Abstract
The aim of this study was to explore the feasibility of Raman spectroscopy combined with computer algorithms in the diagnosis of primary Sjögren syndrome (pSS). In this study, Raman spectra of 60 serum samples were acquired from 30 patients with pSS and 30 healthy controls (HCs). The means and standard deviations of the raw spectra of patients with pSS and HCs were calculated. Spectral features were assigned based on the literature. Principal component analysis (PCA) was used to extract the spectral features. Then, a particle swarm optimization (PSO)-support vector machine (SVM) was selected as the method of parameter optimization to rapidly classify patients with pSS and HCs. In this study, the SVM algorithm was used as the classification model, and the radial basis kernel function was selected as the kernel function. In addition, the PSO algorithm was used to establish a model for the parameter optimization method. The training set and test set were randomly divided at a ratio of 7:3. After PCA dimension reduction, the specificity, sensitivity and accuracy of the PSO-SVM model were obtained, and the results were 88.89%, 100% and 94.44%, respectively. This study showed that the combination of Raman spectroscopy and a support vector machine algorithm could be used as an effective pSS diagnosis method with broad application value.
Similar content being viewed by others
Introduction
Primary Sjögren syndrome (pSS) is a chronic systemic autoimmune disease that primarily affects the exocrine glands, particularly the lacrimal and salivary glands, resulting in symptoms of dry eyes and dry mouth. It is sometimes accompanied by systemic features affecting extraglandular sites such as the joints, blood, kidneys, lungs, vessels, and nerves1. Due to its systemic involvement, pSS can present a variety of clinical manifestations that lead to confusion and delay in diagnosis. Moreover, definitive diagnosis of pSS mainly depends on the clinical manifestations, specific immunological changes, and other special examinations, such as dry eye examination, labial gland biopsy, and parotid gland tomography2. Early multiple examinations for pSS lead to a cumbersome diagnostic process that is costly and complex, and invasive tests such as labial gland biopsy have limitations in clinical application and are difficult to repeat. Therefore, a rapid, efficient and convenient method based on serum is more suitable.
Recently, Raman spectroscopy combined with machine learning algorithms has provided a more rapid and efficient method for the early diagnosis of many diseases3,4,5. Raman spectroscopy is an optical spectroscopic technique based on the inelastic scattering of light. It can be used to detect biological macromolecules, including proteins, lipids, and DNA, in biological samples and provides abundant molecular information at the microscopic level6,7. Therefore, Raman spectroscopy is commonly used in biomolecular detection. Raman spectroscopy can be used to detect changes in diseases at the biomolecular level and aid in the early diagnosis of diseases. It has been widely used in the early screening of Alzheimer's disease8, meningioma9, dengue virus infection5, cervical cancer10, oral cancers11, and so on.
Due to the high dimensionality of spectral data, redundant interference occurs, reducing the accuracy of the model. Therefore, we used the PCA dimensionality reduction method for feature selection to improve the accuracy of the model. This experiment was based on the serum Raman spectrum combined with the support vector machine algorithm, the radial basis kernel function was selected as the kernel function, and the PSO algorithm was the parameter optimization method. Finally, the PSO-SVM classification model was established. Through the classification results of the model, the feasibility of Raman spectroscopy combined with a support vector machine algorithm for the rapid detection of pSS patients and healthy controls (HCs) was verified.
Methods
Patient selection
Thirty patients with pSS who met the American–European classification criteria (AECG) and 30 healthy controls were enrolled in this study. Patients with other autoimmune diseases, malignant tumors, or active infections were excluded from this study. A signed consent form was obtained from all patients. The study was approved by the ethics committee of the People's Hospital of Xinjiang Uygur Autonomous Region.
Sample preparation
Three milliliters of whole blood was collected into tubes without any anticoagulant and centrifuged at 1500g for 10 min to isolate the serum. The serum was then collected into EP tubes and frozen at − 80 °C until detection by Raman spectroscopy. For each measurement, approximately 15 µL of the serum sample was prepared in a quartz cuvette.
Raman spectral data acquisition
All Raman spectra were recorded using a Raman spectrometer (LabRAM HR Evolution Raman Spectrometer, HORIBA Scientific Ltd.) in the range of 400–4000 cm−1. An Ar+ laser with a wavelength of 532 nm and power of 50 mW was used for Raman excitation. Spectra were acquired using a 10 × objective within 3 s. Three spectra per location were recorded in the wavenumber interval of 400–4000 cm−1. To exclude experimental interference and artifactual errors, three Raman spectra of each sample were recorded at different positions in the same plane.
