Abstract
Pinus elliottii used as rootstock instead of homologous rootstock, have been proved to accelerate early growth of the scion (Pinus massoniana), for cultivation of large diameter wood. However, the basal diameter of scions in heterologous grafts was significantly smaller than self-graft 10 years later, according to field investigation, which was opposed to cultivation objectives. Although advantage of heterologous grafts has been reported, less is known about the long term effect of heterologous rootstock on scions of P. massoniana. The aim of present study was to investigate the mechanism of the above difference. Toward this aim, the growth traits and physiological characteristics of scions in the two graft groups were studied, and the underlying mechanism was preliminarily explored through transcriptome sequencing technology. Results showed that scions of heterologous grafts had less TSCA compared to self-grafts, while no significant difference of plant height, number of branches and canopy volume between two graft groups. Besides, scion leaves of heterologous grafts displayed higher antioxidant enzyme activity and lower chlorophyll content. And interactions between rootstocks and scions had also changed the mineral element composition of scion leaves. Compared with homologous grafts, scion leaves of heterologous grafts accumulated more K+, Mg2+ and Zn2+, but less Ca2+,which have been proved to be conducive to the growth of stem diameter of P. massoniana. Moreover, a comparative transcriptome analysis of two graft groups showed that DEGs between them were mainly caused by the specificity of rootstock. GO and KEGG analysis found that heterologous rootstock had different gene expression preferences, and the gene expression level between rootstocks and scions were significantly different, such as auxin auxin-related genes and stress responsive genes. That may imply that auxin pathway played an important role not only in grafting healing process, but also in maintaining the growth between scion and stock. Summary of all above results, we concluded that the long term effect of heterologous rootstock on scions may be unsatisfactory with the later rapidly growth of scion, probably due to delayed graft incompatibility between scion and stock of heterologous grafts. This study may remind us that the long-term growth of the scion deserves attention as well as the healing process, which could also provide a basis for delayed graft incompatibility.
Similar content being viewed by others
Introduction
Grafting is an effective and essential technique used worldwide in the planting industries to increase the yield, enhance biotic and abiotic stress resistance and modify the scion architecture1,2,3,4,5. Many studies have indicated profound influence of rootstocks on scion cultivars. Because the scion-rootstock interaction influences tree physiology, absorption capacity of mineral elements, yield efficiency, maturity and fruit quality4,6,7,8,9,10.
Pinus massoniana (Lamd.) is one of the most widely distributed tree species in the genus of Pinus in China, which plays a pivotal role in ecological environment construction and sustainable forestry production11. A great deal of research has been conducted on germplasm resources, genetic determination and selection, and improved varieties breeding12. Meanwhile, multi-generation clonal seed orchards based on homologous graft have been established13. Nevertheless, the slow growth of P. massoniana at early stages seriously hindered its popularization and application. Interspecific hybridization is difficult because of its cross-incompatibility, long cross breeding cycles, and difficult separation of progeny traits14. Graft is one of the most widely useful methods of asexual reproduction, aim to make genetic improvement of germplasm resources so as to improve offspring and improve their performance15. The key to success of grafting is the compatibility between rootstock and scion16. Therefore, the selection of rootstock is extremely important17. As a member of the double vascular subgenus,
Pinus elliottii (Engelm.) grows faster and has a better trunk structure than P. massoniana. In addition, its afforestation application areas overlap partially with that of P.massonian18. It was also reported that the 2.5-year-old grafted trees were 60.8% and 197.2% larger of diameter than the homologous graft trees, when P. elliottii were used to graft P. massonian by researchers of Guangxi Academy of Forestry13. Thus, the heterologous graft might be a good idea to ameliorate the characteristics of slow growth of Masson's pine at early growth stage. Although the success of heterologous grafts, we found that scions grafted on P. elliottii were not better than those self-grafted tress, 10 years later after graft, particularly in the trunk cross-sectional area (TSCA). It is very necessary to study the causes of this problem, because large diameter wood is the ultimate goal of commercial foresters depend on basic research.
Based on 10 years of heterologous and homologous grafted seedlings, we were able to detect the long term effect of heterologous rootstocks on scion by physiological and biochemical methods, and preliminarily explore the underlying mechanism together with transcriptome sequencing technology. And the results were supposed to have important practical application value and theoretical significance.
