Abstract
Diploid and polyploid species derived from the euploid series x = 11 occur in the genus Psidium, as well as intraspecific cytotypes. Euploidy in the genus can alter the gene copy number, resulting in several “omics” variations. We revisited the euploidy, reported genomic (nuclear 2C value, GC%, and copy number of secondary metabolism genes) and epigenomic (5-mC%) differences in Psidium, and related them to essential oil yield and composition. Mean 2C values ranged from 0.90 pg (P. guajava) to 7.40 pg (P. gaudichaudianum). 2C value is intraspecifically varied in P. cattleyanum and P. gaudichaudianum, evidencing cytotypes that can be formed from euploid (non-reduced) and/or aneuploid reproductive cells. GC% ranged from 34.33% (P. guineense) to 48.95% (P. myrtoides), and intraspecific variations occurred even for species without 2C value intraspecific variation. Essential oil yield increased in relation to 2C value and to GC%. We showed that P. guajava (diploid) possesses two and P. guineense (tetraploid) four copies of the one specific TPS gene, as well as eight and sixteen copies respectively of the conserved regions that occur in eight TPS genes. We provide a wide “omics'' characterization of Psidium and show the outcome of the genome and epigenome variation in secondary metabolism.
Similar content being viewed by others
Introduction
The neotropical and monophyletic genus Psidium, family Myrtaceae, contains approximately 92 species1, hich are taxonomically classified in four sections: Psidium (10 species), Obversifolia (six species), Apertiflora (31 species) and Mitranthes (26 species)2. ~ 60 Psidium species occur in Brazil distributed in all biomes, in several phytogeographic domains, representing great diversification3. Psidium guajava L. (guava tree, Psidium section) is the well-known species of the genus due to its relevance for fruit production4 and medicinal value5,6. Other Psidium species, popularly called “araçás”, are potential genetic resources for breeding programs and medicinal purposes7, as well as they are relevant for taxonomic, ecological and evolutive studies in this genus2.
Besides diploid species, in Psidium occur polyploid species with different ploidy levels commonly derived from the basic chromosome number x = 11. One of the consequences of euploidy in Psidium is the interspecific and intraspecific variation of the nuclear 2C value. Previously, we found that the increase (2C = 0.93 pg – 2C = 5.12 pg) of the nuclear 2C value is outcome of the increase in ploidy level (diploid with 2C = 0.93 pg – 2C = 0.98 pg to octoploid with 2C = 5.12 pg) in seven Psidium species8. In addition to interspecific variation, genomic differences have been identified for P. cattleyanum. This species is considered a polyploid complex due to genetic diversity (intraspecific variation) of the 2n chromosome number and, consequently, the ploidy level (triploid 2n = 3x = 33 to duodecaploid 2n = 12x = 132 chromosomes), nuclear 2C value (tetraploid with 2C = 2.17 pg to duodecaploid with 2C = 5.64 pg), and number of the CMA3 + /DAPI-, 18S rDNA and 5S rDNA sites (triploid with three sites to duodecaploid with 12, 12 and 10 sites)9.
Despite the 2n chromosome number importance for ploidy level determination (euploidy condition), previous data indicate that the nuclear 2C value is an indicator of higher ploidy level in Psidium8,9. Thus, nuclear 2C value can be used especially when dealing with a large number of individuals. Nuclear 2C value is mainly measured using flow cytometry, which is widely used because it is fast, accurate and reproducible10.
In addition to the nuclear 2C value, the AT/GC base composition can also be measured by flow cytometry, expanding the genomic data11. AT/GC base composition knowledge allows inferences about the genome structure and dynamic. So, AT/GC base composition must be incorporated into “omics” data, and consequently used in taxonomic, systematic and evolutive studies. Psidium guajava genome has GC = 39.5%12, and P. cattleyanum transcriptome has ~ 49% in the yellow and red morphotypes13.
