Abstract
Tissue segmentation of histology whole-slide images (WSI) remains a critical task in automated digital pathology workflows for both accurate disease diagnosis and deep phenotyping for research purposes. This is especially challenging when the tissue structure of biospecimens is relatively porous and heterogeneous, such as for atherosclerotic plaques. In this study, we developed a unique approach called ‘EntropyMasker’ based on image entropy to tackle the fore- and background segmentation (masking) task in histology WSI. We evaluated our method on 97 high-resolution WSI of human carotid atherosclerotic plaques in the Athero-Express Biobank Study, constituting hematoxylin and eosin and 8 other staining types. Using multiple benchmarking metrics, we compared our method with four widely used segmentation methods: Otsu’s method, Adaptive mean, Adaptive Gaussian and slideMask and observed that our method had the highest sensitivity and Jaccard similarity index. We envision EntropyMasker to fill an important gap in WSI preprocessing, machine learning image analysis pipelines, and enable disease phenotyping beyond the field of atherosclerosis.
Similar content being viewed by others
Introduction
Atherosclerosis is a chronic inflammatory process resulting in arterial stiffening and plaque formation, and is the leading cause of myocardial infarction, ischemic stroke, and peripheral artery disease1,2. Historically, researchers and pathologists have characterized atherosclerotic plaque through standard histology and light microscopy analysis3,4,5,6. The composition of atherosclerotic plaques is highly variable, with different plaque types having distinct clinical manifestations3,7. For instance, more stable, fibrous-rich atheroma plaques are typically asymptomatic, whereas unstable, thin-cap fibroatheroma plaques are more prone to rupture and thrombus formation underlying cerebral or coronary events5,6. However, the value of atherosclerotic plaque composition in predicting cardiovascular events remains a subject of debate and ongoing research7,8,9.
Histological analysis of atherosclerosis is mainly used in the research setting and can reveal the extent of disease progression as well as underlying etiology. For example, in pre-clinical models of atherosclerosis, the amount of smooth muscle content and collagen extracellular matrix in the plaque often correlates with greater plaque stability8. These features can now be easily captured from stained tissue sections and digitized as high-resolution whole-slide images (WSI). WSI data provides a rich resource for quantitative and qualitative image analysis and has been a focus of digital pathology10. Overall, this has simplified archiving, enabled remote diagnosis, and accelerated both clinical decision-making and research investigations11.
Accurately identifying tissue (foreground) from background is a necessary initial step in WSI analysis in digital pathology workflows. However, the process of WSI acquisition has its own technical challenges. It is often challenging to identify or "mask" the entire non-tissue region from WSI, e.g., due to factors such as dust, translucent tissue, or weak immunostaining, which may lead to incorrect diagnosis12. Since rescanning and manually inspecting slides is labor-intensive, automated algorithms that highlight the tissue-specific areas to be analyzed is a more efficient solution13. These algorithms also adjust for inter- and intra-image aberrations in color, staining and slide preparation through more robust tissue segmentation.
The most straightforward method for identifying tissue on a white background is to set a specified threshold for the grayscale version of the image14, specific color channels15, or the optical density of the red, green, and blue (RGB) color channels16. Alternatively, tissue can be identified from WSIs split into a uniform patch grid that is subjected to grayscale image threshold17,18. Otsu's thresholding19 is a commonly used tissue masking method in WSI analysis studies13,20,21. Several image segmentation methods have been developed to extend Otsu’s method and improve foreground extraction from WSI, such as GrabCut22 and Foreground Extraction from Structural Information (FESI)23. While these methods are easy to implement, it has been difficult to generalize to more variable stains or clinical specimens. A more accurate approach is still needed in digital pathology workflows, especially for specimens containing small empty spaces between extracellular matrix fibers (e.g., collagen) or due to decalcifying procedures, and transparent tissue (e.g., adipose or lipid droplets) in atherosclerotic plaques. Image thresholding techniques based on entropy maximization have been routinely applied to infrared image analysis24,25,26. Similar to infrared images with poor contrast and noise, WSI may also benefit from entropy thresholding to distinguish tissues from the slide background; to our knowledge this approach has not yet been applied to WSI.
