Abstract
Multisite collection and preservation of peripheral blood mononuclear cells (PBMCs) for centralized analysis is an indispensable strategy for large cohort immune phenotyping studies. However, the absence of cross-site standardized protocols introduces unnecessary sample variance. Here we describe the protocol implemented by the Province of Ontario Neurodevelopmental Disorders (POND) Network's immune platform for the multisite collection, processing, and cryopreservation of PBMCs. We outline quality control standards and evaluate the performance of our PBMC processing and storage protocol. We also describe the Child Immune History Questionnaire results, an assessment tool evaluating pre-existing immune conditions in children with neurodevelopmental disorders (NDDs). Cell viability was assessed in samples from 178 participants based on strict quality control criteria. Overall, 83.1% of samples passed quality control standards. Samples collected and processed at the same site had higher quality control pass rates than samples that were collected and subsequently shipped to another site for processing. We investigated if freezer time impacted sample viability and found no difference in mean freezer time between samples that passed and failed quality control. The Child Immune History Questionnaire had a response rate of 87.1%. The described protocol produces viable samples that may be used in future immune phenotyping experiments.
Similar content being viewed by others
Introduction
Neurodevelopmental disorders (NDDs) are a diverse group of conditions defined by communication, behavioural, and cognitive deficits that result from atypical brain development1,2,3. NDDs include but are not limited to Autism Spectrum Disorder (ASD), Attention-Deficit/Hyperactivity Disorder (ADHD), Obsessive–Compulsive Disorder (OCD), Intellectual Disability (ID), Rett syndrome, Down syndrome, and other genetic disorders that influence neurodevelopment. The pathophysiology of NDDs is multifactorial, with studies reporting multiple genetic and environmental risk factors4,5,6,7,8. Furthermore, the clinical presentation of NDDs is heterogeneous, with shared symptoms across disorders and symptom severity varying between and within disorders1,2,3. Understanding the biological basis of this heterogeneity may aid in extending the current diagnostic criteria and provide novel biomarker-based approaches to guiding treatment and research.
There is evidence that the immune response contributes to the development and progression of NDDs. Increased levels of pro-inflammatory markers in the brain and periphery9,10,11, as well as differences in surface receptors, function, and counts of monocytes12,13,14,15 and lymphocytes16,17,18,19, have been reported in NDDs compared to typically developing (TD) individuals. Hyperactivity and deficits in social interactions and communication correlate with higher levels of inflammation in people with NDDs20,21,22,23, suggesting that inflammation and dysregulated immune responses may contribute to the heterogeneity of symptoms seen in NDDs. Stratifying individuals based on immune phenotype and identifying biomarkers specific to behavioural outcomes may be an effective option to precision health approaches for NDDs.
Studies on the immune system’s role in the etiology or symptomology of NDDs have been limited due to small sample sizes, unstandardized approaches to handling biospecimens, and inconsistencies in the types of cells and cellular markers analyzed. The immune research platform of the Province of Ontario Neurodevelopmental Disorder (POND) Network aims to conduct comprehensive immunophenotyping in a large cohort of people with different NDDs from multiple sites across the province. Flow cytometry is the tool of choice to analyze several cellular parameters; however, conducting multisite immunophenotyping is challenging. Studies have demonstrated that multiple factors, including equipment types, calibration, gating, and analysis, contribute to an increase in data variation when using flow cytometry across multiple sites24,25,26,27. Cryopreservation is commonly used to allow for centralized investigation of peripheral blood mononuclear cells (PBMCs), ameliorating the confounding factors present in multisite analyses24,25,26. Studies of long-term cryopreservation have shown that cell viability decreases by less than 1% every year that they are frozen, indicating minimal cell loss over time28. However, it has also been shown that cryopreservation may alter the counts of monocyte and lymphocyte populations and affect the expression of several surface receptors compared to fresh samples29,30,31. The differences in cell counts and surface receptor expression appear consistent across the literature, allowing for their consideration during experimental design29,30,31. These studies demonstrate the feasibility of using cryopreserved PBMCs to facilitate a centralized analysis approach for samples collected at multiple sites. This paper presents a standardized approach used to collect and bank biospecimens for immunophenotyping and assesses the viability of cryopreservation for centralized analysis in a multisite study of TD individuals and individuals with NDDs.