Polyploidy has been considered one of the main genomic changes that results in genetic8 and epigenetic modifications14 influencing the population genetic structure, ecological niche differentiation, diversification and speciation in plants15. About it, Psidium is one outstanding example of the polyploid impact in speciation and geographic distribution8,15. Euploidy (autopolyploidy, true allopolyploidy or segmental allopolyploidy) plays a central role in shaping and restructuring plant genomes15. Regardless of the genomic origin, one euploidy outcome is the increase of the gene copy number, which probably results in phenotypic changes and/or new traits. Also furthering phenotypic variations, genomic changes occur (“genomic shock”) after the euploidy, such as aneuploidy, structural chromosome rearrangements, mobile elements activation or silencing, and DNA sequence change16. Thus, the euploidy and its outcomes are sources of evolutionary novelties. Duplicate genes can follow an evolutive path from an initial state of complete redundancy, in which a copy is probably disposable, to a stable situation17,18 – neutral theory. Still, they may include new gene functions and expression patterns. The duplicated genes can maintain their original or similar function, undergo diversification in function or expression patterns, or a copy can be silenced by mutations or epigenetic mechanisms19. Many duplicate genes can have loci in tandem in the genome or occur regionalized (within a few Mbp)20,21. Additionally, phylogenetic evidence links genome increase (nuclear 2C value) to the increase in the overall percentage of global 5-methylcytosine (5-mC%). 5-mC is an important epigenetic chemical change in DNA that promotes heterochromatinization (chromatin compact level increase) and, consequently, gene expression control22,23.
Leaf essential oils, characteristic of the Myrtaceae family secondary metabolism, are rich in terpene and exhibit quali- and quantitative variations reported24. Terpenes play ecological roles, and essential oils are economically exploited25,26,27 for their biological and phytotherapeutic activities28. Our research group evidenced that seven P. cattleyanum plants exhibited different nuclear 2C values (2C = 3.20 pg – 2C = 6.03 pg), seven monoterpenes and eight sesquiterpenes in essential oils. From these data, the P. cattleyanum plants were discriminated in three cytotypes (nuclear 2C value) related to three chemotypes (monoterpene and sesquiterpene compounds). P. cattleyanum plants with the relatively lower nuclear 2C values (2C = 3.23 pg – 2C = 4.71 pg) produced a lower amount of essential oils composed mostly of hydrogenated monoterpenes. Differently, the plants with relatively higher nuclear 2C values (2C = 5.81 pg and 2C = 6.03 pg) produced a higher amount of essential oils composed mostly of hydrogenated sesquiterpenes, as trans-caryophyllene and alpha-humulene24.
We aimed to revisit the 2n chromosome number (euploidy), measure the nuclear 2C value, GC% and 5-mC%, and determine the copy number of TPS genes in Psidium species of three sections (Psidium, Apertiflora and Obversifolia). In addition, we correlated these genomic and epigenomic data with the yield and composition of the essential oils, evidencing the outcomes of the genetic and epigenetic differences in this phenotype.
Results
Intraspecific and interspecific variation of nuclear 2C value and GC%
The nuclear 2C value of each Psidium access ranged from 0.90 pg (P. guajava) to 7.40 pg (P. gaudichaudianum, Supplementary Table S2). We noticed, for the first time, the nuclear 2C value for P. acidum, P. gaudichaudianum, P. friedrichsthalianum, P. macahense and P. rufum. Psidium guajava, P. oblongatum and P. macahense presented the lowest 2C values. Psidium guajava and P. oblongatum are diploids (2n = 2x = 22 chromosomes). Therefore, possibly P. macahense also has the same 2n chromosome number and ploidy level, since this species has closer mean nuclear 2C value (2C = 0.93 pg) than P. guajava (2C = 0.96 pg) and P. oblongatum (2C = 0.99 pg).
