Abstract
Escherichia coli O157:H7 is a foodborne pathogen, which causes various health conditions in humans, including fatigue, nausea, bloody diarrhoea and in some cases, even death. In 2017, 15.71% of the total imported food products in Saudi Arabia (SA) were meat-based. India and Brazil are two of the top five countries from where SA imports meat. According to the Saudi Food and Drug Authority, in 2017, at least 562, 280, and 50 samples of imported beef, chicken and sheep meat, respectively, were tested for the presence of E. coli O157:H7. Amongst these, E. coli O157:H7 was detected in respectively 6.80% and 2.20% of the tested beef meat samples imported from India and Brazil as well as in respectively 6.96% and 3.57% of the tested chicken samples imported from Brazil and Ukraine. Moreover, the pathogen was detected in 2.13% of the tested sheep meat samples imported from India. The present report provides evidence that imported meat can serve as the carrier of E. coli O157:H7, which may lead to epidemics within the Kingdom of Saudi Arabia.
Similar content being viewed by others
Introduction
Escherichia coli is a Gram-negative, facultative, anaerobic bacterium considered to be a commensal organism in the human body1. However, the E. coli strain O157:H7 is a pathogen that poses a threat to human life by causing several diseases, such as haemolytic–uraemic syndrome (HUS), which may be fatal in some cases2. The primary reservoir of E. coli O157:H7 is meat, although it has also been isolated from fruits and vegetables3,4. The O157:H7 strain was first detected in 1982. Within only two decades (1982–2002), it has been responsible for 73,000 illnesses annually in the United States alone, causing as many as 350 outbreaks5. Illnesses caused by E. coli O157:H7 have been reported in over 30 countries across six continents6.
Escherichia coli strains that produce Shiga toxins (Stx1 and Stx2) are called Shigatoxigenic E. coli (STEC)7, while those that produce Shiga-like toxins (verotoxins) are called verotoxigenic E. coli (VTEC)8. The pathogenicity of STEC is associated with virulence factors such as enterohaemolysin (encoded by hlyA), intimin (encoded by eae) and Stx1 and Stx2 (encoded by stx1 and stx2)7. STEC isolates are further divided into two groups: O157 and non-O1571. O157 isolates belong to the H7 and NM serogroups, whereas non-O157 isolates belong to the O26, O45, O103, O111, O121, and O145 serogroups1,5. Notably, O157, O26, O103, O111 and O145 are also classified as enterohaemorrhagic E. coli (EHEC)9. Interestingly, a comprehensive E. coli O157:H7 clade-typing study (clades 1–9) of 269 HUS patients and 387 asymptomatic carriers (ACs) in Japan between 1999 and 2011 reported that clades 6 and 8 were frequently found in HUS patients10. Furthermore, the norV gene, which codes a nitric oxide reductase (Shiga toxin inhibitor in anaerobic conditions), was found intact in clade 1–3 isolates but not in clade 4–8 isolates10.
In Saudi Arabia (SA), no E. coli O157:H7 outbreak has been reported to date, and the prevalence of this pathogen remains unknown. However, it has been isolated from several local cattle farms11. Reporting outbreaks in SA is challenging because of its inefficient data collection system12. For this reason, since 2003, the Saudi Food and Drug Authority (SFDA) has taken control of all food safety regulations, which has also helped avoid overlapping with other authorities13. As a member of the Gulf Cooperation Council (GCC), SA is required to apply the GCC Standardization Organization’s (GSO) microbiological criteria for foodstuffs [GSO/1016/2015 (E)] E: referring to the English version14. Accordingly, the SFDA labs follow the GSO 2015 guideline stating that all kinds of food must be free from E. coli O157:H7.
Statistical information on food imported into SA over the past decade is limited. A recent study identified the main source of imported meat only in 201713. Approximately 80% of the food available in Saudi Arabian markets is imported, and 15.71% of it is meat-based13. Therefore, the main aim of this study was to compare imported meat contaminated with E. coli O157:H7 with the total meat imported in 2017. To that end, the study evaluated the possibility of detecting E. coli O157:H7 in meat products imported into SA in 2017 using the SFDA’s monitoring system to provide foundational data for creating a database of the O157:H7 serotype.
Methods
Sample collection
The data used in this study were extracted from the laboratory information management system (LIMS) of the SFDA database, an online tool for data management operated by LabVantage Solutions, Inc. Typically, when shipments of imported consumable meat arrive at Saudi port customs, SFDA inspectors collect samples and send them to SFDA labs for analysis. Thereafter, the inspected samples are referred for E. coli O157:H7 detection. The data used in this study pertained to analyses of raw (not ready-to-eat ‘RTE’) products only. Sample’s specific details can be found in Supplementary Table 1, 2, 3 and 4.