Results
Banked biospecimens from 178 participants were analyzed. The participants' demographics are provided in Table 1. 82.6% of the participants had NDD (n = 147), with ASD making up 71.4% of the NDD diagnoses (n = 105). 81.5% of the participants were male, and 18.5% were female, which reflects the sex differences in NDD prevalence. The Child Immune History Questionnaire was available from 87.1% of participants (n = 155). Response and completion rates by diagnosis and questionnaire section can be found in Table 1. The "Infection" section of the questionnaire had a significantly lower response rate in the NDD group than in the TD group (p < 0.01). The proportion of participants who reported having an autoimmune disease or allergy did not differ between TD and NDD (p = 0.15). There was a higher proportion of people who reported having a “significant” infection in the NDD group compared to TD (p = 0.02). There was also a significantly higher proportion of individuals in the NDD group who reported a history of immunosuppressive medication use (p < 0.01).
Viability of banked PBMCs
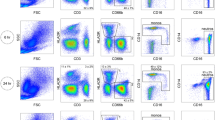
The viability of thawed PBMCs was determined using two different viability assessment methods. The Trypan Blue exclusion test was conducted on 178 PBMC samples, with 93.8% (n = 167) passing. These samples proceeded to a viability assessment using flow cytometry. A frequency distribution of Trypan Blue counts can be found in Fig. 1a. Of the 167 PBMC samples that proceeded to flow cytometry analysis, 88.7% (n = 148) passed the flow cytometry quality control criteria. A frequency distribution of the % alive CD45+ cells can be found in Fig. 1b. The overall quality control pass rate of cryopreserved PBMCs was 83.1% (Table 2).
Quality control data. (A) Frequency distribution of Trypan Blue counts in thawed samples. The percent frequency of post-thaw sample PBMC counts measured using the Trypan Blue exclusion test. Samples containing more than 500,000 cells, indicated by the dotted line, proceeded to quality control phase two. (B) Frequency distribution of alive CD45+ Cells in thawed samples. The percent frequency of CD45+ cells that were negative for the Zombie Red viability stain as a percent of CD45+ cells in the sample. Samples with a survival rate of 70%, indicated by the dotted line, were considered viable for future use.
The viability of banked PBMCs by site is shown in Table 3. Early in sample collection, the transportation method from Site 1 to the processing site was changed from samples being transported on ice, to being transported at room temperature. This change led to an 32.1% increase in sample viability. Fisher’s exact test showed that samples processed on site had a significantly higher proportion of quality control passes than sites where samples were transported prior to processing (p < 0.01).
The mean time (± SE) spent in the freezer before the viability assessment was 286.1 ± 10.6 days. Samples that passed quality control spent 276.0 ± 11.1 days frozen, whereas samples that failed quality control spent 321.0 ± 27.3 days frozen. The difference in the number of days and months spent in the freezer between samples that passed or failed quality control was not significant (p = 0.11).Complete blood count (CBC) reports detailing absolute counts of lymphocytes, monocytes, neutrophils, eosinophils, and basophils were available for 62.9% of participants (n = 112). Of the participants with CBC reports, 89 passed and 23 failed quality control criteria for the Trypan Blue counts and % alive CD45+ cells. The profile of leukocytes available on the CBC reports were examined and showed that there were no differences in absolute counts of leukocytes (prior to banking) between the samples that passed and failed quality control (Supplementary Fig. 1).
Discussion
This report provides an initial assessment of the viability and quality of the biospecimen collection protocol for the POND immune research platform and an analysis of participant immune-related medical history. The biospecimen collection protocol was designed to address the challenges associated with the multisite collection of PBMCs. Additionally, centralized storage and analysis are more cost-effective as less equipment and personnel are required at the collection sites. The Child Immune History Questionnaire was implemented to collect data on conditions that may influence immune status, allowing for their consideration when conducting immune phenotyping.