Interspecific variation in nuclear genome size was confirmed by mean nuclear 2C values of the Psidium species (Table 2). Considering the lowest mean 2C value = 0.93 pg for P. macahense and highest 2C = 4.99 pg for P. gaudichaudianum, we realize a variation equivalent to 2C = 4.06 pg more nuclear DNA. Additionally, the individual nuclear 2C values also show the interspecific variation in nuclear genome size, reaching 2C = 6.50 pg more nuclear DNA, since one access of P. guajava has 2C = 0.90 pg and one access of P. gaudichaudianum has 2C = 7.40 pg.
In addition to interspecific variation, the individual values point to intraspecific variation of the nuclear 2C value, including among relatives. Accesses belonging to P. guajava (2C difference = 0.13 pg among individuals), P. guineense (2C = 0.20 pg) and P. acidum (2C = 0.08 pg) showed less variation of the nuclear 2C value. These nuclear 2C values differences are lower than the 1Cx value of P. guajava (0.475 pg) and P. oblongatum (0.490 pg) determined considering the basic chromosome number (x = 11) of the genus Psidium and the ploidy level of these species (2n = 2x = 22 chromosomes – diploid8). Therefore, the intraspecific variation found among accesses of P. guajava, P. guineense and P. acidum is probably a consequence of secondary metabolites that interfere with the intercalation of the propidium iodide fluorochrome to DNA in the staining step for nuclear suspension preparation for flow cytometry.
We also observed intraspecific variation of the nuclear 2C value among individuals, as well as in the relatives of P. myrtoides, P. cattleyanum and P. gaudichaudianum. The nuclear 2C value difference was: 2C = 0.40 pg between individuals of P. myrtoides, 2C = 5.03 pg of P. cattleyanum, and 2C = 2.76 pg of P. gaudichaudianum. For these species, the nuclear 2C value differences are close to or higher than the reference 1Cx value (1Cx = 0.475 pg P. guajava—1Cx = 0.490 pg P. oblongatum) at the basic chromosome number x = 11 of Psidium. Therefore, these values indicate that individuals of these species have different 2n chromosome numbers among each other, possibly arising from numerical chromosomal changes (euploidy and/or aneuploidy).
According to the mean nuclear 2C values (Table 2) and the data reported for diploid and polyploid Psidium species (Table 1, http://ccdb.tau.ac.il/search/, https://cvalues.science.kew.org/search/angiosperm), we suggest that accesses of the species P. acidum, P. rufum, P. friedrichsthalianum and P. gaudichaudianum are potential polyploids. Thus, individuals of these species probably have more than 2n = 2x = 22 chromosomes. However, chromosome number counting should be conducted to confirm the ploidy level.
Six groups were identified in relation to the individual nuclear 2C value (Table 3). Group I comprises the individuals with the smallest nuclear 2C value (2C = 0.90 pg to 2C = 1.10 pg), which include the diploid species P. guajava, P. oblongatum, and the P. machaense that, based on the 2C and 1Cx values, possible is a diploid species. Group II (2C = 1.80 pg—2C = 2.08 pg) has the tetraploid species P. guineense and P. acidum, as well as all 14 P. guajava × P. guineense hybrids, one individual of P. cattleyanum and seven individuals of Psidium sp. The evaluated P. guajava × P. guineense hybrids have 2C = 1.90 pg, exhibiting the same nuclear 2C value (Table 2). This mean value is equivalent to the tetraploid genomic origin. Different progenies are possible considering the P. guajava × P. guineense crossing: (a) allotriploid hybrids (~ 2C = 1.43 pg) from the fusion of reduced reproductive cells of the two species, (b) allotetraploid hybrids (~ 2C = 1.91 pg) generated from the fusion of non-reduced reproductive cells of P. guajava and reduced P. guineense, (c) allopentaploid hybrids (~ 2C = 2.38 pg) formed by fusing reduced reproductive cells of P. guajava and non-reduced P. guineense, and (d) allohexaploid hybrids (~ 2C = 2.85 pg) arising from the fusion of non-reduced reproductive cells of the two species. Therefore, probably the P. guajava × P. guineense hybrids with 2C = 1.90 pg are the result of the fusion of non-reduced reproductive cells of P. guajava and reduced P. guineense. In addition, we found a P. guajava × P. guineense hybrid (Hib_11) mixoploid with 30% of cells with 2C = 0.95 pg (similar to diploid P. guajava) and 70% of cells with 2C = 1.90 pg (similar to tetraploid P. guineense) (Supplementary Table S2).