E. coli O157:H7 detection
Enrichment
Samples weighing 25 g selected for enrichment were placed in sterilised sample bags. They were then homogenised with 225 mL of modified tryptone soya broth (mTSB) supplemented with novobiocin to obtain a ratio as follows: mTSB + sample of 1/10 (mass to volume). The sample bags were massaged by hand and then incubated at 41.5 °C for 12–18 h. Escherichia coli O157 strain ATCC 43895 and blank were added as positive and negative controls, respectively. After incubation, the samples were subjected to immunomagnetic separation. Subsequently, 50 µL of each sample was streaked out on pre-dried cefixime tellurite sorbitol MacConkey (CT-SMAC) agar plates using sterile loops to obtain many well-isolated colonies and incubated at 37 °C for 18–24 h.
Colony selection
After incubation, at least five presumptive colonies were selected randomly from each plate and placed into polymerase chain reaction (PCR) tubes containing 10 µL of distilled water (dH2O) as a preparation step for DNA extraction.
DNA extraction
The samples were prepared using a PrepMan™ Ultra Sample Preparation Reagent Kit (lot number 1809191) according to the manufacturer’s protocol.
PCR detection
Real-time PCR (RT-PCR) was performed to amplify the O157:H7-specific target DNA sequences using a MicroSEQ™ E. coli O157:H7 Detection Kit (lot number 1804034) according to the manufacturer’s protocol. Non-pathogenic E. coli ATCC 25922, non-O157 'O111 and O26' and Salmonella strains enteritis and arizona were added as negative controls. A 7500 Fast System and Sequence Detection System (SDS) software v1.4.2 were used for the analysis. Each sample was analysed in triplicate. The thermal cycling conditions are displayed in Supplementary Table 5. International Organization for Standardization (ISO) 17025 (2017) and 13136 (2012) were used in SFDA labs and to isolate E. coli O157:H7, respectively15,16.
Statistical analysis
Statistical analyses were performed using Microsoft Office Excel Professional Plus 2019. For pairwise comparisons, the t-test was used to compare between samples to assess differences in the prevalence of E. coli O157:H7. Values of P < 0.05 were considered statistically significant.
Results
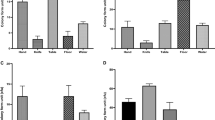
Escherichia coli O157:H7 strains were detected at varying frequencies in imported beef, sheep and chicken meat. The O157:H7 strain was most prevalent in chicken (6.07%) and beef (5.90%), while in sheep (2.00%), with a significant difference (P < 0.05; Table 1).
Regarding chicken, the greatest proportion of samples contaminated with E. coli O157:H7 was imported from Brazil (6.96%), followed by Ukraine (3.57%), while no contaminated samples were imported from Jordan, India, or Tunisia, with a significant difference (P < 0.05). Regarding beef, the greatest proportion of contaminated samples was imported from India (6.80%), followed by Brazil (2.20%). Finally, all sheep meat samples contaminated with E. coli O157:H7 were imported from India (2.1%; Table 1).
The highest frequency of E. coli O157:H7 contamination was found in products imported from Indian companies (30 of 476 samples: eight from company A, five from company B, four from company C, three from company D, two from company E, two from company F and six from other companies; Table 2, Supplementary Table 1, 2 and 3). More beef than sheep meat samples imported from India were screened given the high demand for the former in SA in 201713. Therefore, the prevalence of E. coli O157:H7 in beef samples was higher than in sheep meat samples (6.80% and 2.13%, respectively). Products imported from Brazilian companies were also frequently contaminated (18 of 321 samples: four from company G, two from company H, two from company I, two from company K and eight from other companies; Table 2, Supplementary Table 1, 2 and 3). In this case, however, the prevalence of E. coli O157:H7 in chicken samples was higher than in beef samples (6.96% and 2.20%, respectively). To ensure anonymity, the companies’ names have been replaced with letters A to K.
Discussion
Contaminated raw meat is the source of 90% of foodborne infections17. Thirty-one pathogens, including E. coli O157:H7, were responsible for 10 million annual episodes of foodborne illnesses in the United States4. In the present study, samples of imported raw meat were obtained from imported meats in the ports of SA, and the prevalence of E. coli O157:H7 in these samples was confirmed (Table 1). Meat products imported from India and Brazil were the most frequently contaminated (Table 1).