Materials and methods
Plant materials
This study has complied with relevant institutional, national, and international guidelines and legislation. The collection of materials has been approved by relevant departments. The material were collected from the gene collection area (26°16′ N, 107°31′ E) of P. massoniana national fine variety base in Duyun City, Guizhou Province, China, on September 2019. The scions used in the experiment were all from the same clone of Masson's pine. Thrity 10-year-old trees in heterologous grafts P. massoniana (scion)/P. elliottii (stock) and thirty 10-year-old trees in homologous grafts were selected for growth traits measurement. Mature leaves were collected from these trees for physiological measurement. Phloem tissues of 5 cm above and below the graft union of three trees in each grafted groups were collected for RNAseq. And all collected samples were quickly frozen by liquid nitrogen and stored at − 80 °C.
Measurement of growth parameters
For estimating canopy volume, the tree height was measured from the collar region at the base to the longest shoots at top. Diameter at breast height (DBH) and canopy diameter were both measured by measuring scale in N–S and E–W directions. TCSA was measured using the following formula: TCSA = π (d/2)2, where d = average of cross measurement of trunk in N–S and E–W directions. Canopy volume (CV) was calculated according to the equation Canopy volume = 4/3*π*a2b, Where a: spread (E-W) + spread (N-S)/2, b: 1/2tree height.
Chlorophyll content
The leaf chlorophyll content was estimated by 95% ethanol extraction method19. The extinction values of samples were measured by UV Spectrophotometer at 470 nm, 649 nm and 665 nm for pigment concentrations Chlorophyll a (Ca) and Chlorophyll b (Cb) respectively. And the Chlorophyll content calculation formula was below:
Leaf nutrient analysis
Leaves in each tree were randomly collected from all directions, washed with tape water and followed by double distilled water. The leaves were then dried at 105° ± 1 °C in oven for 0.5 h and then at 60 °C to constant weight.
Nitrogen content in leaves was determined by using Digestion Block method20.Phosphorus in leaves was estimated by vando-molybdo-phosphoric yellow color method21.Total K+ contents in leaves were estimated according to Jackson22. For quantification of Ca2+and K+ in leaves, 1.0 g dry weight was digested in diacid (using HNO3 and HClO4 in the ratio of 9:4). K+ were analysed by a micro-processor based flame photometer. And Ca2+, Mg2+, Zn2+, Cu2+and Fe2+contents were determined by atomic absorption spec-trophotometer.
Biochemical parameters
Determination of SOD, POD, and CAT activities
In each sample, 0.5 g fresh leaf was ground with a pestle in an ice-cold mortar with 4 ml of 50 mM phosphate buffer (pH 7.0). Activations of enzymes were measured in the supernatant of homogenates centrifuged for 20 min at 4 °C at 12,000 rpm. SOD activity was assayed by measuring its ability to inhibit the photochemical reduction of nitro blue tetrazolium23. POD activity was measured as the increase in absorbance at 470 nm caused by guaiacol oxidation24. CAT activity was measured as the decline in absorbance at 240 nm caused by a decrease in H2O2 removal25.
Total soluble proteins and Total soluble sugar
Samples were placed in 1.5 mL microfuge tubes and crushed in 100 mM sodium phosphate buffer (pH 7.8) for protein and carbohydrate extraction. At 4 C, samples were centrifuged for 15 min at 16,000 rpm. The supernatants were removed and analyzed. Protein samples were diluted 1:50 and carbohydrate samples were diluted 1:25 in the sodium phosphate buffer before analysis. Protein levels were determined using a BCA assay following the manufacturer’s directions with absorbance readings at 595 nm and quantified by comparison to BCA standards26. Soluble sugar were measured according to Laurentin and Edwards27 and following the manufacturer’s directions with absorbance readings at 620 nm and quantified by comparison to glucose standards.