In this paper, we propose a fully automated approach for separating foreground (tissue) and background in WSI of HE-stained atherosclerotic plaque samples. Our method, EntropyMasker, is unaffected by changes in scanning or image processing conditions, by using a measure of local entropy and generating corresponding binary tissue masks. This allows for increased scalability across various tissue contexts and implementation in both machine learning and computer vision cell segmentation pipelines, e.g., using CellProfiler27 and slideToolKit28.
Methods
Our method: EntropyMasker
For WSI processing, ideally the slide background is evenly achromatic, with the background mostly one color, e.g., black (fluorescence) or white (brightfield). Unfortunately, in practice, there is often inconsistency of the background color due to variations in image capturing color temperature (e.g., from shades of blue to yellow), resulting in edge artifacts from tile-based scanning methods29. Therefore, the slide background cannot be easily masked by transforming the images from RGB color space to the HSV (Hue-Saturation-Value) and excluding the white color (0°, 0%, 100%).
In information theory30, entropy is defined as the log-base-2 of the number of possible outcomes for a sent/received message. In image analysis, entropy refers to the degree of randomness or complexity of pixels in a defined region or neighborhood of pixels. We defined the local entropy of a specific region as follows:
where \(p_{F} \left( i \right)\) represents the probability of a grayscale pixel, \(i\), in a local footprint region.
In the case of \(p_{F} \left( i \right) = 0\) for some \(i\), the corresponding sum value and \(0log_{2} 0\) is set to \(0\), which is consistent with the limit: \(\mathop {lim}\limits_{{p_{F} \to 0^{ + } }} p_{F} \log \left( {p_{F} } \right) = 0\).
Based on this approach, homogeneous pixels within local neighborhood regions (disk-shaped footprints with radius of 5) have a low probability and contribute less to the local entropy, whereas heterogeneous pixels within local neighborhood regions have a high probability and contribute more to the local entropy. Also, the background of an image has lower entropy due to higher brightness and more homogeneous texture, whereas foreground tissue has higher entropy due to the diversity of pixel intensities in the local neighborhood. In other words, the heterogenous pixels in tissue with high entropy are more informative than homogenous background pixels with low entropy31.
The purpose is to extract the tissue (foreground) from the background. We first calculated local entropies and then we made a histogram of all local entropies. Since the background of the image has a lower entropy value, we first defined the threshold as the local minima of the histogram of the entropy map. Next, we applied this threshold to convert the local entropy map into a binary map which distinguishes background noise from tissue (foreground) in the WSI.
Otsu’s method
Otsu's method is a clustering-based image thresholding algorithm19, commonly utilized in image analysis and digital histopathology applications20,29,32,33,34. Following a bimodal histogram, the technique assumes that the image has two classes of pixels (tissue pixels and background pixels). It determines the best threshold for dividing the two groups such that their total intra-class variance is as low as possible.
Adaptive thresholding
Adaptive thresholding is another widely used method for calculating the threshold value for smaller areas of WSIs35,36,37,38. Typically, these threshold values are calculated in two different ways, either using the mean of the neighborhood area, or a Gaussian weighted sum of neighborhood values.
slideMask method from slideToolKit
The slideToolKit28 is an assistive tool set for the histological quantification of whole-slide images (https://github.com/swvanderlaan/slideToolKit). The slideMask tool in slideToolKit uses convert from ImageMagick (https://imagemagick.org) and a miniature version of the WSI to generate a mask (https://github.com/swvanderlaan/slideToolKit/blob/master/slideMask). The image is first blurred to eliminate dust and speckles. Then Fuzz in ImageMagick39 (which is computed as the root mean squared difference between two colors of the pixels) is used to create a fuzzy, non-stringent selection to identify the white background, which is subsequently substituted with black. The slideMask tool has settings for blur and fuzziness that may be explored and altered. Manual adjustments may be made to generated masks in any image editor (such as free GNU Image Manipulation Program; GIMP38,40). Unwanted regions on the WSI (such as marker stripes or air bubbles beneath the coverslip) may necessitate this procedure.