Group III (2C = 2.72—2C = 4.38 pg) contains most of the individuals evaluated, including the species P. cattleyanum, P. myrtoides, P. rufum and one Psidium sp. Due to the variation in 2n chromosome number and ploidy level of P. cattleyanum and P. myrtoides (Table 1), the group III includes possibly hexaploid and octaploid accesses. Group IV (2C = 4.47 pg—2C = 5.19 pg) has individuals from P. cattleyanum, P. gaudichaudianum, P. friedrichsthalianum and three Psidium sp, which are probably octaploids (Table 1), as well as the only individual from group V of P. cattleyanum. Group VI includes the three individuals with the highest 2C values (2C = 6.83 pg—2C = 7.40 pg), two from P. cattleyanum and one from P. gaudichaudianum. Probably the accesses in group VI have a ploidy level higher than octaploid (2n = 8x = 88 chromosomes).
Psidium cattleyanum showed the highest intraspecific variation of the nuclear 2C value, exhibiting individuals in five of the six groups. Differently, less intraspecific variation was confirmed for the diploid species P. guajava (all individuals in group I), for the tetraploids P. guineense and P. acidum (group II) and for the hexaploid P. myrtoides (group III, Table 3).
The GC% values of the evaluated 137 individuals ranged from 34.33% for an individual of P. guineense to 48.95% for an individual of P. myrtoides. The largest intraspecific variations (~ 10%) were in P. myrtoides and P. cattleyanum, which are species that also show nuclear 2C value intraspecific variation. For the other species, the GC% ranged from 0.50% for P. acidum to 8.74% for P. guineense. The variation in the two half-sib families of P. myrtoides was 2.12% (Family 7) and 10.24% (Family 8). Within families of P. cattleyanum, a small variation was observed (1.36% for Family 7 and 3.47% for Family 3), despite intraspecific nuclear genome size variation in the species of up to 2C = 5.03 pg. The intraspecific variation of GC% was ~ 3.00% for the two families of P. guajava, and of ~ 2.10% P. gaudichaudianum (Table 3).
Seven groups were obtained from comparative GC% analysis. Psidium cattleyanum, P. guajava, P. guineense, P. myrtoides, P. gaudichaudianum, P. guajava × P. guineense hybrids, and Psidium sp. showed individuals in at least two groups, evidencing GC% intraspecific variation. Groups III—VI consisted of diploid and polyploid species. Only one family of P. cattleyanum showed greater stability being allocated only to group VII (Table 3).
5-mC%, yield and chemical composition of the essential oil
We compiled in Supplementary Table S3 the unpublished and published values, for each Psidium individual, of 5-mC%, yield, and the percentage of the chemical compounds identified from the essential oil. 5-mC values varied between 16.34% (P. guajava) to 33.30% (P. myrtoides), and between 0.20% (P. guajava) to 0.95% (P. cattleyanum) for yield of the essential oil. A total of 56 compounds were identified for Psidium species. Of these compounds, 55 are chemically classified between hydrocarbons and oxygenated mono- and sesquiterpenes.