Algorithm description
Support vector machine (SVM) is a powerful supervised learning method capable of transforming data into a high-dimensional space for classification problems12. The SVM algorithm can be used to analyze data from small samples and data with high dimensions. It not only has a good nonlinear fitting ability and high generalization but also has the advantages of obtaining a global optimum through the objective function. At present, SVM has been widely used in the detection of diseases such as diabetes, breast cancer, and lung cancer13,14,15. In the process of SVM modeling, it is more important to choose the appropriate C and g parameters. At the same time, the application of the kernel function can also improve the performance of SVM. Separate collections. In this study, the radial basis kernel function was chosen as the kernel function.
PSO is an evolutionary computation technique for solving optimization problems16. The core idea of PSO is to find the optimal solution through collaboration and information sharing among individuals in the group. PSO was originally developed by Kennedy and Eberhart. It was inspired by research on bird and fish flock movement behaviors. First, a population of random particles is initialized, and then the system is updated at each iteration through searches for the optimal solution. PSO has the advantages of simple operation and a small amount of calculation, which can further reduce the time for optimizing parameters17. PSO-SVM has the advantages of a strong learning ability and sensitivity to small sample data and is widely used in machine learning methods. Therefore, in this study, the PSO-SVM algorithm was used to build a diagnostic model to achieve a rapid distinction between patients with pSS and HCs.
Data analysis
Raman spectra were normalized to [0,1] by the "mapminmax" function in MATLAB r2018a. The normalization process can reduce the effect of laser power fluctuation on the sample data18. To improve the diagnostic accuracy and efficiency of SVM, PCA was used to characterize the serum Raman spectra.
All algorithms were implemented in MATLAB r2018a. SVM classification analysis was performed using the libsvm toolbox created by Lin and Chang.
Informed consent
This study was approved by the ethics committee of the People's Hospital of Xinjiang Uygur Autonomous Region (in these studies). Informed consent was obtained from all participants before participating in the interview study. All methods were carried out in accordance with relevant guidelines and regulations (e.g., Helsinki guidelines).
Results
Spectral comparison
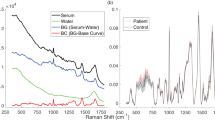
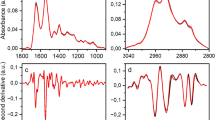
The means of the raw spectra of patients with pSS and HCs were calculated (Fig. 1). A comparison of the Raman spectra showed that five peaks [proline (959 cm−1), phenylalanine (1003 cm−1), carotenoids (1155 cm−1), tryptophan (1355 cm−1), and beta-carotene (1514 cm−1)] were different between patients with pSS and HCs. The spectral features of these substances were assigned based on the available literature (Table 1)19,20,21. Compared to those of HCs, the Raman peak intensities of proline, phenylalanine, carotenoids, tryptophan and beta-carotene were lower in patients with pSS.
Feature extraction
After feature extraction, principal component analysis (PCA) was used for dimensionality reduction. PCs with an overall contribution of 90% will generally be retained. In this study, the total contribution of these 29 PCs was 99.99%. In addition, the most significant three PCs were extracted to plot the principal component scatter plots of the training and test sets (Fig. 2). There is a degree of variation between patients with pSS and healthy controls.
Model evaluation
Forty-two samples (21 from patients with pSS and 21 from HCs) were randomly selected as the training set, and 18 samples (9 from patients with pSS and 9 from HCs) were selected as the test set. Classification of pSS patients and HCs was performed using an SVM classifier. In the SVM model, PSO-SVM was employed to optimize the penalty parameter C and Gaussian width g. In PSO-SVM, the search range of C was set to [2−8,28], the search range of g was set to [2−8,28], and the size step was set to 0.3. The PSO parameter local search ability was set to 1.5, and the overall search ability was set to 1.7; the maximum evolutionary number (MaxGen) was set to 200, and the maximum population size (sizepop) was 20. The radial basis function (RBF) was chosen as the kernel function for the SVM. The accuracy, sensitivity, and specificity of the PSO-SVM classification model were 88.89%, 100%, and 94.44%, respectively. In addition, to further illustrate the classification capability of the model, as shown in Table 2, we used the confusion matrix to evaluate the performance of the PSO-SVM algorithm.