The prevalence of E. coli O157:H7 was the highest in raw meat products imported from India, posing a threat to public health in the Kingdom of Saudi Arabia (Table 2). According to Shinde et al. (2020), E. coli O157:H7 was frequently isolated from healthy Indian cattle on both organised and non-organised farms in and around the Pune District in India during 2015. This can be explained by the fact that new generations of cattle may carry the pathogen but may not present any symptoms, thus appearing as heathy livestock; however, the consumption of meat from such asymptomatic carriers of E. coli O157:H7 may affect humans, representing a severe public health concern. Furthermore, subsequent studies in the same region revealed the presence of E. coli O157:H7 isolates resistant to a number of common antibiotics used for livestock animals against this pathogen, including cefotaxime, streptomycin, penicillin G, kanamycin, ampicillin, tetracycline, gentamycin and piperacillin. These findings, in addition to our results, emphasise the need for the further of assessment of imported meat, specifically from India, to ensure public health safety. In another recent study in China, clinical isolates of E. coli exhibited high resistance to conventional antibiotics for livestock, including sulfamethoxazole, trimethoprim/sulfamethoxazole, tetracycline, nalidixic acid and ampicillin18.
Amongst samples of meat imported from Brazil, E. coli O157:H7 was detected at different frequencies in products from several companies (Table 2). The prevalence of E. coli O157:H7 in samples from only specific companies (G, H, I, J, K and others), but not others, indicates internal contamination through air during rearing at the livestock farms19, slaughter20, or processing5 (Fig. 1). According to Santos et al. (2018), the prevalence of STEC in Brazilian food products was approximately 9.50%, which was primarily attributed to the development of multi-resistance to antibiotics in these strains. Notably, Brazil is the second largest exporter and the third major producer of beef worldwide1.
The detection of E. coli O157:H7 in samples of meat imported from one company each in Ukraine and UAE also indicates unhygienic handling that led to contamination (Table 2), highlighting the need for the revision of processing and packaging steps in these regions5.
Of note, the present report only includes results from products that have been undergone E. coli O157:H7 testing from the port of SA. Many shipments may have been excluded from the examination for approval and owners may have only been asked to produce a list of essential documents13. In addition, to import food products into SA, the SFDA mandates a registration certificate authorised by the Saudi health ministry, an industry certificate authorised by the commerce ministry and a quality certificate (e.g. International Organization for Standardization 9001 or 22000, Good Manufacturing Practice and Hazard Analysis Critical Control Point)13. Therefore, to ensure public safety, the SFDA has announced a list of countries from where the import of food into SA is prohibited (available at https://www.sfda.gov.sa/en/list_countries).
Summery
The presence of E. coli O157:H7 in samples of imported raw meat highlights the need for more regular surveillance at the borders of SA before the products are made available on the market for consumption by the public. Our results underscore the necessity of more stringent control protocols for the approval of imported food products, particularly from India and Brazil, which are the major suppliers of meat to SA. Moreover, the detected E. coli O157:H7 isolates should be tested against antibiotics that are commonly used to treat livestock. For the future investigations and as an alternative method, we suggest tracking different sources of E. coli O157 contaminations by clade typing10.
Data availability
The All data analysed are reported in this manuscript, and specific reference numbers of samples at SFDA database are listed in supplementary 1, 2 and 3. There is no other data to be provided.
Abbreviations
- SA:
-
Saudi Arabia
- SFDA:
-
Saudi Food and Drug Authority
- HUS:
-
Haemolytic–uremic syndrome
- STEC:
-
Shigatoxigenic E. coli
- VTEC:
-
Verotoxigenic E. coli
- EHEC:
-
Enterohaemorrhagic E. coli
- GCC:
-
Gulf Cooperation Council
- GSO:
-
GCC Standardization Organisation
- mTSB:
-
Modified Tryptone Soya Broth
- ISO:
-
International Organization for Standardization
- CT-SMAC:
-
Cefixime tellurite sorbitol MacConkey agar
- dH2O:
-
Distilled water
- RT-PCR:
-
Real-time polymerase chain reaction
- SDS:
-
Sequence detection system
- LIMS:
-
Laboratory information management system
References
dos Santos, E. C. C. et al. Escherichia coli O26 and O113: H21 on carcasses and beef from a slaughterhouse located in Mato Grosso. Brazil. Foodborne Pathog. Dis. 15, 653–659 (2018).
Luna, S. et al. Outbreak of E. coli O157: H7 infections associated with exposure to animal manure in a rural community—Arizona and Utah, June–July 2017. Morb. Mortal. Wkly. Rep. 67, 659 (2018).
Haramain, S. E. & Yagoub, S. O. The antimicrobial susceptibility and detection of virulence genes of Escherichia coli O157: H7 isolated from leafy green vegetables. Eur. J. Biol. Biotechnol. 2, 70–74 (2021).
Riley, L. W. Extraintestinal foodborne pathogens. Annu. Rev. Food Sci. Technol. 11, 275–294 (2020).