Total RNA extraction and RNA library construction
Sample preparation
Total RNA was extracted from the above samples using the TRIZOL kit (Invitrogen) and then treated with DNaseI to remove DNA. Before preparation of the RNA libraries, total RNA samples were quantified. 1% agarose gel was used to monitor degradation and contamination of the RNA samples. The mRNA was enriched with magnetic beads of Oligo (dT) beads after the quality test. The fragmentation buffer was added to break the mRNA into short pieces. Using mRNA as template, cDNA was synthesized by reverse transcription using six base random primers. Illuminape library was established after double stranded cDNA purified, terminal repair. After passing quality inspection, Illumina PE library was constructed and 2 × 150 bp cDNA library was sequenced. The raw data generated in this study have been uploaded to Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/). The accession number is BioProject ID: PRJNA792704.
Data analysis
The quality of the original sequencing data of each sample was evaluated by fastx _ toolkit_0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/); Seqprep (https://github.com/jstjohn/SeqPrep) was used for quality control on the original sequencing data, the reads with linker, N ratio greater than 0.1% and low-quality sequences were removed to obtain clean reads. Bowtie2 (https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.3.5.1/) was used to compare the full-length transcriptome of the third generation of P.massoniana obtained for mapping and RSEM for transcription. In this quantity, the obtained read Count value is FPKM to analyze gene expression levels. The gene differential expression analysis of multiple samples (≥ 2) was carried out by DESeq2 (http://bioconductor.org/packages/stats/bioc/DESeq2/) , which was defined as differentially expressed genes (DEGs) according to the screening criteria of|log2.Fold_change|> 1 and q.value < 0.005. Differential expression gene (DEGs) were assigned to gene ontology (GO) enrichment analysis using the GOseq R package, and GO terms with corrected P-values < 0.05 were considered significantly enriched. The statistical enrichment of DEGs in
KEGG pathways (http://www.genome.jp/kegg/) was tested using KOBAS software.
Data analysis
All data were analyzed with SPSS Statistical Software (Version24.0). Student’s t-test was used to compare means between two graft groups. Data visualization and GSEA analysis were performed using the Majorbio Cloud Platform (https://cloud.majorbio.com ) and GraphPad Prism 8 (https://www.graphpad.com/scientific-software/prism/).
Results
Morphological characteristic changes of scion between two grafted groups
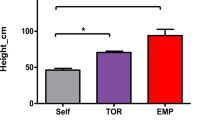
Less TSCA of scions was found in heterologous grafts compared to homologous graft (Table 1). Plant height, number of branches, and canopy volume did not differ significantly between two graft groups. It showed that the long-term impact of heterogenous rootstock on scions was mainly reflected in the transverse growth of scions.
Photosynthetic pigments concentration of scion leaves
Lower chlorophyll a and total chlorophyll content were both found in scion leaves of heterologous grafts compared to homologous grafts (Fig. 1). However, chlorophyll b, chlorophyll a/b ratio had no significantly difference between two graft groups. Higher photosynthetic pigment content may mean more photosynthetic products under the same conditions, which may be more conducive to the lateral growth of plants when maintaining almost the same other growth traits.
Scion leaf nutrient analysis
Scion leaves were found to be similar statistically in terms of total N and total P content, indicating that these two elements were little influenced by rootstock. However, scion leaves of heterologous grafts significantly accumulated more Mg2+, but lower K+ and Ca2+ when compared to homologous grafts (Fig. 2). In addition, the scion leaves micro-nutrients were also affected by heterologous rootstock (Fig. 3).Both Cu2+ and Fe3+ were not significantly different, while more Zn2+ accumulation in scion leaves of heterologous grafts than homologous grafts.
Thus, the content of different ions in scion leaves differed between two graft groups due to rootstocks, because different rootstocks had different absorption preferences for ions.
Biochemical parameters
Much higher superoxide dismutase (SOD), polyphenoloxidase (POD), and catalase(CAT) activity and lower malondialdehyde (MDA) content of scion leaves in heterologous grafts were found than those in homologous grafts (Fig. 4), indicating that higher resistance of against oxidative stress had been induced by heterologous rootstocks. Additionally, scion leaves of total soluble sugar and protein content were also more than leaves of homologous grafts, which indicated that these substances were easily affected by heterologous rootstocks. Because antioxidant enzyme systems and soluble substances were often associated with stress resistance, we could speculate that heterologous rootstocks improved the stress resistance of scions compared to homologous rootstocks in the same living environment. And the increase in resistance could be maintained for many years after grafting.
