Abstract
Porcine circovirus 4 (PCV4) is considered a novel PCV, firstly found in China in 2019 and later discovered in Korea. This present study investigated the prevalence and genetic characteristics of PCV4 from high pig-density areas in Thailand during 2019–2020. From 734 samples, three samples (0.4%) from aborted fetuses and porcine respiratory disease complex (PRDC) cases were found positive for PCV4, two of the PCV4-positive samples were coinfected with both PCV2 and PRRSV, and the other PCV4-positive sample was found coinfected with PCV2. In situ hybridization (ISH) revealed the presence of PCV4 in the bronchial epithelial cells and in lymphocytes and histiocyte-like cells in the lymphoid follicles of the PRDC-affected pig. The complete Thai PCV4 genome had over 98% nucleotide identity with other PCV4 strains and was closely related to the Korean and Chinese PCV4b strains. Importantly, the amino acid residue at position 212 of the Cap gene is recommended for differentiating PCV4a (212L) from PCV4b (212M) based on currently available PCV4 genome sequences. These findings provide important clues for the pathogenesis, epidemiology, and genetic characteristics of PCV4 in Thailand.
Similar content being viewed by others
Introduction
To date, four species of porcine circoviruses (PCVs) have been identified, including PCV1, PCV2, PCV3, and PCV41. PCV1 is nonpathogenic in pigs. PCV2 causes various symptoms collectively called porcine circovirus-associated diseases (PCVADs) or porcine circovirus diseases (PCVDs)1,2,3. PCV3 has been found in pigs with multiple clinical signs; however, the pathogenesis is still unknown1,4,5. Recently, a novel PCV4 has been identified in China and Korea both in clinically healthy and infected pigs with several clinical presentations, including respiratory and enteric signs and skin lesions suggestive of porcine dermatitis and nephropathy syndrome (PDNS)6,7,8,9,10,11. Therefore, despite having limited information on PCV4, the virus should not be overlooked. Furthermore, due to the lesson learned from past experiences with other swine viruses in Asia12, many emerging viruses have spread among countries due to the consequences of globalization through international trade and travel, both officially and unofficially. Therefore, the recent findings of PCV4 in China and Korea would raise awareness of the virus to the Asian swine practitioners to investigate this novel pathogen in their areas as part of the focus area “prevent and detect” to understand the disease distribution and its impact. In this study, we investigated from the samples obtained during 2019–2020 for the presence of PCV4 in Thailand and its genetic characterization.
Results
Prevalence and geographical distribution of PCV4 during 2019–2020
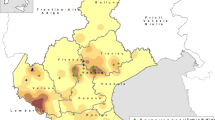
To investigate the existence of PCV4, several types of swine samples (n = 734) from different geographical regions mainly located in the high pig density areas of Thailand were used. At sample levels, the overall positive rates of PCV4 were 0.4% (3/734). Notably, three PCV4-positive samples from clinically infected pigs with abortion and respiratory signs were found coinfected with PCV2 (3/3) and PRRSV (2/3) (Table 1). Among the 145 pig farms in these 18 provinces, 2.07% (3/145) were PCV4-positive. The geographic distribution of PCV4-positive farms was shown in Fig. 1.
Genetic characteristics of PCV4
In this study, all PCV4-positive samples were selected and sequenced for genetic characterization and phylogenetic analysis. The results showed that Thai PCV4 strains were 1770 nt in length, with 891 nt of ORF1 (Rep) and 687 nt of ORF2 (Cap). The complete genome sequences of the Thai PCV4 strains were aligned against those of reference viruses from China and Korea (Table 2). The results showed that the complete genome of the 3 Thai PCV4 strains shared 100% nucleotide identity to each other and shared 98.3–99.0% nucleotide identity with other reference PCV4 strains (Table 2). An amino acid sequence analysis indicated that the Rep and Cap genes of the Thai PCV4 strains shared similarity of 98.6%-100% and 97.8%-99.1%, respectively, when compared to each other and the reference strains. The Thai PCV4 strains showed the highest similarity to PCV4 NM2 from China with 99% nucleotide identity (complete genome), 100% Rep amino acid identity, and 98.6% Cap amino acid identity. Based on the phylogenetic analysis of complete genome nucleotide sequences of 41 PCV4 strains, the viruses have undergone evolution, resulting in two main distinct branches (PCV4a and PCV4b) (Fig. 2). The results showed that the Thai PCV4 strains belonged to PCV4b and clustered in the same branch with PCV4/KU-02010 strain found in Korea (Fig. 2). In the present study, PCV4 strains identified in China belong to both PCV4a and PCV4b genotypes. Interestingly, in the Korean and Thai swine farms, only PCV4b strains were detected. Moreover, the phylogenetic trees based on Cap and Rep genes were constructed for assessing genetic relationships (Supplementary Figure 1). The findings demonstrated that different genomic regions of PCV4 yielded similar outcomes.