All proposed methods were implemented in the Python programming language using OpenCV41 and other required packages (see Data and code availability for more details).
Athero-express biobank study
Patient population
Atherosclerotic plaques were obtained from patients undergoing an arterial endarterectomy procedure and included in the Athero-Express Biobank Study (AE), an ongoing biobank study at the University Medical Centre Utrecht (Utrecht, The Netherlands) and the St. Antonius Hospital (Nieuwegein, The Netherlands)3. The medical ethical committees of the respective hospitals approved the AE which was registered under number TME/C-01.18. This study complies with the Declaration of Helsinki, and all participants provided informed consent. However, considering national laws plaque-material are considered ‘waste biomaterial’ and are always allowed to be used without any personal information regardless of informed consent. In this study we also considered this ‘waste biomaterial’, hence no clinical information, e.g., age, is given here nor when sharing data of the relevant samples (n = 3). The study design of the AE was described before3, but in brief: during surgery blood and plaques are obtained, stored at − 80 °C and plaque material is routinely used for histological analysis3,42.
Whole-slide staining and scanning
The standardized (immuno)histochemical analysis protocols used in the AE biobank have been described previously3,42. In short, 4‐micron cross‐sections of the paraffin‐embedded segments were cut using a microtome, and 8 different stains were applied for endothelial cells (CD34), macrophages (CD68), elastic Van Gieson (EvG), fibrin, red blood cells (glycophorin C, GLYCC), hematoxylin and eosin (HE) for nuclei, collagen (picrosirius red, SR), and smooth muscle cells (SMCs, α‐actin) on consecutive slides. We set up ExpressScan to obtain whole-slide images (WSIs) by scanning stained slides at 40× using a Roche Ventana iScan HT (https://diagnostics.roche.com/global/en/products/instruments/ventana-iscan-ht.html) or Hamamatsu C12000-22 Digital slide scanner (https://www.hamamatsu.com/eu/en/product/life-science-and-medical-systems/digital-slide-scanner/index.html). WSIs were stored digitally as z-stacked .TIF (Roche) at 0.25 micron/pixel or .ndpi (Hamamatsu) at 0.23 micron/pixel brightfield microscopy images10. The slides numbers used for this project are listed in Supplemental Table 1.
Manual tissue annotation and masking
The ground truth tissue areas for each WSI were annotated manually at 40 × magnification using QuPath43. If the tissue had disintegrated into several disjointed fragments during preparation or staining the annotators marked them with a single enclosing polygon. Binary masks of these tissue annotations were then generated using groovy scripts in QuPath.
Statistical analyses
We assessed the algorithms using several metrics: The Jaccard index, sensitivity, False positive Rate (FPR) and pixel accuracy, comparing the algorithms' output to the ground truth binary reference masks of the 97 images in the test set.
Jaccard index was applied to assess the performance of our masking algorithms, by measuring the overlap area of target masks with the ground truth masks annotated by experts and masks of the algorithm outputs divided by the area of the union of both masks. Mathematically, sensitivity, false positive rate and pixel accuracy can be expressed as:
Evaluation and results
We developed EntropyMasker, a novel masking method using entropy-filtering (Fig. 1). First, we compared our method against three conventional thresholding methods: Otsu’s, adaptive thresholding using the mean of neighborhood area as the threshold, and adaptive thresholding using the Gaussian weighted sum of neighborhood values as the threshold. We also evaluated these methods against the previously developed slideMask tissue masking method in the slideToolKit28.
Schematic overview of the EntropyMasker method for masking whole-slide images. Atherosclerotic plaque tissue samples were collected from arterial endarterectomy surgeries, sectioned and mounted on tissue slides as part of the Athero-Express Biobank Study. The slides were subjected to histological staining for overall morphology (hematoxylin & eosin; HE), collagen deposition (Picrosirius red; SR), fibrin deposition (Fibrin), elastin fibers (Elastica Van Gieson; EVG) or immunohistochemical staining for cell-type specific markers: alpha-smooth muscle actin (a-SMA), macrophages (CD68), endothelial cells (CD34), or erythrocytes (glycophorin C). Whole-slide images of each slide were generated using high-resolution scanners and images were stored in a database (~ 25,000 images). Red, Green, Blue (RGB) color images were converted to grayscale images and grayscale pixels were used to calculate entropy threshold map for histogram analysis, and tissue segmentation was performed to create tissue mask of WSIs for subsequent data analysis and benchmarking. Schematic was created with BioRender.com.