Correlation between measurement indexes
Aim to investigate the correlativity between scion growth traits and these physiological and biochemical indexes, a correlation analysis was conducted (Fig. 5). The result showed that antioxidant enzymes exhibited strong positive correlations with one another, whereas strong negative correlations with TSCA. That was to say that higher antioxidant enzymes activity of leaves may not be conducive to the lateral growth of the stem. TSCA had strongly negative correlation with Mg and soluble protein, but positive correlation with MDA. MDA showed positive correlation with K and Ca. Ca showed positive correlation with K, but negative correlation with Zn. These results indicated that proper MDA content could be good for stem diameter by affecting absorption of mineral elements. Total chlorophyll correlated positively with TSCA, but negatively with Zn, soluble sugar and antioxidant enzymes, which meant that antioxidant enzymes and soluble sugars may indirectly regulate photosynthesis by affecting the content of photosynthetic pigments to play an important role in TSCA. To put it simply, there was a complex network among mineral elements accumulation, photosynthetic pigment, soluble sugars and proteins as well as antioxidant enzymes activity to regulate the growth of scion. Heterologous rootstock had affected the synthesis and metabolism of scion, result in different growth trait.
Illumina sequencing, mapping and quantitative assessment of transcriptomes
RNA-Seq results were summarized in Table 2. A total of 44 million raw reads were generated per sample. After quality control, 6.23 to 8.77 Gb clean bases with a Q30 ratio over 89.97% and a GC content over 44.67% were obtained from each of the 12 libraries. Based on these results, the sequencing data had a high degree of coverage in the genome, and the data were accurate and sufficient for further bioinformatics analysis. The heat map analysis of correlation between samples showed that the correlation between Heterog_r group and other groups was low, which was also caused by the rootstock species specificity (Fig. 6A). According to PCA analysis (Fig. 6B), the scions grafted on heterologous rootstocks had greater intra-group differences than homologous grafts. DegSeq2 was used to screen the differentially expressed genes and the number of differentially expressed genes was counted (Fig. 6C). Among them, there were 989 DEGs between Heterog_s and Homog_s, with 271 up-regulated and 718 down-regulated genes. 1239 DEGs with 660 up-regulated genes and 579 down-regulated genes were found between Homog_r and Homog_s. 14,501 DEGs between Heterog_r and Heterog_s were found, with 5787 up-regulated genes and 8714 down-regulated genes. The number of DEGs between Heterog_r and Homog_r was 10,725, with 7213 up-regulated genes and 3512 down-regulated genes. The venendiagram of DEGs (Fig. 6D) of the four groups above was carried out. All above results indicated that more different genes between scion and rootstock were found in heterologous grafts, and more genes expression had been effected by heterologous rootstock, which then finally affected the growth of scion.
Functional annotations of DEGs
GO and GSEA enrichment analysis
To evaluate gene function of these DEGs, DEGs between grafted scions and rootstocks were annotated using GO and GSEA method (Fig. 7A–D). And DEGs between Homog_s and Homog_r significantly enriched in auxin-activated signaling pathway (GO:0009734) were upregulated in Homog_s. While GSEA enrichment analysis of DEGs between Heterg_s vs Heterg_r showed that biological processes such as response to abscisic acid (GO:0009737) were upregulated in Heterg_r. DEGS between Homog_s and Heterg_s, Homog_r and Heterg_r were also analyzed. And results showed biological processes of polysaccharide metabolic process (GO:0005976), cofactor biosynthetic process (GO:0051188) and sulfur compound metabolic process (GO:0006790) were both upregulated in Heterg_s between Homog_s and Heterg_s, which may indicate more metabolic pathways have changed in Heterg_s caused by heterologous grafting. Then DEGS of rootstocks in DNA-templated transcription initiation (GO:0006352),positive regulation of nucleobase-containing compound (GO:0045935) and other molecular functions were upregulated in Homog_r, but cellular amino acid catabolic process (GO:0009063), response to water deprivation (GO:0009414) and other biological processes were upregulated in Heterg_r. All above indicated different rootstocks had different gene expression preferences and interactions between rootstocks and scions may be more complex than we imagine.