Other studies have examined the effects of long-term cryopreservation on PBMCs28,29. A study assessing the viability of PBMCs cryopreserved from multiple sites for up to 12 years found that the median viability was 90%, with a minimum viability of 82%28. Additionally, a study assessing cryopreservation of immune cells in whole blood found viability of 58 ± 3% after four years; however, most of the loss was attributed to neutrophils29. The authors reported a monocyte viability of 68 ± 8% and a lymphocyte viability of > 90%29. It is important to note that in our described protocol granulocytes are isolated and stored separately from other PBMC populations. The ranges provided by both studies are comparable to the overall PBMC viability of 83.1% in the present study, determined by the described quality control standards.
Site-wise comparisons indicate that immediate processing provides better sample viability than transporting samples from a collection site to a processing site. Additionally, transportation conditions appear to be a significant factor in sample viability. Changing the ttransportation protocol from on ice to room temperature greatly increased sample viability of site 1.
To date, attempts to characterize a comprehensive immune phenotype of those with NDDs are limited16,18. These studies used flow cytometry to analyze PBMC populations in children with ASD and TD controls, with one study using fresh blood samples and the other using cryopreserved PBMCs16,18. While both studies found immune cell population differences between ASD and TD, their findings are limited to young children between 5 and 7 years old16,18. Expanding the age ranges is important. The POND immunophenotyping platform recruits individuals between 1 and 23 years old with various NDDs.
Sample processing and cryopreservation present two important limitations. The first limitation is due to the processing involved in sample preparation. As the volume of blood collected varies and cells are lost with each wash cycle, absolute counts of cell populations are not comparable to counts obtained using fresh blood samples. Therefore, PBMC populations were reported as %CD45+ cells or standardized to cells per million. Additionally, it has been reported that cryopreserved PBMCs yield different cell counts of monocyte and lymphocyte populations when compared to fresh blood28,29. However, these variations in monocyte and lymphocyte counts appear to be consistent across the literature and are therefore an inherent limitation of cryopreservation which needs to be considered during study design29,30. The collection of CBC reports in conjunction with PBMCs will be used in future studies to further assess potential differences between cryopreserved counts and fresh blood counts. The second identified limitation is that cryopreservation has been shown to reduce the mean fluorescent intensity of specific cell markers on monocytes, lymphocytes, and granulocytes, making less abundant markers more difficult to detect29. Furthermore, cryopreservation reduces the reliability of surface markers of activation CD45RA and CD62L despite the cells remaining viable32. The surface markers with reduced reliability appear to be consistent across the literature31,32. If unreliable surface markers are of interest, flow cytometry should be conducted on fresh blood samples instead of on cryopreserved samples. This limits the ability to conduct a multisite analysis of these markers, and therefore should be addressed during study design. In summary, the POND Network immune platform has designed and implemented a protocol for multisite sample collection and single site storage and provided an assessment of the viability and quality of samples procured. The protocol allows for the collection of biospecimens from individuals with a variety of NDDs across the Province of Ontario and provides a framework for reliable immunophenotyping. This study will allow for an extensive analysis of immunophenotyping in NDDs and provide insight into immune biomarkers linked to the clinical presentation of NDDs.
Methods
Participants
Participants were recruited through the POND Network (pond-network.ca) at 5 sites: Holland Bloorview Kids Rehabilitation Hospital, Toronto; The Hospital for Sick Children, Toronto; Queens University, Kingston; Lawson Health Research Institute, London; and McMaster Children's Hospital, Hamilton. Primary caregivers and participants provided either written informed consent or assent after a complete description of the study was provided in accordance with the Tri Council Policy Statement, the Declaration of Helsinki, and the International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use (ICH)–Good Clinical Practice (GCP) guidelines and institutional policy. Approval for collection and sample banking were obtained from the research ethics boards of each institution including Holland Bloorview Research Ethics Board, Sick Kids Research Ethics Board, General Research Ethics Board at Queen’s University, Western University’s Health Sciences Ethics Board, and Hamilton Integrated Research Ethics Board.