Application of quantitative WSI analysis pipelines requires access to uniformly processed and well-characterized tissue biobanks. The Athero-Express study3 is a large-scale vascular tissue biobank comprising over 3600 carotid and femoral endarterectomy surgical specimens, which include detailed clinical outcomes and follow-up. This has led to several prospective studies to correlate local atherosclerotic plaque composition with future local and systemic vascular outcomes, using histology7,8,9,10, RNA44,45, genetics46, and protein47,48 data. In the histology-based analyses, local plaque indices such as plaque hemorrhage and neovascularity were shown to correlate with more adverse vascular outcomes7,9. We evaluated each method using 97 randomly selected WSIs from Athero-Express (Supplemental Table 1 for a minimalistic baseline table and Supplemental Table 2 for details on usage) at 40X magnification on layer 6 (pixel spacing at layer 6 is around 8.36 µm) consisting of 8 different stains: HE, picrosirius red, Fibrin, EVG, a-SMA, CD34, CD68, and GLYCC. We did not apply our method to layer 1 due to memory constraints.
The average Jaccard index, also referred to as the Intersection over Union (IoU) metric, of the adaptive thresholding method where its threshold value is the weighted sum of neighborhood values where weights are a gaussian window was 0.7702 and the average sensitivity was 0.8597 (Table 1, Supplemental Table 3 includes results for all stains and methods). This masking method produced the highest pixel accuracy among all the traditional methods which was 0.9485. Otsu’s method had a much lower false positive rate than any of the approaches examined at the cost of much lower average Dice scores and sensitivity levels. The quality of the outcome of the slideMask algorithm was unevenly distributed resulting in a relatively low Jaccard index (IoU) across all the tested methods (Fig. 2). The average false positive rate was 0.0153, which is the lowest of all masking methods in comparison, and its average pixel accuracy was 0.9133. Our automatic local entropy-based masking method, EntropyMasker, had the best average Jaccard index (IoU) 0.8837 and the highest sensitivity 0.9685, outperforming Otsu’s method, two adaptive methods and slideMask masking method in both metrics. The average false positive rate was 0.0176 which is much lower than Otsu’s method, and two adaptive methods.
Evaluation metrics for different masking methods. Comparison of EntropyMasker with other masking methods: Otsu, slideMask, and adaptive thresholding methods, using manually annotated tissue sections as ground truth. 97 images from 59 individual patient samples were randomly selected and used to evaluate each method by calculating the (A) Jaccard index (intersection over union metric), (B) sensitivity, (C) false positive rate, and (D) pixel accuracy.
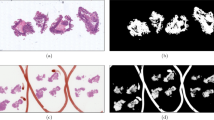
The results of comparing the 5 masking methods for an H&E-stained whole-slide image are shown in Fig. 3 where generated tissue masks are overlaid by pseudocolor (green). To show the generalizability of our automatic entropy method, we tested on whole-slide images with 8 different types of staining. Examples of masking results of our proposed method are shown on Fig. 4 where generated tissue masks are overlaid by pseudocolor (green).
Comparison of results of 5 masking methods for a randomly selected H&E-stained whole-slide image. (A) Expert generated annotation (ground truth) using Qupath Software (Version 0.3.2)43. (B) Our EntropyMasker method. (C) Otsu’s method. (D) Adaptive method where the threshold value is the mean of neighborhood area. (E) Adaptive method where the threshold value is the weighted sum of neighborhood values where weights are a gaussian window. (F) slideMask method. All the generated tissue masks are overlaid by pseudocolor (green).
Generalizability across the different antibody-specific or histology stains. Images of the upper part are the original whole-slide images at layer 6 with 8 different types of staining methods respectively and images of the lower part are the examples of masking results of our EntropyMasker method where generated tissue masks are overlaid with pseudocolor (green). (A) H&E; (B) Glycophorin C; (C) Sirius red; (D) Fibrin; (E) CD68; (F) CD34; (G) Smooth muscle alpha-actin; (H) EVG.