GSEA analysis of different grafting groups, (A) auxin-activated signaling pathway (GO:0009734) between Homog_s and Homog_r, (B) response to abscisic acid (GO:0009737) between Heterg_s and Heterg_r, (C) polysaccharide metabolic process (GO:0005976) between Homog_s and Heterg_s, (D) response to water deprivation (GO:0009414) between Homog_r and Heterg_r.
KEGG enrichment analysis between rootstock and scion of two grafted groups
KEGG pathway enrichment analysis was employed to further understand the main biochemical metabolic and signal transduction pathways between two grafted groups (Fig. 8A,B). Ten pathways significantly enriched in upregulated DEGs between Homog_s and Homog_r, with most DEGs being involved in Flavonoid biosynthesis, Plant hormone signal transduction, Phenylpropanoid biosynthesis, Flavone and flavonol biosynthesis, Brassinosteroid biosynthesis, alpha-Linolenic acid metabolism, Phenylalanine metabolism, Stilbenoid, diarylheptanoid and gingerol biosynthesis, Ascorbate and aldarate metabolism and Tyrosine metabolism. While eleven pathways significantly enriched in upregulated DEGs between Heterg_s and Heterg_r, with most DEGs being involved in Cysteine and methionine metabolism, beta-Alanine metabolism, Histidine metabolism, Selenocompound metabolism, Propanoate metabolism, Pentose and glucuronate interconversions, Valine, leucine and isoleucine degradation, Tryptophan metabolism, Glycerolipid metabolism, Sphingolipid metabolism and Glycolysis/Gluconeogenesis.
Go and KEGG enrichment analysis of DEGs caused by graft
Due to the interaction between rootstock and scion, intersection of DEGs between Heterg_r and Heterg_s and DEGs between Homog_r and Homog_s, namely common DEGs caused by heterologous and homologous grafts, were also analyzed by GO (Fig. 9) and KEGG (Fig. 10). The results indicated that two grafts both affect genes expression between scion and rootstock, such as glycogen (starch) synthase activity, auxin-activated signaling pathway, amyloplast and so on. Flavonoid biosynthesis, Plant hormone signal transduction and Endocytosis pathways were significantly enriched by KEGG.
DEGs involved in Auxin signal transduction
Since auxin plays an important role during grafting reconnection, we wanted to investigate whether this hormone also affected the growth between scion and rootstock at a later stage. Thus, the auxin signal transduction pathway of plant hormones was primarily studied (Fig. 11). It was easily found that Heterg_s and Heterg_r did not exhibit consistent upregulation or downregulation of auxin-related genes compared to self-graft, which was mainly caused by reverse gene expression of Heterg_r. The same gene expression pattern between Heterg_s and Homog_s showed that Heterg_r had little effect on the auxin related genes expression of Heterg_s. But the different gene expression pattern of Heterg_r may be affected by scion, for the complex interaction between rootstock and scion.
Discussion
In modern agriculture, grafting is widely used to improve biotic and abiotic stress tolerance, modify plant architecture, induce precocious flowering and rejuvenate old perennial varieties, boost yield, and more28,29. It is critical to the tree growth that the rootstock-graft interactions are compatible, including the root characteristics4,6,30, absorption and translocation of water and nutrients5,31,32 and different signals passing through the graft union1,4. In the present study, we measured the growth and development traits, physiology and biochemistry of P. massoniana scion as well as transcriptomic difference to study the long-term effect of heterologous rootstock on scion growth. Unexpectedly our results showed that the P. elliottii rootstock had no significantly positive effect on the development and physiology of the P. massoniana scion compared to homologous graft 10 years after graft. Although heterologous grafting scion had more content of soluble sugar, soluble protein and higher antioxidant enzymes activity, their TCSA were lower than homologous grafted scion, which was not good for timber forest cultivation.
Rootstock had a meaningful impact on nutrient absorption and the nutrient uptake efficiency varied with the rootstocks28,32. In heterologous graft groups rootstocks changed the content of mineral elements in leaves, with lower content of K+ and Ca2+ and higher content of Zn2+ and Mg2+compared to homologous graft groups, which was consistent with the previous researches that nutrition efficiency could be influenced by the rootstock33,34. Difference in leaf nutrient concentration of different rootstocks might be due to alteration of root morphology and hydraulic conductance. Because it have been reported that rootstocks had differential capacity of hydraulic conductance, which was positively correlated with nutrient accumulation6,34. As for P. massoniana, leaf calcium was positively correlated with thoracic diameter, but leaf zinc was inversely correlated with diameter31, which was consistent with our correlation analysis.