Phylogenetic tree based on the complete genome sequences of 3 Thai PCV4 strains and other reference strains. The Thai PCV4 sequences obtained in this study were marked with solid black circles. The colored background represented the country of origin of the PCV4 viruses. The phylogenetic tree was constructed using the neighboring-joining method with a p-distance model and bootstrapping at 1000 replicates.
Nucleotide sequence comparison and amino acid sequence analysis of PCV4
An analysis of nucleotide and amino acid sequences of the Cap and Rep genes against reference PCV4 strains demonstrated that Thai PCV4 genomes displayed nucleotide identities of 97.6%-98.9% amino acid and 98.4%-100% amino acid in Cap gene and Rep genes, respectively compared to those of reference viruses. The Thai PCV4 strains shared amino acid identities of 97.8%-99.1% and 98.6%-100% in Cap gene and Rep gene compared to those of reference viruses. There were no amino acid deletions or insertions. Compared with prototype PCV4 strain HNU-AHG1-2019 (accession no. MK986820), Thai PCV4 genomes had 20 nucleotide substitutions (Table 3). For deduced amino acid analysis, one amino acid substitution (Q155K) was seen in Rep gene, while 3 amino acid substitutions (N27S, I80V, and I96V) were found in Cap gene (Fig. 3). Among them, there was one unique amino acid substitution (I80V) in the Thai PCV4 strains of Cap gene compared with other PCV4 strains. Compared with the representative strains (Fig. 3), the Cap gene of PCV4a contains specific amino acid patterns of 27S and 212L, while certain PCV4b strains have unique amino acid patterns of 27N and 212M. Interestingly, the sequences from the present study revealed that the Thai PCV4 strains were grouped in PCV4b, even though they showed amino acid 27S in the Cap gene, and possessed the same amino acid at position 212M, similar to the two PCV4 strains (KU-02010 and KU-02011) from Korea. The findings suggest that amino acid variation at position 212 of Cap gene could be used for differentiating PCV4a (212L) from PCV4b (212M) (Fig. 3).
Comparison of Cap amino acid sequences of PCV4 isolates in this study. Dots are used to denote the residues that are consistent with HNU-AHG1 (MK986820). The red boxes indicate the amino acid at positions 27 and 212, previously proposed for differentiation of PCV4a and PCV4b. Mutations at residues position 80 of Thai PCV4 strains are shown with the green box.
Detection of PCV4 by in situ hybridization
The PCV4-positive pig (19RBR247) with respiratory illness was submitted for necropsy. Prominently, lungs with enlarged tracheobronchial lymph nodes were diffusely mottled and failed to collapse with dark red to purple consolidation and diffuse fibrinous attachment in the pleura (Fig. 4A). Microscopically, lungs revealed moderate to severe diffuse pulmonary and interlobular edema with mild multifocal hyperplasia of bronchus-associated lymphoid tissue, and moderate to severe diffuse broncho-interstitial pneumonia. Tracheobronchial lymph nodes showed moderate multifocal lymphoid depletion. To investigate the tissue localization of PCV4, lung and tracheobronchial lymph node were tested using in situ hybridization. The results showed that PCV4-ISH-positive signals were mainly observed in the cytoplasm of bronchial epithelium (Fig. 4B). Moreover, a few positive signals were detected in the lymphocytes and histiocyte-like cells of the tracheobronchial lymph node (Fig. 4C). No hybridization signals were seen in internal negative control of lung (Fig. 4D) and tracheobronchial lymph node tissue sections (Fig. 4E).
Gross and microscopic lesions and in situ hybridization (ISH) using PCV4-specific probe targeting Cap gene of PCV4. Grossly, lung failed to collapse with dark red to purple consolidation and diffuse fibrinous attachment in the pleura (A). Lung: PCV4-positive cells were characterized by pink to brilliant red in the cytoplasm of bronchial epithelial cells (arrows) (B). Tracheobronchial lymph node: positive signals were observed in lymphocytes and histiocyte-like cells in the lymphoid follicle (arrows) (C). No hybridization signals were seen in the internal negative control of lung and tracheobronchial lymph node (D,E).