Since we used two different scanners, we also evaluated the EntropyMasker performance using different image formats (.ndpi or .TIF), which did not influence the Jaccard index (Supplemental Fig. 1). Also, to mitigate the influence of the porosity of the tissue, we evaluated these methods on both high (20X) and low (5x) resolution WSIs, however the performance was unaffected (Supplemental Table 4).
The computation speed for Otsu’s method, adaptive methods, slideMask and proposed EntropyMasker method on a 3.6 GHz Quad-Core Intel Core i7 processor, 32 GB 2400 MHz DDR4 memory iMac are 0.08, 0.11, 0.25 and 0.34 s per one million pixels, respectively.
Conclusions and discussion
Overall, our automatic entropy masking method, EntropyMasker, performed well on atherosclerotic plaque cross-sectional WSIs derived from 8 different types of stains. With a relatively high average Jaccard index our method was able to separate out the foreground and background accurately and consistently from these complex images (Fig. 2 and 4).
In comparison, we observed the popular masking method, Otsu’s, and our previous method, slideMask, both tended to miss some tissue areas along the tissue boundary. This likely results from the relatively lower intensity of these areas, where Otsu’s method sets a threshold in the middle of two peaks, thus partially omitting the higher intensity pixels compared to the threshold (Fig. 2). The slideMask method computes the root mean squared difference between two colors, however this becomes problematic when the intensity of tissue area pixels is relatively large (Fig. 2).
We also found that the adaptive methods tend to produce ‘porosity’ during the masking, since these methods miss small regions of the image which are supposed to be regarded as tissue (Fig. 2). This large scale missing tissue makes Otsu’s, slideMask and adaptive thresholding methods impractical for use in many image processing workflows10,28, especially when applied to atherosclerotic plaque images. Given the limited availability and heterogeneous nature of these plaques, an ideal masking method will retain all the plaque components, which can be used for automatic cellular segmentation tasks to discover tissue or compartment-specific markers for disease progression and phenotyping. This is also a valuable feature of EntropyMasker, which can be implemented at the preprocessing stage of various machine learning pipelines for other diseases and research areas which require a complete WSI masking step49,50,51.
Despite the accurate and consistent performance of EntropyMasker, it is worth pointing out some known limitations. In comparison to the other methods that we evaluated, our method has the highest sensitivity, and thus tends to over-include a few pixels outside the edges of tissue (Fig. 3). While these regions represent a small fraction of the total tissue areas that we tested, this should be more carefully considered when evaluating smaller tissue sections11. Also, our method had slightly lower pixel accuracy than the adaptive methods, which may be important for tasks involving more subtle intensity differences in defining target regions of the tissue52. Compared to the other methods discussed, ours is slightly slower (0.34 s per one million pixels compared to 0.11 s for Otsu’s) which may pose an issue when examining thousands of images, although this should be negligible since our method is scalable on any high-performance compute cluster.
In conclusion, we demonstrated the effectiveness of our proposed method for tissue masking on human atherosclerotic plaque WSIs of different types of stains including HE, CD34, CD68, EVG, smooth muscle cell α‐actin, picrosirius red, fibrin and Glycophorin C. By evaluating our method against other popular masking methods and those recently developed, we also demonstrated that our entropy-based masking method is scalable and had optimal performance across these WSIs. Given its scalability in WSI processing, we envision EntropyMasker to be readily adopted across various disease contexts and implemented in existing pipelines as an input to both deep learning and cell counting tasks.
Data availability
The full histological data used in this study are available here https://doi.org/10.34894/GN4YOS including the high- and low-resolution whole-slide images.
All documented code and the tutorial to run EntropyMasker can be found here https://github.com/CirculatoryHealth/EntropyMasker.
References
Ross, R. Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 340, 115–126 (1999).
Lusis, A. J. Atherosclerosis. Nature 407, 233–241 (2000).