More researches showed complex physiological metabolites were affected during graft union formation5,7,29. Grafting, as a wounding stress, triggers antioxidant defense systems4, resulting a higher level of reactive oxygen species (ROS) or a less efficient detoxification system on incompatible scion/rootstock interfaces35. Initial healing of the graft union does not ensure long-term compatibility, because some stock/scion combinations incompatibility may appear only after several years35,36. In this article, higher antioxidant enzymes activity, soluble protein were found in heterologous grafting scion leaves, which could indicate that incompatibility may exist between rootstock and scion in P. massoniana/P. elliottii heterologous graft system due to a belated response to the auxin and carbohydrate imbalance37,38 caused by phloem graft union irregularities. Meanwhile, the study also revealed that rootstock-scion interactions persist throughout the composite plant's lifespan, even if graft compatibility was satisfactory38.
It has also been reported that grafting induces differences in the transcriptome profile of grafted parents in plants, such as grapevines39,40. However, much attention had been absorbed on the connection of the graft union. Until recently, little was known about the long-term changes between rootstocks and scions. RNA-Seq was used in this study to identify DEGs in heterologous and homologous grafted tissues. Functional annotation and enrichment analysis of DEGs revealed that the overrepresented genes were involved in physiological metabolism (flavonoid biosynthetic process, auxin-activated signaling pathway, cellular response to auxin stimulation) between scion and rootstock in homologous graft group, while response to water deprivation and abscisic acid between scion and rootstock in heterologous graft group. These transcriptomic results suggested that stress about water existed between scion and rootstock in heterologous grafting group due to delayed graft incompatibility, with the later rapidly growth of scion41. And this could be partly proved by the higher antioxidant enzymes activity, soluble protein and soluble sugar in leaves.
Then we focused on the auxin-activated signaling pathway in homologous graft. Results showed that AUX1、IAA13、IAA9 genes, bZIP transcription factor were differentially expressed between scion and rootstock in homologous graft group when compared to heterologous graft. All those auxin related genes may contribute to maintain the local auxin concentration above and below the graft port42,43 and regulate the normal function of nutrients and water transportation, resulting in more stem growth. Thus, IAA may also play an important role in maintaining the function of grafted plant, not only at the beginning of graft union reconnection. When compared with Heterg_s and Homog_r, water deprivation and abscisic acid response were both upregulated in Heterg_r. Together with physiological data, we could conclude that P. elliottii as rootstock to graft P. massoniana was more sensitive to water deprivation than P. massoniana. The observed reactions (higher antioxidant enzyme activities, soluble sugars, and protein) in scions were mainly due to upregulation of enriched genes between rootstock and scion in Heterologous graft.
Conclusion
Complex interactions between rootstock and scion have changed mineral element composition and antioxidant enzyme activity in scion as well as transcription changes. Based on the results of compared transcriptome analysis, there may be late incompatibility between rootstock and scion of Heterologous graft. The delayed incompatibility was mainly caused by auxin imbalance between rootstocks and scions and large moisture requirement for rapid growth of scion in later period.
Data availability
The RNA sequence generated in this study have been uploaded to Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/).The accession number is BioProject ID: PRJNA792704.
References
Baron, D., Esteves Amaro, A. C., Pina, A. & Ferreira, G. An overview of grafting re-establishment in woody fruit species. Sci. Hortic. 243, 84–91 (2019).
Bletsos, F. A. & Olympios, C. M. Rootstocks and grafting of tomatoes, peppers and eggplants for soil-borne disease resistance, improved yield and quality. Eur. J. Plant Sci. Biotechnol. 2(1), 62–73 (2008).
Vegetable grafting: Principles and practices. (CABI, 2017).
Frioni, T. et al. Grafting cv. Grechetto gentile vines to new M4 rootstock improves leaf gas exchange and water status as compared to commercial 1103P rootstock. Agronomy 10, 708 (2020).
Musa, I. et al. Effects of grafting on morphophysiological and yield characteristic of eggplant (Solanum melongena L.) grafted onto wild relative rootstocks. Plants 9, 1583 (2020).