Discussion
PCV2 and PCV3 are at least the two porcine circoviruses causing major threats to the global swine industry1,2,3. The finding of PCV4 should not be neglected since Asia is the major world hub of pig production, and the virus itself may originate here and spread to other regions or vice versa. To date, PCV4 was discovered only in the Asian continent, China, Korea, and recently, Thailand, but not yet in others6,7,8,9,10,11,13,14.
In the present study, the results showed that only 3 of the 734 samples were tested positive for PCV4, with a positive rate of 0.4% (3/734) and 2.07% (3/145) of farm levels. Interestingly, extremely low prevalence of PCV4 was demonstrated in Thailand compared to previous reports6,7,9,10,11 suggesting low transmission and infection rate within the pig herds. However, nearly 80% of the swine farms tested were mainly located in the high pig density areas of Thailand, which might not reflect the whole picture on PCV4 infection status of the country. Thus, in the future, large-scale field sampling of all regions could provide more information about PCV4 epidemiology in Thailand. Therefore, the threat of PCV4 to the Thai swine industry is skeptical and yet to be elucidated.
Recently, pigs inoculated with rescued PCV4 alone showed histopathological changes in several organs, but no obvious clinical signs found15. In the present study, we found that three PCV4-positive samples from clinically infected pigs were found coinfected with PCV2 (3/3) and PRRSV (2/3). These results apparently suggest that absence of other factors, such as coinfections, PCV4 infection might remain asymptomatic. However, the synergistic effect of PCV4 infection might exacerbate the disease severity and be associated with PRDC in clinically infected pigs as found in the present study. Therefore, co-infections and some unknown factors, yet to be elucidated, might involve PCV4 pathogenesis and its clinical outcomes.
According to in situ hybridization results, the virus presence was found in the bronchiolar epithelial cells and in lymphocytes and histiocyte-like cells in the lymphoid follicles consistently with the characteristics of PCV2 infection16. Whether PCV4 infection in bronchiolar epithelial cells and tracheobronchial lymph node induces the observed broncho-interstitial pneumonia and the lymphoid depletion should be further studied. The contribution of PCV4 to the pathogenesis of PRDC and immune modulation should be of interest. These findings also suggest that a marked tropism of PCV4 for bronchial epithelial cells may impair the epithelial barrier function, thus predisposing the infected pigs to secondary infections and PRDC. The case history of the PCV4 positive cases found that the PCV4-infected pigs were found coinfected with PCV2, PRRSV, and/or Streptococcus spp. Additionally, bronchial epithelial infection might contribute to viral shedding dynamics in the nasal secretion. Notably, it has been reported that the highest positive rates of PCV4 were detected in nasal swabs followed by serum samples6. Therefore, nasal swabs might be a better target specimen for PCV4 detection and surveillance. Further investigation is needed.
For genetic analysis, Thai PCV4 strains shared up to 98% nucleotide identity with other reference PCV4 strains. The high genetic stability of PCV4 is consistent with the previous reports7,11. It is noted that the Thai PCV4 strains were closely related to the Chinese PCV4 sub-cluster. Although the virus had high genetic stability, some genetic variations could be observed among pig populations from different countries that probably specific to the geographic origin17. Additionally, the Thai PCV4 strains were highly related to each other, possibly, due to a single point introduction with low infection rate or restricted gene flow in the neighboring districts of Western Thailand, suggesting that the novel virus might be confined to these areas affecting the prevalence and genetic diversity.