Verhoeven, B. A. N. et al. Athero-express: differential atherosclerotic plaque expression of mRNA and protein in relation to cardiovascular events and patient characteristics. Rationale and design. Eur. J. Epidemiol. 19, 1127–1133 (2004).
Burke, A. P., Kolodgie, F. D., Farb, A., Weber, D. & Virmani, R. Morphological predictors of arterial remodeling in coronary atherosclerosis. Circulation 105, 297–303 (2002).
Burke, A. P. et al. Coronary risk factors and plaque morphology in men with coronary disease who died suddenly. N. Engl. J. Med. 336, 1276–1282 (1997).
Farb, A. et al. Coronary plaque erosion without rupture into a lipid core. A frequent cause of coronary thrombosis in sudden coronary death. Circulation 93, 1354–1363 (1996).
Willem, E. H. et al. Composition of carotid atherosclerotic plaque is associated with cardiovascular outcome: A prognostic study. Circulation 121, 1941–1950 (2010).
van Lammeren, G. W. et al. Time-dependent changes in atherosclerotic plaque composition in patients undergoing carotid surgery. Circulation 129, 2269–2276 (2014).
Vrijenhoek, J. E. P. et al. Sex is associated with the presence of atherosclerotic plaque hemorrhage and modifies the relation between plaque hemorrhage and cardiovascular outcome. Stroke 44, 3318–3323 (2013).
Mekke, J. M. et al. Glycophorin C in atherosclerotic plaque is associated with major adverse cardiovascular events after carotid endarterectomy. bioRxiv https://doi.org/10.1101/2021.07.15.21260570 (2021).
Baxi, V., Edwards, R., Montalto, M. & Saha, S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod. Pathol. 35, 23–32 (2022).
Bándi, P., Balkenhol, M., van Ginneken, B., van der Laak, J. & Litjens, G. Resolution-agnostic tissue segmentation in whole-slide histopathology images with convolutional neural networks. PeerJ 7, e8242 (2019).
Bandi, P. et al. From detection of individual metastases to classification of lymph node status at the patient level: The CAMELYON17 challenge. IEEE Trans. Med. Imaging 38, 550–560 (2019).
Hart, S. N. et al. Classification of melanocytic lesions in selected and whole-slide images via convolutional neural networks. J. Pathol. Inform. 10, 5 (2019).
Yu, K.-H. et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 7, 12474 (2016).
Xia, T., Kumar, A., Feng, D. & Kim, J. Patch-level tumor classification in digital histopathology images with domain adapted deep learning. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2018, 644–647 (2018).
Liu, Y. et al. Artificial intelligence-based breast cancer nodal metastasis detection: Insights into the black box for pathologists. Arch. Pathol. Lab. Med. 143, 859–868. https://doi.org/10.5858/arpa.2018-0147-oa (2019).
Coudray, N. et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567. https://doi.org/10.1038/s41591-018-0177-5 (2018).
Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66. https://doi.org/10.1109/tsmc.1979.4310076 (1979).
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309 (2019).
Vanderbeck, S., Bockhorst, J., Komorowski, R., Kleiner, D. E. & Gawrieh, S. Automatic classification of white regions in liver biopsies by supervised machine learning. Hum. Pathol. 45, 785–792 (2014).
Rother, C., Kolmogorov, V. & Blake, A. GrabCut. in ACM SIGGRAPH 2004 Papers on - SIGGRAPH ’04 (ACM Press, 2004). https://doi.org/10.1145/1186562.1015720.
Bug, D., Feuerhake, F. & Merhof, D. Foreground extraction for histopathological whole slide imaging. Informatik aktuell https://doi.org/10.1007/978-3-662-46224-9_72 (2015).
Feng, D., Wenkang, S., Liangzhou, C., Yong, D. & Zhenfu, Z. Infrared image segmentation with 2-D maximum entropy method based on particle swarm optimization (PSO). Pattern Recogn. Lett. 26, 597–603. https://doi.org/10.1016/j.patrec.2004.11.002 (2005).