Relationship of the mean nuclear 2C value, GC% and 5-mC% values, yield and chemical composition of the essential oil
The nuclear 2C and GC% values were positively correlated (correlation of 0.51). These values also had positive correlation with 5-mC% (correlation of 0.35 and 0.36, respectively) and with essential oil yield (correlation of 0.72 and 0.70, respectively). The variables related to genome (nuclear 2C value and GC%) and to epigenome (5-mC%) correlated negatively with (E)-Nerolidol (correlation of − 0.64, − 0.45 and − 0.55, respectively), β-Bisabolol (correlation of − 0.61, − 0.46 and − 0.35, respectively) and the group of oxygenated sesquiterpenes (− 0.87, − 0.70 and − 0.46, respectively). The α-Pinene and the hydrocarbon monoterpene group were positively correlated with nuclear 2C value (correlation of 0.54 and 0.49, respectively) and 5-mC% (correlation of 0.60 and 0.53, respectively). The nuclear 2C value along with GC% correlated positively with the β-caryophyllene (correlation of 0.70 and 0.65, respectively), α-Copaene (correlation of 0.57 and 0.56, respectively) and the hydrocarbon sesquiterpene group (correlation of 0.42 and 0.52, respectively). These genomic data correlated negatively with 14-Hydroxy-epi-(E)-Caryophyllene (correlation of − 0.50 and − 0.46, respectively) and Selin-11-en-4a-ol (correlation − 0.44 and − 0.38, respectively), which belong the oxigenated sesquiterpene group. In addition, the β-Selinene and Hinesol correlated negatively with nuclear 2C value (correlation of − 0.35 and − 0.42 respectively).
TPS gene copy number
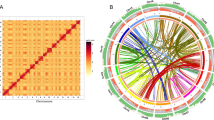
Copy number of TPS genes was determined in diploid P. guajava (2n = 2x = 22 chromosomes and 2C = 0.95 pg) and tetraploid P. guineense (2n = 4x = 44 chromosomes and 2C = 1.90 pg). We expected the copy number to be exactly double in the tetraploid P. guineense. The genes showed distinct marking patterns when comparing the hybridization signal number in relation to the ploidy of the species. For the specific region used, we detected two hybridization signals in P. guajava and four in P. guineense nuclei (Fig. 1).
Copy number of TPS genes. P. guajava possesses one copy of the specific TPS gene (a), while P. guineense shows two copies of this gene (b). From the probe in conserved motifs, we evidenced eight fluorescence signals in P. guajava (c) and sixteen in P. guineense (d), being that strong fluorescence signals are related to cluster genes. Based on these results, we showed the polyploidy impact in the gene copy number in Psidium.
From the general primer of the conserved motives, the P. guajava nuclei showed four strong signals and four weak signals. P. guineense nuclei exhibited eight strong and eight weak signals (Fig. 1). The presence of weak signals was considered as DNA sequences that have a relatively homology in relation to the probe, corresponding to regions that had the same origin, but that accumulated differences in the gene sequence. In addition, the weak hybridization signals can be resulted by occurrence of a single gene copy. Differently, the strong hybridization signals correspond to gene copies in tandem repeats, which form clusters that amplify the fluorescence signal.
Therefore, we confirmed that the copy number of the TPS genes was directly related to the ploidy level of the species with double of signals found in the tetraploid P. guineense. This result also shows the genome evolution of these Psidium species.
Discussion
We report the nuclear 2C and GC% values, TPS gene copy number, as well as 5-mC% and essential oil yield and composition of a considerable number of Psidium species and individuals. The GC% values, with the exception of P. guajava12, are unpublished for all Psidium, as well as the nuclear 2C values for the P. acidum, P. gaudichaudianum, P. friedrichsthalianum, P. macahense and interspecific hybrids (P. guajava L. × P. guineense). 5-mC% are also unpublished for P. cattleyanum, P. guineense, P. myrtoides, P. gaudichaudianum, P. friedrichsthalianum, P. oblongatum and one access of the genus. In addition, the determination of the TPS gene copy number evidenced other genomic outcomes of the polyploidy beyond the nuclear 2C and GC% interspecific variation. The results bring advances about the structure, organization and evolution of the genome and epigenome of Psidium species in inter- and intraspecific contexts, including analyses of related individuals. The genomic and epigenomic data were contextualized with the yield and diversity of compounds in the leaf essential oils, which are rich in mono- and sesquiterpenes that have ecological and economic importance for the Myrtaceae family.