Additionally, there are currently no fully established guidelines for classifying PCV4 genotypes, and the temporary proposals made so far have not been consistent. This is in contrast with PCV2 genotyping that a unified classification scheme based on Cap gene was proposed18. For PCV4 genotyping, there have been several studies proposing different criteria and markers to differentiate PCV4 genotypes, such as 2-group classification (PCV4a and PCV4b) and 3-group classification (PCV4a, PCV4b, and PCV4c)10,19,20,21. Among these studies, the residue at 27 and 212 of capsid gene were also proposed to be used in distinguishing PCV4a (27S and 212L) from PCV4b (27N and 212M)19,20. Recently, PCV4c was proposed with specific amino acid pattern 27N, 28R, and 212M. However, in some genotyping criteria, PCV4c might be classified into PCV4b10,20. Therefore, the use of differing nomenclatures can create ambiguity and misinterpretation of results. Further investigation is required to establish the standardized criteria for genotyping PCV4, given the current paucity of sequence information. As mentioned above, previous reports showed that the amino acid at the position 27 of the Cap gene could be used as a marker to distinguish between PCV4a (27S) and PCV4b (27N)19,20. However, the results in this study showed that the amino acid residue at position 27 cannot be used to differentiate between PCV4a and PCV4b since the Thai PCV4b strains and the Korean PCV4b strains (KU-02010 and KU-02011) showed N27S (Fig. 3). The novel findings of this study suggest that amino acid variation at position 212 could be used for differentiating PCV4a (212L) from PCV4b (212M) for 2-group classification. Moreover, one unique amino acid substitution (I80V) was also found in the Thai PCV4 strains in Cap gene. The amino acid substitution occurred at residue 72–88 in B-cell epitopes on PCV4 capsid gene20. It may alter antigenic properties of the viruses and their immunogenic modifications caused by genetic mutation. However, the effects of genetic mutation on pathogenicity need further investigation. In terms of the genetic variation of PCV4 and its genotypes, a study conducted in various regions in China found that PCV4a was the most common genotype followed by PCV4b10,11,19,20,21. However, in pig populations from Korea and Thailand, only the PCV4b genotype was detected8. It is plausible to consider that PCV4 strains isolated from various geographic regions or pig populations may potentially demonstrate genetic variations, which may imply the existence of geographical or host specificity in the virus.
To date, PCV4 was discovered only in the Asian continent, China, Korea, and recently Thailand6,7,8,9,10,11,13,14. This study described the presence of PCV4 in Thailand from the retrospective samples and firstly demonstrated the viral tropism in the bronchial epithelial cells. The three Thai PCV4 strains demonstrated their genetic similarity with the Korean and Chinese PCV4b strains. However, the prevalence of PCV4 in Thailand is extremely low, and clinical involvement of PCV4 remains unclear. Apparently, it should be noted that the amino acid residue at position 212 of the Cap gene should be used for differentiating PCV4a and PCV4b. Further studies are needed to determine the role of PCV4 infection related to clinical signs and its impact on the Thai swine farms. The classification of PCV4 genotypes requires further investigation and clarification due to the limited available PCV4 sequences.
Materials and methods
Sample collection and viral DNA extraction
Seven hundred thirty-four samples from 145 swine farms submitted for diagnostic purposes at the Chulalongkorn University, Veterinary Diagnostic Laboratory (CU-VDL), and Diagnostic Laboratory of Large Animal Hospital and Students Training Center during January 2019- December 2020 were used for this study. The samples consisted of serum (n = 426), tissue (n = 188), fetus (n = 75), semen (n = 25), feces (n = 16), colostrum (n = 2), and oral fluid (n = 2) that were mainly collected from the high pig density areas in the Western, Central, and Eastern parts of Thailand.
Total viral DNA was extracted using IndiMag Pathogen kit of viral RNA/DNA (Indical Bioscience, Germany) following the manufacturer's instruction. The extracted DNA was stored at -80 °C until used.
Molecular detection of PCV4
TaqMan® real-time PCR was performed to detect PCV4 targeting the replicase gene (rep) of PCV4 using two newly designed primer pairs and probe (Supplementary Table S1). Briefly, PCR reactions were performed in a total 20 µl reaction containing 0.4 µM of forward and reverse primers, 0.2 µM of probes, 10 µl of Luna® Universal Probe qPCR master mix (NEB, MA, USA), and 3 µl of extracted DNA. The PCR condition consisted of initial denaturation at 95 °C for 60 s followed by 45 cycles of 95 °C for 15 s and 60 °C for 30 s using Quantstudio5 real-time system (Applied Biosystems, USA). Positive control plasmid was synthesized by inserting a full length of PCV4 rep gene (891 bp) into the pUC18 vector by GenScript Company (Nanjing, China). Detection limit of the PCV4 TaqMan® real-time PCR was 200 copies/µl of standard plasmid DNA.
Complete genome amplification and sequencing
The complete genomes of PCV4-positive samples were amplified using two primer sets (Supplementary Table S1). The PCR reactions were performed in 25 µl reaction mixtures containing 3 µl of extracted DNA, 0.5 µM of forward and reverse primers, and 12.5 µl of Q5® High-Fidelity 2 × master mix. The PCR thermal profile involved an initial denaturation of 98 °C for 30 s followed by 35 cycles of 98 °C for 10 s, 72 °C for 30 s, 72 °C for 50 s, and final extension at 72 °C for 2 min. The PCR products were purified using Nucleospin™ Gel and PCR clean-up (MACHEREY–NAGEL, Germany) and submitted for sequencing by a barcode-tagged sequencing platform (Celemic, Seoul, Korea). The obtained nucleotide sequences were further analyzed and assembled with SeqMan, and Editseq software v.5.03 (DNASTAR Inc., Madison, Wisconsin, USA), then deposited in GenBank under accession no. ON854861-ON854863.