Zhang, H., Qian, W., Wan, M. & Zhang, K. Infrared image enhancement algorithm using local entropy mapping histogram adaptive segmentation. Infrared Phys. Technol. 120, 104000. https://doi.org/10.1016/j.infrared.2021.104000 (2022).
Deng, H., Sun, X., Liu, M., Ye, C. & Zhou, X. Infrared small-target detection using multiscale gray difference weighted image entropy. IEEE Trans. Aerosp. Electr. Syst. 52, 60–72. https://doi.org/10.1109/taes.2015.140878 (2016).
Stirling, D. R. et al. Cell profiler 4: Improvements in speed, utility and usability. BMC Bioinform. 22, 433 (2021).
Nelissen, B. G. L., van Herwaarden, J. A., Moll, F. L., van Diest, P. J. & Pasterkamp, G. SlideToolkit: An assistive toolset for the histological quantification of whole slide images. PLoS ONE 9, e110289 (2014).
Anghel, A. et al. A high-performance system for robust stain normalization of whole-slide images in histopathology. Front. Med. 6, 193 (2019).
Tsallis, C. Beyond Boltzmann–Gibbs–Shannon in physics and elsewhere. Entropy 21(7), 696 (2019).
Lin, J. Divergence measures based on the Shannon entropy. IEEE Trans. Inf. Theory 37, 145–151. https://doi.org/10.1109/18.61115 (1991).
Arunachalam, H. B. et al. Viable and necrotic tumor assessment from whole slide images of osteosarcoma using machine-learning and deep-learning models. PLoS ONE 14, e0210706 (2019).
Guo, Z. et al. Publisher correction: A fast and refined cancer regions segmentation framework in whole-slide breast pathological images. Sci. Rep. 10, 8591 (2020).
Romo-Bucheli, D., Janowczyk, A., Gilmore, H., Romero, E. & Madabhushi, A. Automated tubule nuclei quantification and correlation with oncotype DX risk categories in ER+ breast cancer whole slide images. Sci. Rep. 6, 32706 (2016).
Valkonen, M. et al. Metastasis detection from whole slide images using local features and random forests. Cytometry A 91, 555–565 (2017).
Wang, P., Li, P., Li, Y., Xu, J. & Jiang, M. Classification of histopathological whole slide images based on multiple weighted semi-supervised domain adaptation. Biomed. Signal Process. Control 73, 103400. https://doi.org/10.1016/j.bspc.2021.103400 (2022).
Kothari, S., Phan, J. H. & Wang, M. D. Eliminating tissue-fold artifacts in histopathological whole-slide images for improved image-based prediction of cancer grade. J. Pathol. Inform. 4, 22. https://doi.org/10.4103/2153-3539.117448 (2013).
Lu, C. & Mandal, M. Automated analysis and diagnosis of skin melanoma on whole slide histopathological images. Pattern Recogn. 48, 2738–2750. https://doi.org/10.1016/j.patcog.2015.02.023 (2015).
ImageMagick Studio LLC. ImageMagick. ImageMagick https://imagemagick.org/.
GIMP. GIMP https://www.gimp.org/.
Bradski. The openCV library. Dr. Dobb’s Journal: Software Tools for the Professional.
van Lammeren, G. W. et al. Atherosclerotic plaque vulnerability as an explanation for the increased risk of stroke in elderly undergoing carotid artery stenting. Stroke 42, 2550–2555 (2011).
Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Depuydt, M. A. C. et al. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ. Res. 127, 1437–1455 (2020).
Slenders, L. et al. Intersecting single-cell transcriptomics and genome-wide association studies identifies crucial cell populations and candidate genes for atherosclerosis. Eur. Heart J. Open https://doi.org/10.1093/ehjopen/oeab043 (2021).
van der Laan, S. W. et al. Genetic susceptibility loci for cardiovascular disease and their impact on atherosclerotic plaques. Circ. Genom. Precis. Med. 11, e002115 (2018).
Peeters, W. et al. Adipocyte fatty acid binding protein in atherosclerotic plaques is associated with local vulnerability and is predictive for the occurrence of adverse cardiovascular events. Eur. Heart J. 32, 1758–1768 (2011).
de Kleijn, D. P. V. et al. Local atherosclerotic plaques are a source of prognostic biomarkers for adverse cardiovascular events. Arterioscler. Thromb. Vasc. Biol. 30, 612–619 (2010).