A sub-cohort of children and young adults with NDDs and TD participants that contributed immune samples between January 2017 and July 2019 were enrolled. Eligible participants were between the ages of 1 and 23 years with a diagnosis including ASD, ADHD, OCD, and ID, in addition to TD controls. Diagnoses were confirmed using standardized assessment tools. ASD was confirmed using the Autism Diagnostic Observation Schedule (ADOS) and the Autism Diagnostic Interview-Revised (ADI-R), ADHD was confirmed using the Parent Interview for Child Symptoms (PICS), and OCD was confirmed using the Children's Yale-Brown Obsessive–Compulsive Scale (CY-BOCS)33,34,35,36. Exclusion criteria for the immune platform were as follows: fever greater than 38 °C in the preceding 3 days, vaccination in the preceding 7 days, oral or intravenous steroids in the preceding 14 days, oral or intravenous antibiotics in the preceding 21 days, chemotherapy, intravenous immunoglobulins of monoclonal antibodies in the preceding 12 months and known primary or secondary immunodeficiency.
Immune metrics
The immune medical history of participants was assessed using the Child Immune History Questionnaire (Supplemental File 1) to collect information about the participants' immune status. The questions were grouped into three sections for analysis: autoimmune/allergy, childhood infection, and medication. Autoimmune/allergy-related questions ascertained if the participant had environmental, food, or other allergies, asthma, autoimmune cytopenia, or other autoimmune diseases. The childhood infection questions collected information about the participant's history of infections, including oral thrush, ear infections, skin warts, chickenpox, or other self-described “significant” infections that required antibiotic use. Lastly, the medication section questions determined if the participant had previously used immunosuppressants or oral/IV steroids. Sections were considered incomplete if at least one question was left blank. Participants with incomplete responses were removed using case-wise deletion.
Biospecimen collection
Venous blood was drawn from participants: (1) in a 10 mL heparinized blood tube for the isolation of PBMCs, granulocytes and plasma (367,874, Becton, Dickinson and Company, Mississauga, ON); (2) in two 5 mL silica-coated serum tubes for the isolation of serum (366,434 G, Becton, Dickinson and Company, Mississauga, ON); (3) 4 mL EDTA treated tube for complete blood counts (367,844, Becton, Dickinson and Company, Mississauga, ON). Anonymized samples (1 and 2) were transported from the blood collection site to the nearest processing lab, where biospecimens were cryopreserved. The EDTA samples (3) were transported to 3rd party medical labs for CBC analysis. The date and time of the blood draw were recorded. Biospecimen samples were processed with minimal delay after collection. Samples collected in Hamilton and London areas were immediately transported to the processing lab at McMaster University. Samples collected in Toronto were immediately transported to the processing lab at The Hospital for Sick Children. Samples collected in Kingston were immediately processed in the lab on site.
PBMC and granulocyte isolation
PBMCs and granulocytes were isolated using Ficoll Paque Plus (17-1440-03, GE Healthcare, Mississauga, ON). Ficoll Paque Plus was pipetted into the bottom of the processing tube, below the blood, according to the manufacturer's protocol with minor alterations. Blood layers were separated by centrifugation at 514 RCF for 25 min with the brake off. PBMCs were collected from the buffy coat interphase and washed with phosphate buffered saline (PBS) (see Supplementary Protocol 1). Following PBMC isolation, RBCs and granulocytes were isolated from the Ficoll Paque separation. Red blood cells were lysed, and pelleted granulocytes were washed in PBS. Concentrations of PBMCs and granulocytes were determined using a hemocytometer. Cells were resuspended in 10% dimethyl sulfoxide (DMSO) (Caledon, 4100-1-05) human AB (hAB) (VWR, CA45001-062) serum freezing media and frozen with 1 mL aliquots of 5–10 × 106 cells/mL per vial. PBMCs and granulocytes were frozen at − 80 °C using a cell freezer for storage37.
Biospecimen banking
Samples were stored locally and shipped to the POND McMaster Biobank (Hamilton, ON). Documentation of the biospecimen collection and aliquots was submitted electronically and in paper format with each shipment. Samples were stored in the POND McMaster Biobank at St. Joseph's Healthcare in Hamilton, ON. Biobank sample IDs were assigned to individual blood sample aliquots, and an up-to-date inventory of samples is maintained. Upon research proposal approval from platform leads, POND associated labs and collaborators request samples to be sent for research purposes.