Phylogenetic analysis
For pairwise comparison and genetic characterization of PCV4, the complete nucleotide sequences were aligned using the Clustal W algorithm of BioEdit 7.2.5 (https://bioedit.software.informer.com/) with the reference PCV4 strains from the GenBank database. Phylogenetic trees were reconstructed with MEGA version 10.2.6 using the neighbor-joining algorithm (NJ) with 1000 bootstrap replicates22.
Currently, guidelines for identifying PCV4 genotypes are not yet fully established, but they have been temporarily proposed and were not consistent. Generally, PCV4 strains have been classified into two main genotypes, PCV4a and PCV4b10,19,20. However, a single study has proposed that PCV4 might be classified into three genotypes, including PCV4c21. To simplify the analysis, in this present study, PCV4 strains were classified into two main genotypes, PCV4a and PCV4b. The phylogenetic analyses were carried out using the complete genome, Rep gene, and Cap gene, following the approach previously described10.
Detection of PCV4 in tissues using in situ hybridization
Of the three PCV4-positive samples, the formalin-fixed paraffin-embedded (FFPE) tissue samples were available for only one case, 19RBR247, and thus used for virus localization analysis. The FFPE tissues were stained with hematoxylin and eosin (HE) for histopathology study. To identify PCV4 tissue localization, in situ hybridization (ISH) was performed according to a previously described protocol with some modifications23. Briefly, PCV4-specific probe targeting 110 bp of the Cap gene of PCV4 was constructed using a PCR DIG Probe Synthesis Kit (Roche Diagnostics, Basel, Switzerland) following the manufacturer’s instructions. The sections were incubated overnight at 40 °C in a moist chamber with PCV4-specific probe. The PCV4-specific DIG probe was detected by using 1:200 anti-DIG AP Fab fragments (Roche Diagnostics, Basel, Switzerland). Hybridization signals were detected as pink to brilliant dark red colorimetric staining of Permanent Red (LPR) (Dako, Glostrup, Denmark) with 50% hematoxylin counterstaining. Slides incubated without the DIG probe were used as a negative control.
Data availability
The datasets generated and analysed during the current study are available in the NCBI genomes repository under the accession numbers ON854861-ON854863.
References
Opriessnig, T., Karuppannan, A. K., Castro, A. & Xiao, C. T. Porcine circoviruses: Current status, knowledge gaps and challenges. Virus Res. 286, 198044. https://doi.org/10.1016/j.virusres.2020.198044 (2020).
Segales, J. Porcine circovirus type 2 (PCV2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Res. 164, 10–19. https://doi.org/10.1016/j.virusres.2011.10.007 (2012).
Klaumann, F. et al. Current knowledge on porcine circovirus 3 (PCV-3): A novel virus with a yet unknown impact on the swine industry. Front. Vet. Sci. 5, 315. https://doi.org/10.3389/fvets.2018.00315 (2018).
Ouyang, T. et al. Recent progress on porcine circovirus type 3. Infect. Genet. Evol. 73, 227–233. https://doi.org/10.1016/j.meegid.2019.05.009 (2019).
Sirisereewan, C., Thanawongnuwech, R. & Kedkovid, R. Current understanding of the pathogenesis of porcine circovirus 3. Pathogens 11, 64. https://doi.org/10.3390/pathogens11010064 (2022).
Zhang, H. H. et al. Novel circovirus species identified in farmed pigs designated as porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 67, 1057–1061. https://doi.org/10.1111/tbed.13446 (2020).
Ha, Z. et al. Retrospective surveillance of porcine circovirus 4 in pigs in Inner Mongolia, China, from 2016 to 2018. Arch. Virol. 166, 1951–1959. https://doi.org/10.1007/s00705-021-05088-w (2021).
Nguyen, V. G. et al. Molecular-based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound. Emerg. Dis. https://doi.org/10.1111/tbed.14017 (2021).