Nirschl, J. J. et al. A deep-learning classifier identifies patients with clinical heart failure using whole-slide images of H&E tissue. PLoS ONE 13, e0192726. https://doi.org/10.1371/journal.pone.0192726 (2018).
Lipkova, J. et al. Deep learning-enabled assessment of cardiac allograft rejection from endomyocardial biopsies. Nat. Med. 28, 575–582 (2022).
Bouteldja, N. et al. Deep learning-based segmentation and quantification in experimental kidney histopathology. J. Am. Soc. Nephrol. 32, 52–68 (2021).
Khened, M., Kori, A., Rajkumar, H., Krishnamurthi, G. & Srinivasan, B. A generalized deep learning framework for whole-slide image segmentation and analysis. Sci. Rep. https://doi.org/10.1038/s41598-021-90444-8 (2021).
Acknowledgements
We also acknowledge support from the Netherlands CardioVascular Research Initiative of the Netherlands Heart Foundation (CVON 2011/B019 and CVON 2017-20: Generating the best evidence-based pharmaceutical targets for atherosclerosis [GENIUS I&II]), the ERA-CVD program ‘druggable-MI-targets’ (Grant No: 01KL1802), and the Chan Zuckerberg Initiative Foundation Data Insights program (Grant No: DI-092).We would also like to thank all the (former) employees involved in the Athero-Express Biobank Study of the Departments of Surgery of the St. Antonius Hospital Nieuwegein and University Medical Center Utrecht for their continuing work. We would like to thank (in no particular order) Marijke Linschoten, Arjan Samani, Petra H. Homoed-van der Kraak, Tim Bezemer, Tim van de Kerkhof, Joyce Vrijenhoek, Evelyn Velema, Ben van Middelaar, Sander Reukema, Robin Reijers, Joëlle van Bennekom, and Bas Nelissen. Lastly, we would like to thank all participants of the Athero-Express Biobank Study; without you these studies would not be possible.
Funding
Funding for this research was provided by National Institutes of Health (NIH) grant nos. R00HL125912 and R01HL14823 (to CLM), a Leducq Foundation Transatlantic Network of Excellence (‘PlaqOmics’) grant no. 18CVD02 (to CLM and SWvdL), and the EU H2020 TO_AITION Grant no. 848146 (to SWvdL).
Author information
Authors and Affiliations
Contributions
Y.S., J.M.M., S.W.vdL., and C.L.M. conceptualized the study. Y.S., J.M.M., C.A.G., and S.W.vdL. contributed to the data curation. Y.S. performed the formal analysis. S.W.vdL. and C.L.M. provided funding support. Y.S. contributed to the primary investigation. C.A.G., S.W.vdL., and C.L.M. contributed to project administration. J.M.M., G.J.dB., D.P.V.dK., G.P., A.V., and S.W.vdL. contributed the Athero-Express resources. Y.S., C.A.G., and S.W.vdL. contributed to software development. G.J.dB., D.P.V.dK., G.P., C.A.G., S.W.vdL., and C.L.M. contributed to project supervision. F.C. contributed to validation studies. Y.S., S.W.vdL., and C.L.M. contributed to visualization. Y.S. contributed to writing the original draft. All authors (Y.S., F.C., J.M.M., G.J.dB., D.P.V.dK., G.P., A.V., C.A.G., S.W.vdL., and C.L.M.) contributed to review and editing the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
Dr. Sander W. van der Laan has received Roche funding for unrelated work. Dr Craig A. Glastonbury has stock options in BenevolentAI and is a paid consultant for BenevolentAI, unrelated to this work. Dr. Clint L. Miller has received AstraZeneca funding for unrelated work. All other authors declare that they have no competing interests relevant to the contents of this paper to disclose.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, Y., Cisternino, F., Mekke, J.M. et al. An automatic entropy method to efficiently mask histology whole-slide images. Sci Rep 13, 4321 (2023). https://doi.org/10.1038/s41598-023-29638-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-29638-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.