Rangel, J. M., Sparling, P. H., Crowe, C., Griffin, P. M. & Swerdlow, D. L. Epidemiology of Escherichia coli O157: H7 outbreaks, united states, 1982–2002. Emerg. Infect. Dis. 11, 603 (2005).
Rahal, E. A., Kazzi, N., Nassar, F. J. & Matar, G. M. Escherichia coli O157: H7—Clinical aspects and novel treatment approaches. Front. Cell. Infect. Microbiol. 2, 138 (2012).
Mesele, F. & Abunna, F. Escherichia coli O157: H7 in foods of animal origin and its food safety implications. Adv. Biol. Res. (Rennes) 13, 134–145 (2019).
Shinde, D. B., Singhvi, S., Koratkar, S. S. & Saroj, S. D. Isolation and characterization of Escherichia coli serotype O157: H7 and other verotoxin-producing E. coli in healthy Indian cattle. Vet. World 13, 2269 (2020).
Beutin, L., Jahn, S. & Fach, P. Evaluation of the ‘GeneDisc’real-time PCR system for detection of enterohaemorrhagic Escherichia coli (EHEC) O26, O103, O111, O145 and O157 strains according to their virulence markers and their O-and H-antigen-associated genes. J. Appl. Microbiol. 106, 1122–1132 (2009).
Iyoda, S. et al. Phylogenetic clades 6 and 8 of enterohemorrhagic Escherichia coli O157: H7 with particular stx subtypes are more frequently found in isolates from hemolytic uremic syndrome patients than from asymptomatic carriers. In Open Forum Infectious Diseases vol. 1 ofu061 (2014).
Bosilevac, J. M. et al. Prevalence of Escherichia coli O157: H7 and Salmonella in camels, cattle, goats, and sheep harvested for meat in Riyadh. J. Food Prot. 78, 89–96 (2015).
Al-Mazrous, Y. Y. Food poisoning in Saudi Arabia. Saudi Med. J 25, 11–14 (2004).
Alrobaish, W. S., Vlerick, P., Luning, P. A. & Jacxsens, L. Food safety governance in Saudi Arabia: Challenges in control of imported food. J. Food Sci. 86, 16–30 (2021).
GSO. Microbiological criteria for foodstuffs. GCC Standardization Organization (2015).
ISO 13136. Microbiology of food and animal feed—Real-time polymerase chain reaction (PCR)-based method for the detection of food-borne pathogens—Horizontal method for the detection of Shiga toxin-producing Escherichia coli (STEC) and the determination of O157, O111, O26, O103 and O145 serogroups. The International Organization for Standardization (2012).
ISO 17025. General requirements for the competence of testing and calibration laboratories. The International Organization for Standardization (2017).
Hadi, A.-A.J. & Jabbar, A.-K.N. Isolation and identification of zoonotic importance bacteria from meat, meat products and Human in Diyala. Iraq. Int. J. plant Res. 20, 1 (2020).
Yassin, A. K. et al. Antimicrobial resistance in clinical Escherichia coli isolates from poultry and livestock. China. PLoS One 12, e0185326 (2017).
Chmielowiec-Korzeniowska, A., Trawińska, B., Tymczyna, L., Bis-Wencel, H. & Matuszewski, Ł. Microbial contamination of the air in livestock buildings as a threat to human and animal health–a review. Ann. Anim. Sci. 21, 417–431 (2021).
Irshaid, F. I., Jacob, J. H. & Alhawamdeh, S. I. The Slaughtering and Dressing Procedures of Livestock Inside the Butcher Shops Generate High Levels of Bacterial Contamination. J. Basic Appl. Sci. 14, 165–173 (2018).
Acknowledgements
We thank Dr. Abdullah Alowaifeer (Chief Executive of Reference Laboratories and Director of Chemistry Reference Laboratory at SFDA) for his contribution in data analysis, and Mr. Abdullah Alajlan (Section Head of Microbiology Department) for the useful discussions and advice on data presentation, and Mr. Fahad Allabidi for transforming the report data from SFDA system to be readable tables.
Disclaimer
The views expressed in this paper are those of the author(s) and not do not necessarily reflect those of the SFDA or its stakeholders. Guaranteeing the accuracy and the validity of the data is a sole responsibility of the research team.
Author information
Authors and Affiliations
Contributions
M.A.A. and S. M. I. A. conceived the study. M.A.A., S. M. I. A. and M. I. M. collected and analysed the data. M.A.A. drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alhadlaq, M.A., Mujallad, M.I. & Alajel, S.M.I. Detection of Escherichia coli O157:H7 in imported meat products from Saudi Arabian ports in 2017. Sci Rep 13, 4222 (2023). https://doi.org/10.1038/s41598-023-30486-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-30486-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.