Dubey, A. & Sharma, R. M. Effect of rootstocks on tree growth, yield, quality and leaf mineral composition of lemon (Citrus limon (L.) Burm.). Sci. Hortic. 200, 131–136 (2016).
Irisarri, P., Errea, P. & Pina, A. Physiological and molecular characterization of new apricot cultivars grafted on different prunus rootstocks. Agronomy 11, 1464 (2021).
Souza, L. S., Diniz, R. P., de Jesus Neves, R., Alves, A. A. C. & de Oliveira, E. J. Grafting as a strategy to increase flowering of cassava. Sci. Hortic. 240, 544–551 (2018).
Tecchio, M. A., da Silva, M. J. R., Callili, D., Hernandes, J. L. & Moura, M. F. Yield of white and red grapes, in terms of quality, from hybrids and Vitis labrusca grafted on different rootstocks. Sci. Hortic. 259, 108846 (2020).
Zhang, Z., Cao, B., Gao, S. & Xu, K. Grafting improves tomato drought tolerance through enhancing photosynthetic capacity and reducing ROS accumulation. Protoplasma 256, 1013–1024 (2019).
Wu, F., Zhu, P. H. & Ji, K. S. Responses of masson pine (Pinus massoniana) distribution patterns to future climate change. J. Nanjing For. Univ. (Nat. Sci. Edition) 46, 196–204 (2022).
Ji, K. S. et al. Progresses and achievements of genetic improvement on Masson pine (Pinus massoniana) in China. J. Nanjing For. Univ. (Nat. Sci. Edition) 46, 10–22 (2022).
Yang, Z. Q. & Wei, Y. R. Study on construction technology of clone grafting seed orchard of Masson's Pine. Guangxi For. Sci., pp 56–61 (2001).
Luo, Q. Q. et al. Analysis and optimization of growth and Morphological and qualitative Character variation of Masson’s Pine clones. J. Northeast For. Univ. 009, 050 (2022).
Fullana-Pericàs, M., Conesa, M. À., Pérez-Alfocea, F. & Galmés, J. The influence of grafting on crops’ photosynthetic performance. Plant Sci. 295, 110250 (2020).
Chen, Z. et al. Transcriptome changes between compatible and incompatible graft combination of Litchi Chinensis by digital gene expression profile. Sci. Rep. 7, 3954 (2017).
Hou, Y. et al. Metabolite profiling and transcriptome analyses provide insight into the regulatory network of graft incompatibility in litchi. Front. Genet. 13, 1059333 (2023).
Gao, W. et al. Diurnal dynamics of photosynthetic characteristics of pinus massonia and pinus elliottii saplings under similar habitat. J. Central South Univ. For. Technol. 32, 34–39 (2012).
Sartory, D. P. & Grobbelaar, J. U. Extraction of chlorophyll a from freshwater phytoplankton for spectrophotometric analysis. Hydrobiologia 114, 177–187 (1984).
Tinggi, U. & Maher, W. Determination of trace elements in biological tissues by aluminum block digestion and spike-height flame atomic absorption spectrometry. Microchem. J. 33, 304–308 (1986).
Jackson, M.L. vandomolybdophosphoric yellow colour method. in Soil Chemical Analysis. p 452 (Prentice Hall of India Pvt Ltd. 1973).
Jackson, M.L. K and Na contents. in Soil Chemical Analysis. p 452 (Prentice Hall of India Pvt Ltd. 1980).
Pedersen, P. L. Mitochondrial adenosine triphosphatase. J. Bioenerg. Biomembr. 6, 243–275 (1975).
Doerge, D. R., Divi, R. L. & Churchwell, M. I. Identification of the colored guaiacol oxidation product produced by peroxidases. Anal. Biochem. 250, 10–17 (1997).
Buhle, E. L. & Aebi, U. Specific labeling of protein domains with antibody fragments. J. Ultrastruct. Res. 89, 165–178 (1984).
Rekowski, A. et al. Determination of soluble wheat protein fractions using the Bradford assay. Cereal Chem. 98, 1059–1065 (2021).
Laurentin, A. & Edwards, C. A. A microtiter modification of the anthrone-sulfuric acid colorimetric assay for glucose-based carbohydrates. Anal. Biochem. 315, 143–145 (2003).