Discussion
pSS is a chronic systemic autoimmune disease characterized by lymphocyte proliferation and progressive exocrine gland damage22. Since the onset of dry syndrome is insidious, the clinical manifestations of patients are different, and the severity of the disease also varies greatly, so the early and clear diagnosis of the disease has important clinical significance to improve the prognosis of patients. However, because the pathogenesis of primary Sjögren syndrome is not yet completely clear, there is still no clear diagnostic standard, and the diagnostic standard used in clinical practice is actually a classification standard23. Therefore, the diagnosis of pSS needs to be confirmed by experienced specialists to prevent a large number of missed diagnoses and misdiagnoses. In addition, the main method to diagnose pSS is through a labial gland biopsy, but this method is invasive and less accepted by patients. Moreover, a labial gland biopsy has certain limitations, and the results are often inconsistent with clinical manifestations and laboratory test results in the early stage of the disease. In addition, the prevalence of pSS is as high as 3–4% in the elderly population, and these patients are often unable to tolerate a labial biopsy. Parotid angiography, parotid ultrasound and MRI are also helpful in the diagnosis of pSS24, but they are not included in the guidelines due to the lack of standardization of these testing techniques. Therefore, the search for new, rapid and noninvasive tests has been a hot research topic in this field.
Raman spectroscopy is a vibrational spectroscopy technique based on the Raman scattering principle25. Relevant studies have demonstrated the feasibility of Raman spectroscopy in different disease fields, and achieved high accuracy in many diagnoses26,27,28. Li M et al. provided a non-invasive and rapid technology for the screening of gastric cancer patients based on serum Raman spectroscopy combined with one-dimensional convolutional neural network, random forest and other machine learning methods29. Hyunku Shin et al. used a variety of deep learning algorithms combined with surface-enhanced Raman spectroscopy (SERS) to achieve early diagnosis of lung cancer and achieved good results30. Similarly, in this exploratory study, we demonstrated that Raman spectroscopy techniques combined with support vector machine algorithms can be used as an effective diagnostic method for pSS. Furthermore, we found that Raman spectroscopy can detect changes in biomolecular composition induced by pathological changes occurring between pSS and HCs, which was consistent with previous study31. In the experiment, due to the weak Raman signal in the detection, it is easily interfered by the fluorescent background, and the signal-to-noise ratio of the spectrum is low, which makes it difficult to distinguish different types of molecular spectral information32,33. Therefore, we need to use advanced pattern recognition algorithms to improve the classification accuracy. Principal component analysis (PCA), an unsupervised feature extraction algorithm that can reduce the dimensionality of Raman spectral data34, has been widely used by many researchers for the extraction of Raman spectral features. Similarly, pattern recognition requires powerful classifiers. In recent years, support vector machine (SVM) have been widely used in the field of pattern recognition with obvious effects. In this study, particle swarm optimization (PSO)-SVM was selected as the method of parameter optimization to rapidly classify patients with pSS and healthy controls.
In this study, we found some differences in the Raman spectra of serum from patients with pSS and HCs. Compared to those in HCs, the proline, carotenoids, and tryptophan peaks were of lower intensity in patients with pSS. This may indicate that pSS patients experience metabolic changes that result in less proline, carotenoids, and tryptophan than HCs. Studies have shown that metabolic levels of proline and tryptophan are significantly altered by the effects of pSS35. And carotenoids can be converted into vitamin A36, which in appropriate concentrations can in turn improve the immune function of cells37. The main etiology of pSS is associated with abnormal immune function38, which represents a possible deficiency of vitamin A in patients with pSS. Based on the differences in serum spectra, the accuracy rate of the PSO-SVM classification model reached 94.44%. Thus, it can be used to rapidly discriminate patients with pSS and HCs. As there was a limited sample size in this study, we plan to collect more samples to validate the results of this exploratory experiment in the future and evaluate the effect of serum Raman spectroscopy for pSS diagnosis.
Conclusion
In this study, we used Raman spectroscopy combined with the PSO-SVM algorithm to rapidly diagnose pSS based on serum samples obtained from pSS patients and healthy controls. The spectral data were reduced using PCA, and the first 29 PCs were taken as input. Through the evaluation metrics of the model, we found that PSO-SVM performed stably, with model specificity, sensitivity and accuracy results of 88.89%, 100% and 94.44%, respectively. This study showed that Raman spectroscopy combined with a support vector machine algorithm could be used as an effective pSS diagnosis method.
