.9.1 in conjunction with the STRING App36.
Ethics
This study received approval from the IRB of E-Da Hospital, Taiwan (EMRP41101N (RI), EMRP31102N). Written informed consent was obtained from all study subjects in accordance with the Declaration of Helsinki. All operations were performed in accordance with the relevant guidelines and regulations.
Data availability
DNA methylation microarray data have been deposited in the Gene Expression Omnibus (GEO) database, under accession number GSE153475.
References
Maccani, M. A. & Marsit, C. J. Epigenetics in the placenta. Am. J. Reprod. Immunol. 62, 78–89. https://doi.org/10.1111/j.1600-0897.2009.00716.x (2009).
Choumenkovitch, S. F. et al. Folic acid intake from fortification in United States exceeds predictions. J. Nutr. 132, 2792–2798. https://doi.org/10.1093/jn/132.9.2792%JTheJournalofNutrition (2002).
Schjenken, J. E. et al. Endocrine disruptor compounds-a cause of impaired immune tolerance driving inflammatory disorders of pregnancy?. Front. Endocrinol. (Lausanne) 12, 607539. https://doi.org/10.3389/fendo.2021.607539 (2021).
Suen, J. L. et al. Environmental factor-mediated transgenerational inheritance of Igf2r hypomethylation and pulmonary allergic response via targeting dendritic cells. Front. Immunol. 11, 603831. https://doi.org/10.3389/fimmu.2020.603831 (2020).
Latini, G., Verrotti, A. & De Felice, C. DI-2-ethylhexyl phthalate and endocrine disruption: A review. Curr. Drug Targets Immune Endocr. Metabol. Disord. 4, 37–40. https://doi.org/10.2174/1568008043340017 (2004).
Wang, I. J., Karmaus, W. J., Chen, S. L., Holloway, J. W. & Ewart, S. Effects of phthalate exposure on asthma may be mediated through alterations in DNA methylation. Clin. Epigenetics 7, 27. https://doi.org/10.1186/s13148-015-0060-x (2015).
Tekola-Ayele, F. et al. DNA methylation loci in placenta associated with birthweight and expression of genes relevant for early development and adult diseases. Clin. Epigenetics 12, 78. https://doi.org/10.1186/s13148-020-00873-x (2020).
Nugent, B. M. & Bale, T. L. The omniscient placenta: Metabolic and epigenetic regulation of fetal programming. Front. Neuroendocrinol. 39, 28–37. https://doi.org/10.1016/j.yfrne.2015.09.001 (2015).
Chang, R. C. et al. DNA methylation-independent growth restriction and altered developmental programming in a mouse model of preconception male alcohol exposure. Epigenetics 12, 841–853. https://doi.org/10.1080/15592294.2017.1363952 (2017).
Liu, J. et al. Placental DNA methylation abnormalities in prenatal conotruncal heart defects. Front. Genet. 13, 878063. https://doi.org/10.3389/fgene.2022.878063 (2022).
Zhou, Q., Xiong, Y., Qu, B., Bao, A. & Zhang, Y. DNA methylation and recurrent pregnancy loss: A mysterious compass?. Front. Immunol. 12, 738962. https://doi.org/10.3389/fimmu.2021.738962 (2021).
Bahado-Singh, R. O., Vishweswaraiah, S., Aydas, B. & Radhakrishna, U. Placental DNA methylation changes and the early prediction of autism in full-term newborns. PLoS ONE 16, e0253340. https://doi.org/10.1371/journal.pone.0253340 (2021).
Strakovsky, R. S. & Schantz, S. L. Impacts of bisphenol A (BPA) and phthalate exposures on epigenetic outcomes in the human placenta. Environ Epigenet 4, dvy022. https://doi.org/10.1093/eep/dvy022 (2018).
Ravaei, A., Emanuele, M., Nazzaro, G., Fadiga, L. & Rubini, M. Placental DNA methylation profile as predicting marker for autism spectrum disorder (ASD). Mol. Med. 29, 8. https://doi.org/10.1186/s10020-022-00593-3 (2023).
Xia, B. et al. In utero and lactational exposure of DEHP increases the susceptibility of prostate carcinogenesis in male offspring through PSCA hypomethylation. Toxicol. Lett. 292, 78–84. https://doi.org/10.1016/j.toxlet.2018.04.022 (2018).
Nadeem, A. et al. Exposure to the plasticizer, Di-(2-ethylhexyl) phthalate during juvenile period exacerbates autism-like behavior in adult BTBR T + tf/J mice due to DNA hypomethylation and enhanced inflammation in brain and systemic immune cells. Prog. Neuropsychopharmacol. Biol. Psychiatry 109, 110249. https://doi.org/10.1016/j.pnpbp.2021.110249 (2021).
Üstündağ, Ü. V. & Emekli-Alturfan, E. Wnt pathway: A mechanism worth considering in endocrine disrupting chemical action. Toxicol. Ind. Health 36, 41–53. https://doi.org/10.1177/0748233719898989 (2020).
Zhang, L. F. et al. Differential expression of the Wnt/β-catenin pathway in the genital tubercle (GT) of fetal male rat following maternal exposure to di-n-butyl phthalate (DBP). Syst. Biol. Reprod. Med. 57, 244–250. https://doi.org/10.3109/19396368.2011.577509 (2011).
Agin, A. et al. Environmental exposure to phthalates and dementia with Lewy bodies: Contribution of metabolomics. J. Neurol. Neurosurg. Psychiatry 91, 968–974. https://doi.org/10.1136/jnnp-2020-322815 (2020).