Flow cytometry and PBMC viability measures
A two-step sample viability check was used for quality control. Single aliquots of PBMCs were thawed in warm RPMI 1640 medium (SLM-240-B, Sigma-Aldrich, Oakville, ON). In step one, live cells were counted on a hemocytometer using the Trypan Blue exclusion test38. Samples with less than or equal to 5 × 105 cells measured using the Trypan Blue exclusion test were considered to fail quality control. Samples that failed the Trypan Blue exclusion test were not used in subsequent flow cytometry. Samples that passed the trypan blue exclusion test criteria proceeded to step two where they were washed with fluorescence-activated cell sorting (FACS) wash (0.5% (w/v) BSA, 5 mM EDTA (pH 7.4–7.6), for 500 mL 2.5 g BSA, 5 mL of 0.5 M EDTA) prior to staining. 5 × 105 cells were stained with Zombie Red Fixable Viability Kit (423,109, Biolegend, Cedarlane, Burlington, ON) and fluorochrome-conjugated antibodies against CD45 (BV510), each diluted 1:100 in a final volume of 50 µL of FACS wash, for 30 min at room temperature. Cells were then fixed using 1-step Fix/Lyse Solution (1X) for 10 min. PBMCs were washed with FACS wash and filtered through 0.45 µm mesh to ensure single-cell suspension before analysis on an LSR II flow cytometer (Becton, Dickinson and Company, Mississauga, ON). Cells stained Zombie Red positive were considered dead cells, while Zombie Red negative cells were considered viable. Samples with a Zombie Red viability of 70% or less were considered to fail quality control.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 8.4.3. Analyses were conducted to assess differences in responses to the Child Immune Questionnaire, and differences between samples that passed and failed quality control. Fisher's Exact Tests were used to compare responses to the Child Immune History Questionnaire between TD and NDD, and to compare quality control pass rate by site. The Shapiro–Wilk Goodness-of-fit test was applied to continuous variables to assess normality. If the data were normally distributed, then the parametric t-test or one-way ANOVA was used. If the data was not normally distributed, then the non-parametric Mann–Whitney U test was used. All tests were 2-tailed, and p < 0.05 was considered significant.
Data availability
All research data is available upon request from the corresponding author.
References
Mullin, A. P. et al. Neurodevelopmental disorders: Mechanisms and boundary definitions from genomes, interactomes and proteomes. Transl. Psychiatry 3, e329. https://doi.org/10.1038/tp.2013.108 (2013).
Colvert, E. et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry 72, 415–423. https://doi.org/10.1001/jamapsychiatry.2014.3028 (2015).
Grzadzinski, R., Huerta, M. & Lord, C. DSM-5 and autism spectrum disorders (ASDs): an opportunity for identifying ASD subtypes. Mol. Autism 4, 1–6. https://doi.org/10.1186/2040-2392-4-12 (2013).
Trost, B. et al. Genome-wide detection of tandem DNA repeats that are expanded in autism. Nature 586, 80–86. https://doi.org/10.1038/s41586-020-2579-z (2020).
Zarrei, M. et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genomic Med. 4, 1–13. https://doi.org/10.1038/s41525-019-0098-3 (2019).
Pinto, D. et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 94, 677–694. https://doi.org/10.1016/j.ajhg.2014.03.018 (2014).
Jiang, H. Y. et al. Maternal infection during pregnancy and risk of autism spectrum disorders: A systematic review and meta-analysis. Brain Behav. Immun. 58, 165–172. https://doi.org/10.1016/j.bbi.2016.06.005 (2016).
Atladóttir, H. Ó. et al. Maternal infection requiring hospitalization during pregnancy and autism spectrum disorders. J. Autism Dev. Disord. 40, 1423–1430. https://doi.org/10.1007/s10803-010-1006-y (2010).
Masi, A. et al. Cytokine aberrations in autism spectrum disorder: A systematic review and meta-analysis. Mol. Psychiatry 20, 440–446. https://doi.org/10.1038/mp.2014.59 (2015).
Ricci, S. et al. Altered cytokine and BDNF levels in autism spectrum disorder. Neurotox. Res. 24, 491–501. https://doi.org/10.1007/s12640-013-9393-4 (2013).
Siniscalco, D., Schultz, S., Brigida, A. & Antonucci, N. Inflammation and neuro-immune dysregulations in autism spectrum disorders. Pharmaceuticals 11, 2–14. https://doi.org/10.3390/ph11020056 (2018).