Vidoy-Mercado, I. et al. Reinvigoration/rejuvenation induced through micrografting of tree species: Signaling through graft union. Plants 10, 1197 (2021).
Williams, B., Ahsan, M. U. & Frank, M. H. Getting to the root of grafting-induced traits. Curr. Opin. Plant Biol. 59, 101988 (2021).
López-Serrano, L. et al. Pepper rootstock and scion physiological responses under drought stress. Front. Plant Sci. 10, 38 (2019).
Baron, D., Amaro, A. C. E., Macedo, A. C., Boaro, C. S. F. & Ferreira, G. Physiological changes modulated by rootstocks in atemoya (Annona x atemoya Mabb.): Gas exchange, growth and ion concentration. Braz. J. Bot. 41, 219–225 (2018).
Sharma, R. M., Dubey, A. K., Awasthi, O. P. & Kaur, C. Growth, yield, fruit quality and leaf nutrient status of grapefruit (Citrus paradisi Macf.): Variation from rootstocks. Sci. Hort. 210, 41–48 (2016).
Liang, J. et al. Grafting improves nitrogen-use efficiency by regulating the nitrogen uptake and metabolism under low-nitrate conditions in cucumber. Sci. Hortic. 289, 110454 (2021).
Nawaz, M. A. et al. Nitrogen use efficiency of watermelon grafted onto 10 wild watermelon rootstocks under low nitrogen conditions. Agronomy 8, 259 (2018).
Gainza, F., Opazo, I. & Muñoz, C. Graft incompatibility in plants: Metabolic changes during formation and establishment of the rootstock/scion union with emphasis on Prunus species. Chilean J. Agric. Res. 75, 28–34 (2015).
Ermel, F. F., Kervella, J., Catesson, A. M. & Poessel, J. L. Localized graft incompatibility in pear/quince (Pyrus communis/Cydonia oblonga) combinations: Multivariate analysis of histological data from 5-month-old grafts. Tree Physiol. 19, 645–654 (1999).
Errea, P., Garay, L. & Marín, J. A. Early detection of graft incompatibility in apricot (Prunus armeniaca ) using in vitro techniques. Physiol. Plant. 112, 135–141 (2001).
Giral, M. et al. Kidney and recipient weight incompatibility reduces long-term graft survival. JASN 21, 1022–1029 (2010).
Franck, N. et al. Contrasting grapevines grafted into naturalized rootstock suggest scion-driven transcriptomic changes in response to water deficit. Sci. Hortic. 262, 109031 (2020).
Xu, C. et al. Transcriptomic analysis of melon/squash graft junction reveals molecular mechanisms potentially underlying the graft union development. Peer J. 9, e12569 (2021).
Aloni, B. et al. Possible mechanisms for graft incompatibility between melon scions and pumpkin rootstocks. Acta Hortic. 782, 313–324 (2008).
Blakeslee, J. J., Spatola Rossi, T. & Kriechbaumer, V. Auxin biosynthesis: spatial regulation and adaptation to stress. J. Exp. Bot. 70, 5041–5049 (2019).
Dubreuil, C., Jin, X., Grönlund, A. & Fischer, U. A local auxin gradient regulates root cap self-renewal and size homeostasis. Curr. Biol. 28, 2581-2587.e3 (2018).
Acknowledgements
The authors thank the experimental materials provided by Duyun National Germplasm Resource Bank. The authors thank the editor and the anonymous reviewers for their useful comment on a previous version of the manuscript.
Funding
This work was supported by funds of National Natural Science Foundation of China (NSFC.32060353); Science and Technology Talent Platform Project of Guizhou Province [2018]5261.
Author information
Authors and Affiliations
Contributions
Q.L. carried out the experiments, performed the data analyses and drafted the manuscript.F.X. performed the data analyses and participated in sample collection. X.W. and Y.Z. made revisions to the manuscript. Y.Y. participated in sample collection and experients. All authors approved the final revision to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, Q., Wang, X., Zhao, Y. et al. Transcriptome and physiological analyses reveal new insights into delayed incompatibility formed by interspecific grafting. Sci Rep 13, 4574 (2023). https://doi.org/10.1038/s41598-023-31804-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-31804-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.