Yen, P. L., How, C. M. & Hsiu-Chuan Liao, V. Early-life and chronic exposure to di(2-ethylhexyl) phthalate enhances amyloid-β toxicity associated with an autophagy-related gene in Caenorhabditis elegans Alzheimer’s disease models. Chemosphere 273, 128594. https://doi.org/10.1016/j.chemosphere.2020.128594 (2021).
Mathew, L. et al. The associations between prenatal phthalate exposure measured in child meconium and cognitive functioning of 12-month-old children in two cohorts at elevated risk for adverse neurodevelopment. Environ. Res. 214, 113928. https://doi.org/10.1016/j.envres.2022.113928 (2022).
Kim, J. I. et al. Association of phthalate exposure with autistic traits in children. Environ. Int. 157, 106775. https://doi.org/10.1016/j.envint.2021.106775 (2021).
Zhang, Y. et al. Downregulating the canonical Wnt/β-catenin signaling pathway attenuates the susceptibility to autism-like phenotypes by decreasing oxidative stress. Neurochem. Res. 37, 1409–1419. https://doi.org/10.1007/s11064-012-0724-2 (2012).
Lei, J., Deng, Y. & Ma, S. Downregulation of TGIF2 is possibly correlated with neuronal apoptosis and autism-like symptoms in mice. Brain Behav. 12, e2610. https://doi.org/10.1002/brb3.2610 (2022).
Palomer, E. et al. Epigenetic repression of Wnt receptors in AD: A role for Sirtuin2-induced H4K16ac deacetylation of Frizzled1 and Frizzled7 promoters. Mol. Psychiatry 27, 3024–3033. https://doi.org/10.1038/s41380-022-01492-z (2022).
Moreland, T. & Poulain, F. E. To stick or not to stick: The multiple roles of cell adhesion molecules in neural circuit assembly. Front. Neurosci. 16, 889155. https://doi.org/10.3389/fnins.2022.889155 (2022).
Gecz, J. & Thomas, P. Q. Disentangling the paradox of the PCDH19 clustering epilepsy, a disorder of cellular mosaics. Curr. Opin. Genet. Dev. 65, 169–175. https://doi.org/10.1016/j.gde.2020.06.012 (2020).
Taylor, S. C. et al. The role of synaptic cell adhesion molecules and associated scaffolding proteins in social affiliative behaviors. Biol. Psychiatry 88, 442–451. https://doi.org/10.1016/j.biopsych.2020.02.012 (2020).
Choi, J. Y. et al. Elevated cerebrospinal fluid and plasma N-cadherin in Alzheimer disease. J. Neuropathol. Exp. Neurol. 79, 484–492. https://doi.org/10.1093/jnen/nlaa019 (2020).
Vezzoli, E. et al. Inhibiting pathologically active ADAM10 rescues synaptic and cognitive decline in Huntington’s disease. J. Clin. Invest. 129, 2390–2403. https://doi.org/10.1172/jci120616 (2019).
Wei, Z. et al. Maternal exposure to di-(2-ethylhexyl)phthalate alters kidney development through the renin-angiotensin system in offspring. Toxicol. Lett. 212, 212–221. https://doi.org/10.1016/j.toxlet.2012.05.023 (2012).
Zhu, Y. P. et al. Di-n-butyl phthalate (DBP) reduces epithelial-mesenchymal transition via IP3R in hypospadias during maternal exposure. Ecotoxicol. Environ. Saf. 192, 110201. https://doi.org/10.1016/j.ecoenv.2020.110201 (2020).
Wu, C. F. et al. Cohort profile: The Taiwan Maternal and Infant Cohort Study (TMICS) of phthalate exposure and health risk assessment. Int. J. Epidemiol. 47, 1047–1047j. https://doi.org/10.1093/ije/dyy067 (2018).
Tsai, T. L. et al. Association between prenatal exposure to metals and atopic dermatitis among children aged 4 years in Taiwan. JAMA Netw. Open 4, e2131327. https://doi.org/10.1001/jamanetworkopen.2021.31327 (2021).
Du, P. et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 11, 587. https://doi.org/10.1186/1471-2105-11-587 (2010).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. https://doi.org/10.1101/gr.1239303 (2003).
Funding
This study was supported by grants from the National Science and Technology Council, Taiwan (MOST111-2314-B-037-004- and 110-2314-B-037-047-MY3 to MTW; MOST108-2314-B-194-001, 107-2314-B-194-001, and 106-2314-B-194-002 to MWYC), Taiwan's National Health Research Institutes (NHRI-EX112-11202PI), and the Integrative Project of National Health Research Institutes and Kaohsiung Medical University (NHRIKMU-111-I005). This study was also supported by the Research Center for Precision Environmental Medicine at Kaohsiung Medical University and the Center for Innovative Research on Aging Society (CIRAS) at National Chung Cheng University under the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Author information
Authors and Affiliations
Contributions
M.T.T., W.-L.H., C.-F.W. and S.-S.L. performed experiments. J.-T.L., Y.-M.C., S.S., Z.-Y.L. and G.-L.L. performed analysis. F.-C.K., C.-H.H. and M.-T.W. collected patient samples. M.T.T., S.S., G.-L.L. and M.W.Y.C. wrote the manuscript. M.-T.W., S.J.L., C.-H.H. and M.W.Y.C. initiated and supervised the study. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tran, M.T.M.T., Kuo, FC., Low, JT. et al. Prenatal DEHP exposure predicts neurological disorders via transgenerational epigenetics. Sci Rep 13, 7399 (2023). https://doi.org/10.1038/s41598-023-34661-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34661-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.