Enstrom, A. M., Onore, C. E., Van De Water, J. A. & Ashwood, P. Differential monocyte responses to TLR ligands in children with autism spectrum disorders. Brain Behav. Immun. 24, 64–71. https://doi.org/10.1016/j.bbi.2009.08.001 (2009).
Sweeten, T. L., Posey, D. J. & McDougle, C. J. High blood monocyte counts and neopterin levels in children with autistic disorder. Am. J. Psychiatry 160, 1691–1693. https://doi.org/10.1176/appi.ajp.160.9.1691 (2003).
Jyonouchi, H., Geng, L. & Davidow, A. L. Cytokine profiles by peripheral blood monocytes are associated with changes in behavioral symptoms following immune insults in a subset of ASD subjects: An inflammatory subtype?. J. Neuroinflammation 11, 1–13. https://doi.org/10.1186/s12974-014-0187-2 (2014).
Jyonouchi, H. & Geng, L. Associations between monocyte and T cell cytokine profiles in autism spectrum disorders: Effects of dysregulated innate immune responses on adaptive responses to recall antigens in a subset of ASD children. Int. J. Mol. Sci. 20, 4731. https://doi.org/10.3390/ijms20194731 (2019).
Basheer, S. et al. Immune aberrations in children with Autism Spectrum Disorder: A case-control study from a tertiary care neuropsychiatric hospital in India. Psychoneuroendocrinology 94, 162–167. https://doi.org/10.1016/j.psyneuen.2018.05.002 (2018).
Ashwood, P. et al. Altered T cell responses in children with autism. Brain Behav. Immun. 25, 840–849. https://doi.org/10.1016/j.bbi.2010.09.002 (2011).
Ashwood, P. et al. In search of cellular immunophenotypes in the blood of children with autism. PLoS One 6, e19299. https://doi.org/10.1371/journal.pone.0019299 (2011).
Saresella, M. et al. An autistic endophenotype results in complex immune dysfunction in healthy siblings of autistic children. Biol. Psychiatry 66, 978–984. https://doi.org/10.1016/j.biopsych.2009.06.020 (2009).
Jyonouchi, H. et al. Serum micrornas in ASD: Association with monocyte cytokine profiles and mitochondriarespiration. Front. Psychiatry 10, 1–18. https://doi.org/10.3389/fpsyt.2019.00614 (2019).
Jyonouchi, H., Geng, L., Streck, D. L., Dermody, J. J. & Toruner, G. A. MicroRNA expression changes in association with changes in interleukin-1ß/interleukin10 ratios produced by monocytes in autism spectrum disorders: Their association with neuropsychiatric symptoms and comorbid conditions (observational study). J. Neuroinflammation 14, 229–229. https://doi.org/10.1186/s12974-017-1003-6 (2017).
D’Eufemia, P. et al. Abnormal intestinal permeability in children with autism. Acta Paediatr. 85, 1076–1079. https://doi.org/10.1111/j.1651-2227.1996.tb14220.x (1996).
De Magistris, L. et al. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J. Pediatr. Gastroenterol. Nutr. 51, 418–424. https://doi.org/10.1097/MPG.0b013e3181dcc4a5 (2010).
Maecker, H. T., McCoy, J. P. & Nussenblatt, R. Standardizing immunophenotyping for the human immunology project. Nat. Rev. Immunol. 12, 191–200. https://doi.org/10.1038/nri3158 (2012).
Finak, G. et al. Standardizing flow cytometry immunophenotyping analysis from the human immunophenotyping consortium. Sci. Rep. 6, 20686–20686. https://doi.org/10.1038/srep20686 (2016).
Owens, M. A., Vall, H. G., Hurley, A. A. & Wormsley, S. B. Validation and quality control of immunophenotyping in clinical flow cytometry. J. Immunol. Methods 243, 33–50. https://doi.org/10.1016/S0022-1759(00)00226-X (2000).
Perfetto, S. P., Ambrozak, D., Nguyen, R., Chattopadhyay, P. K. & Roederer, M. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat. Protoc. 7, 2067–2079. https://doi.org/10.1038/nprot.2012.126 (2012).