Data availability
The datasets generated and analyzed during the current study are not publicly available due to data privacy laws but are available from the corresponding author on reasonable request.
References
Vitali, C., Minniti, A., Pignataro, F., Maglione, W. & Del Papa, N. Management of Sjögren’s syndrome: Present issues and future perspectives. Front. Med. 8, 676885 (2021).
Del Papa, N. & Vitali, C. Management of primary Sjögren’s syndrome: Recent developments and new classification criteria. Ther. Adv. Musculoskelet. Dis. 10(2), 39 (2018).
Parlatan, U. et al. Raman spectroscopy as a non-invasive diagnostic technique for endometriosis. Sci. Rep. 9(1), 1–7 (2019).
Zheng, X. et al. Rapid and low-cost detection of thyroid dysfunction using Raman spectroscopy and an improved support vector machine. IEEE Photonics J. 10(6), 1–12 (2018).
Khan, S. et al. Analysis of dengue infection based on Raman spectroscopy and support vector machine (SVM). Biomed. Opt. Express 7(6), 2249–2256 (2016).
Butler, H. J. et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 11(4), 664–687 (2016).
Paidi, S. K. et al. Raman spectroscopy and machine learning reveals early tumor microenvironmental changes induced by immunotherapy. Cancer Res. 81(22), 5745–5755 (2021).
Ryzhikova, E. et al. Raman spectroscopy of blood serum for Alzheimer’s disease diagnostics: Specificity relative to other types of dementia. J. Biophotonics 8(7), 584–596 (2015).
Mehta, K., Atak, A., Sahu, A. & Srivastava, S. An early investigative serum Raman spectroscopy study of meningioma. Analyst 143(8), 1916–1923 (2018).
Rubina, S., Amita, M., Bharat, R. & Krishna, C. M. Raman spectroscopic study on classification of cervical cell specimens. Vib. Spectrosc. 68, 115–121 (2013).
Sahu, A., Sawant, S., Mamgain, H. & Krishna, C. M. Raman spectroscopy of serum: An exploratory study for detection of oral cancers. Analyst 138(14), 4161–4174 (2013).
Long, Y. et al. PSO-SVM-based online locomotion mode identification for rehabilitation robotic exoskeletons. Sensors 16(9), 1408 (2016).
Thaiyalnayaki, K. Classification of diabetes using deep learning and SVM techniques. Int. J. Curr. Res. Rev. 13(01), 146 (2021).
Vijayarajeswari, R., Parthasarathy, P., Vivekanandan, S. & Basha, A. A. Classification of mammogram for early detection of breast cancer using SVM classifier and Hough transform. Measurement 146, 800–805 (2019).
Nanglia, P., Kumar, S., Mahajan, A. N., Singh, P. & Rathee, D. A hybrid algorithm for lung cancer classification using SVM and neural networks. ICT Express 7(3), 335–341 (2021).
Shen, L., Huang, X. & Fan, C. Double-group particle swarm optimization and its application in remote sensing image segmentation. Sensors 18(5), 1393 (2018).
Zhao, S. & Zhao, Z. A comparative study of landslide susceptibility mapping using SVM and PSO-SVM models based on Grid and Slope Units. Math. Probl. Eng. 2021, 1–15 (2021).
Martyna, A. et al. Improving discrimination of Raman spectra by optimising preprocessing strategies on the basis of the ability to refine the relationship between variance components. Chemom. Intell. Lab. Syst. 202, 104029 (2020).
Li, X. et al. Different classification algorithms and serum surface enhanced Raman spectroscopy for noninvasive discrimination of gastric diseases. J. Raman Spectrosc. 47(8), 917–925 (2016).
Xiao, R. et al. Non-invasive detection of hepatocellular carcinoma serum metabolic profile through surface-enhanced Raman spectroscopy. Nanomed. Nanotechnol. Biol. Med. 12(8), 2475–2484 (2016).
Movasaghi, Z., Rehman, S. & Rehman, I. U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 42(5), 493–541 (2007).
Zhao, J. et al. Research status and future prospects of extracellular vesicles in primary Sjögren’s syndrome. Stem Cell Res. Ther. 13(1), 1–11 (2022).