Tian, R. B. et al. Molecular detection and phylogenetic analysis of porcine circovirus 4 in Henan and Shanxi Provinces of China. Transbound. Emerg. Dis. 68, 276–282. https://doi.org/10.1111/tbed.13714 (2021).
Xu, T. et al. Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene 808, 145991. https://doi.org/10.1016/j.gene.2021.145991 (2022).
Hou, C. Y. et al. Phylogenetic analysis of porcine circovirus 4 in Henan Province of China: A retrospective study from 2011 to 2021. Transbound. Emerg. Dis. 69, 1890–1901. https://doi.org/10.1111/tbed.14172 (2022).
Kedkovid, R., Sirisereewan, C. & Thanawongnuwech, R. Major swine viral diseases: An Asian perspective after the African swine fever introduction. Porcine Health Manag. 6, 20. https://doi.org/10.1186/s40813-020-00159-x (2020).
Franzo, G. et al. Lack of porcine circovirus 4 genome detection in pig samples from Italy and Spain. Pathogens 9, 433. https://doi.org/10.3390/pathogens9060433 (2020).
Vargas-Bermudez, D. S., Mogollon, J. D. & Jaime, J. The prevalence and genetic diversity of PCV3 and PCV2 in Colombia and PCV4 survey during 2015–2016 and 2018–2019. Pathogens 11, 633. https://doi.org/10.3390/pathogens11060633 (2022).
Niu, G. et al. Porcine circovirus 4 rescued from an infectious clone is replicable and pathogenic in vivo. Transbound. Emerg. Dis. 69, e1632–e1641. https://doi.org/10.1111/tbed.14498 (2022).
Huang, Y. Y. et al. Porcine circovirus 2 inclusion bodies in pulmonary and renal epithelial cells. Vet. Pathol. 45, 640–644. https://doi.org/10.1354/vp.45-5-640 (2008).
Faustini, G., Drigo, M., Menandro, M. L., Pasotto, D. & Giovanni, F. Phylodynamic analysis of current Porcine circovirus 4 sequences: Does the porcine circoviruses evolutionary history repeat itself?. Transbound. Emerg. Dis. 69, e3363–e3369. https://doi.org/10.1111/tbed.14638 (2022).
Franzo, G. & Segales, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 13, e0208585. https://doi.org/10.1371/journal.pone.0208585 (2018).
Wang, D., Mai, J., Yang, Y., Xiao, C. T. & Wang, N. Current knowledge on epidemiology and evolution of novel porcine circovirus 4. Vet. Res. 53, 38. https://doi.org/10.1186/s13567-022-01053-w (2022).
Wu, M. et al. Genetic variation analysis of porcine circovirus type 4 in South China in 2019 to 2021. Viruses 14, 1736. https://doi.org/10.3390/v14081736 (2022).
Xu, T. et al. First molecular detection and genetic analysis of porcine circovirus 4 in the Southwest of China during 2021–2022. Front. Microbiol. 13, 1052533. https://doi.org/10.3389/fmicb.2022.1052533 (2022).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. https://doi.org/10.1093/molbev/mst197 (2013).
Piewbang, C. et al. Naturally acquired feline bocavirus type 1 and 3 infections in cats with neurologic deficits. Transbound. Emerg. Dis. 69, e3076–e3087. https://doi.org/10.1111/tbed.14664 (2022).
Acknowledgements
The author (CS) would like to thank the Second Century Fund (C2F), Chulalongkorn University, for providing his PhD scholarship. In addition, we thank the staff of Veterinary Diagnostic Laboratories, Faculty of Veterinary Science, Chulalongkorn University for their technical assistance. Additionally, this project is supported by the National Research Council of Thailand (NRCT): R. Thanawongnuwech NRCT Senior scholar 2022 #N42A650553.
Author information
Authors and Affiliations
Contributions
C.S. and R.T. planned the experiments. C.S., T.C., C.P., S.J. and R.K. carried out the experiments. C.S. and R.K. analyzed and interpreted the results. C.S., R.K. and R.T. drafted and revised the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sirisereewan, C., Nguyen, T.C., Piewbang, C. et al. Molecular detection and genetic characterization of porcine circovirus 4 (PCV4) in Thailand during 2019–2020. Sci Rep 13, 5168 (2023). https://doi.org/10.1038/s41598-023-32382-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-32382-1
This article is cited by
-
Generation of porcine circovirus type 4 virus-like particles and their use to detect serum antibodies
Archives of Virology (2024)
-
First detection of porcine circovirus 4 (PCV-4) in Europe
Virology Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.