Kleeberger, C. A. et al. Viability and recovery of peripheral blood mononuclear cells cryopreserved for up to 12 years in a multicenter study. Clin. Diagn. Lab. Immunol. 6, 14–19. https://doi.org/10.1128/cdli.6.1.14-19.1999 (1999).
Verschoor, C. P., Kohli, V. & Balion, C. A comprehensive assessment of immunophenotyping performed in cryopreserved peripheral whole blood. Cytom. Part B Clin. Cytom. 94, 662–670. https://doi.org/10.1002/cyto.b.21526 (2018).
Draxler, D. F., Madondo, M. T., Hanafi, G., Plebanski, M. & Medcalf, R. L. A flowcytometric analysis to efficiently quantify multiple innate immune cells and T Cell subsets in human blood. Cytometry A 91, 336–350. https://doi.org/10.1002/CYTO.A.23080 (2017).
Costantini, A. et al. Effects of cryopreservation on lymphocyte immunophenotype and function. J. Immunol. Methods 278, 145–155. https://doi.org/10.1016/S0022-1759(03)00202-3 (2003).
Ivison, S. et al. A standardized immune phenotyping and automated data analysis platform for multicenter biomarker studies. JCI Insight 3, 1–20. https://doi.org/10.1172/JCI.INSIGHT.121867 (2018).
Lord, C. et al. The autism diagnostic observation schedule-generic: A standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 30, 205–223. https://doi.org/10.1023/A:1005592401947 (2000).
Lord, C., Rutter, M. & Le Couteur, A. Autism diagnostic interview-revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 24, 659–685. https://doi.org/10.1007/BF02172145 (1994).
Ickowicz, A. et al. The parent interview for child symptoms: A situation-specific clinical research interview for attention-deficit hyperactivity and related disorders. Can. J. Psychiatry 51, 325–328. https://doi.org/10.1177/070674370605100508 (2006).
Scahill, L. et al. Children’s Yale-Brown obsessive compulsive scale: Reliability and validity. J. Am. Acad. Child Adolesc. Psychiatry 36, 844–852. https://doi.org/10.1097/00004583-199706000-00023 (1997).
Disis, M. L. et al. Maximizing the retention of antigen specific lymphocyte function after cryopreservation. J. Immunol. Methods 308, 13–18. https://doi.org/10.1016/j.jim.2005.09.011 (2006).
Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 111, A3.B.1-A3.B.3. https://doi.org/10.1002/0471142735.ima03bs111 (2015).
Acknowledgements
We gratefully acknowledge the participants and the families for helping us conduct and complete this study. This research was conducted with the support of the Ontario Brain Institute (OBI), Canada. The OBI was created to become an internationally recognized center of excellence in brain and neuroscience research. This independent non-profit corporation, funded partially by the Ontario government, is dedicated to improving approaches to the prevention, early diagnosis, treatment and management of neurological, and psychiatric disorders. The opinions, results, and conclusions are those of the authors and no endorsement by the Ontario Brain Institute is intended or should be inferred.
Funding
Funding was provided by Ontario Brain I (Grant No. POND).
Author information
Authors and Affiliations
Contributions
J.A.F., D.M.E.B. and R.W. designed the study. S.C., G.T. and C.M. conducted the study, did the quality control, data analysis and wrote the manuscript. J.A.F. and D.M.E.B. supervised the study. E.A., R.S. and R.N. participated as clinical leads and recruited participants. All authors edited the manuscript and approved its submission.
Corresponding author
Ethics declarations
Competing interests
JAF serves on the Scientific Advisory Board for MRM Health NL and has received consulting/speaker fees from Alphasights, Novozymes, Klaire Labs, Takeda Canada, Rothman, Benson, Hedges Inc, and WebMD. RN has received grant funding from F. Hoffmann-La Roche Ltd, Otsuka Canada Pharmaceutical, Inc., and Maplight Therapeutics. All other authors have no conflicts to report.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cleary, S., Teskey, G., Mathews, C. et al. Assessment of a multisite standardized biospecimen collection protocol for immune phenotyping in neurodevelopmental disorders. Sci Rep 13, 6971 (2023). https://doi.org/10.1038/s41598-023-33380-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-33380-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.