Vitali, C. et al. Classification criteria for Sjögren’s syndrome: A revised version of the European criteria proposed by the American-European Consensus Group. Ann. Rheum. Dis. 61(6), 554–558. https://doi.org/10.1136/ard.61.6.554 (2002).
Knopf, A., Mansour, N., Chaker, A., Bas, M. & Stock, K. Multimodal ultrasonographic characterisation of parotid gland lesions—A pilot study. Eur. J. Radiol. 81(11), 3300–3305 (2012).
Lussier, F., Thibault, V., Charron, B., Wallace, G. Q. & Masson, J.-F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. TrAC Trends Anal. Chem. 124, 115796 (2020).
Ma, D. et al. Classifying breast cancer tissue by Raman spectroscopy with one-dimensional convolutional neural network. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 256, 119732 (2021).
Hong, Y. et al. Label-free diagnosis for colorectal cancer through coffee ring-assisted surface-enhanced Raman spectroscopy on blood serum. J. Biophotonics 13(4), e201960176 (2020).
Koster, H. J. et al. Fused Raman spectroscopic analysis of blood and saliva delivers high accuracy for head and neck cancer diagnostics. Sci. Rep. 12(1), 18464 (2022).
Li, M. et al. A novel and rapid serum detection technology for non-invasive screening of gastric cancer based on Raman spectroscopy combined with different machine learning methods. Front. Oncol. 11, 665176 (2021).
Shin, H. et al. Early-stage lung cancer diagnosis by deep learning-based spectroscopic analysis of circulating exosomes. ACS Nano 14(5), 5435–5444 (2020).
Xue, L. et al. Diagnosis of pathological minor salivary glands in primary Sjogren’s syndrome by using Raman spectroscopy. Lasers Med. Sci. 29, 723–728 (2014).
Xia, L. et al. Identifying benign and malignant thyroid nodules based on blood serum surface-enhanced Raman spectroscopy. Nanomed. Nanotechnol. Biol. Med. 32, 102328 (2021).
Meng, C. et al. Serum Raman spectroscopy combined with Gaussian—Convolutional neural network models to quickly detect liver cancer patients. Spectrosc. Lett. 55(2), 79–90 (2022).
Vrábel, J., Pořízka, P. & Kaiser, J. Restricted Boltzmann machine method for dimensionality reduction of large spectroscopic data. Spectrochim. Acta Part B 167, 105849 (2020).
Fernández-Ochoa, Á. et al. Discovering new metabolite alterations in primary Sjögren’s syndrome in urinary and plasma samples using an HPLC-ESI-QTOF-MS methodology. J. Pharm. Biomed. Anal. 179, 112999 (2020).
Meléndez-Martínez, A. J. An overview of carotenoids, apocarotenoids, and vitamin A in agro-food, nutrition, health, and disease. Mol. Nutr. Food Res. 63(15), 1801045 (2019).
Fernandes, G. Beta-carotene supplementation: Friend or foe?. J. Lab. Clin. Med. 129(3), 285–287 (1997).
Nocturne, G. & Mariette, X. Advances in understanding the pathogenesis of primary Sjögren’s syndrome. Nat. Rev. Rheumatol. 9(9), 544–556 (2013).
Acknowledgements
This work was supported by the Key Research and Development Project of Xinjiang Uygur Autonomous Region (2022B03002-1), the Distinguished Young Talents Project of Natural Science Foundation of Xinjiang Uygur Autonomous Region (2022D01E11), Xinjiang Uygur Autonomous Region Youth Science Foundation Project(2022D01C695) and Tianshan Talent-Young Science and Technology Talent Project(NO.2022TSYCJC0033 and 2022TSYCCX0060).
Author information
Authors and Affiliations
Contributions
X.C. and X.W. were responsible for experimental design and part of the paper writing, C.C. and C.L. were responsible for literature research, Y.S. and Z.L. were responsible for part of the paper writing and experimental design, X.L. and C.C. were responsible for experimental design guidance, and L.W. and J.S. were responsible for experimental design and writing guidance.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, X., Wu, X., Chen, C. et al. Raman spectroscopy combined with a support vector machine algorithm as a diagnostic technique for primary Sjögren’s syndrome. Sci Rep 13, 5137 (2023). https://doi.org/10.1038/s41598-023-29943-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29943-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.