Based on the previous study of the our research group and the basic chromosome set of Psidium (x = 11), the 1C value varied from 0.465 pg (P. cauliforum—diploid species) to 0.640 pg (P. longipetiolatum—octaploid species), and the increase in ploidy culminated in the increase in nuclear DNA content8. Intraspecific variation in 2C value has been demonstrated for P. cattleyanum (2C = 2.00 to 2C = 7.03 pg), corroborating reports of naturally occurring individuals with varied chromosome numbers and ploidy levels for the species, which has cytotypes with 2n = 46, 55, 58 and 8229; 33 (3x), 44 (4x), 55 (5x), 66 (6x), 77 (7x), 88 (8x), 99 (9x), 110 (10x) and 132 (12x)9 (Table 1). Thus, the 2n chromosome number variation in this species can also be supported by the 2C nuclear value variation. Furthermore, we infer that euploidy occurs in the genus not only in P. cattleyanum, but also in P. gaudichaudianum and P. friedrichsthalianum due to the amplitude of 2C nuclear value variation.
The oosphere and the reproductive nucleus of the pollen grain are usually reduced reproductive cells (n—haploid). However, non-reduced and/ou aneuploid reproductive cells can be formed due to errors in anaphase I or II during meiosis, by the non-disjunction of chromosomes32, and/or by the non-occurrence of cytokinesis I or II. Thus, reproductive cells with different euploidy and/or aneuploidy can be generated. Individuals of P. cattleyanum and P. gaudichaudianum, including relatives, showed expressive variations in nuclear 2C value (possibly reflecting the 2n chromosome number) that may have resulted from the unilateral or bilateral fusion of reduced and unreduced reproductive cells. The unreduced reproductive cells may come from one (unilateral) or both parents (bilateral) in cross-fertilization or self-fertilization33. In addition, numerical chromosomal variations (euploidy and aneuploidy) can occur in cells of meristematic regions resulting in mixoploid tissues and/or individuals. Thus, the male and/or female reproductive organs of the flowers can have meiocytes with different chromosome numbers compared to the sporophyte. In this context, it is important to highlight the occurrence of a mixoploid individual (Hib_11), not yet reported in Psidium. The mixoploidy can compromise the stability and fertility of plants in the field and, thus, the use of these plants for breeding purposes is not very desirable34.
In general, polyploid species of Psidium present a greater geographical distribution compared to diploids8, with the exception of P. guajava because it is widely exploited and cultivated and, therefore, present in the most varied regions and biomes35. This fact was pointed out by our research group, considering species P. guajava, P. guineense, P. myrtoides, P. cattleyanum, P. longipetiolatum, P. oblongatum and P. cauliflorum8. In addition, the geographical distribution of P. cattleyanum cytotypes is influenced by the ploidy level. P. cattleyanum cytotypes with higher ploidy levels were identified in regions where the environmental conditions are more adverse, with higher temperatures, higher incidence of solar radiation and lower precipitation9,15,36. Therefore, the polyploid condition of the species studied here, may be favorable for expansion of their geographic distribution, both by natural and anthropic action. Hence, exploitation and utilization of these natural resources is relevant for breeding programs and for familiar production.
We verified inter- and intraspecific variations of GC% for the diploid and polyploid species. Although variations occurred, the overall mean 38.92% CG in Psidium is close to the mean value of 38.06% obtained by means of 22 diploid Eucalyptus species and three of the genus Corymbia, also species of the Myrtaceae family37.
Due to 2C value and GC% variations in Psidium, especially in families, and their influence on secondary metabolism, we suggest, in a practical context, the individual pre-selection of plants to compose an experimental project, breeding program, germplasm bank or cultivation. In this sense, we recommended that the pre-selected accesses or individuals of Psidium should be vegetatively propagated, generating new individuals with the same 2n chromosome number, 2C nuclear value and GC% (genomic stability). On the other hand, inter- and intraspecific genomic diversity is important as a source of genetic resources for breeding.
We verified an increase in essential oil yield in Psidium due to the larger genome, evidencing the impact of the genomic changes (2n chromosome number and 2C nuclear value) in the secondary metabolism, which is a trait of ecological and economic importance. Experimentally, the tetraploid induction (2n = 4x = 72 chromosomes) in Lippia integrifolia (family Verbenaceae) increases the essential oil yield compared to diploids (2n = 2x = 36 chromosomes), in addition to larger leaves and trichomes, structures related to essential oil yield38. Additionally, we showed by FISH that the polyploidy increases the copy number of the orthologs of two TPS genes related to essential oil biosynthesis of Psidium species. Therefore, the polyploidy, also evidenced by 2C nuclear value, affects the essential oil yield in Psidium from the diploid species (P. guajava 2n = 2x = 22 chromosomes) and the hitherto reported closest species P. guineense (2n = 2x = 44). The impact of the polyploidy in the essential oil traits can be related with the diversification and size of TPS gene family in Psidium species. The evolution of the TPS genes in the Myrtaceae family genomes have reported the largest TPS gene family in plants (Eucalyptus spp. having up to 100 genes)26,39 and occurrence of lineage-specific pathways and products. Although the essential oil of Psidium species exhibits a great diversity in its chemotypes conditioned to environmental and genetic variations24,27,40,41, the evolution of TPS genes in Myrtaceae neotropical fresh fruits remain unknown.
The increase of the values of 2C nuclear, CG% and 5-mC% was related to the decrease in (E)-Nerolidol and β-Bisabolol. Therefore, in addition to the genome effect (2n chromosome number, 2C nuclear value and GC%), chemical changes of the cytosine (5-mC) also influences composition of the essential oils. So, we showed the influence of the epigenetic control in the compound biosynthesis of the secondary metabolism in Psidium. The higher abundance of oxygenated sesquiterpenes was related to the occurrence of smaller genomes, with lower CG% and 5-mC%, indicating the genomic and epigenomic influence in this chemical class. In previous studies, the presence of oxygenated sesquiterpenes was clearly increased at the expense of hydrocarbons sesquiterpenes in spring in P. guajava genotypes42. Together, these data, which were reported for the first time, show the influence of genome and epigenome on essential oil yield and in specific compounds, suggesting for epigenetic control for terpene in Myrtaceae.
Conclusion
From genome and epigenome to secondary metabolism, we provided data about the diversity of the Psidium species. We characterize the Psidium germplasm in relation to the 2n chromosome number, 2C nuclear and GC% values, TPS gene copy number and 5-mC%, generating knowledge about species previously studied and also about others not yet evaluated. In addition, we also explore the secondary metabolism, evidence the phenotypic divergences between Psidium species and individuals, and confirm our hypothesis about the influence of the genome and epigenome. Therefore, this work provides an important characterization of the genus Psidium, bringing information and evidence that can be incorporated in further studies, especially in phenotypic responses related to characters of economic interest.
Material and methods
Plant material
We collected leaf samples from ten Psidium species: Psidium acidum (DC.) Landrum, P. cattleyanum Sabine, P. guajava L., P. guineense Sw., P. myrtoides O.Berg, P. gaudichaudianum Proença & Faria, P. friedrichsthalianum (O.Berg) Nied, P. macahense O.Berg, P. oblongatum O.Berg, and P. rufum Mart. ex DC. Leaves were also collected from hybrids of P. guajava x P. guineense. Individuals not identified by species were kept and denominated as genus Psidium (Psi). Brazilian region of occurrence, phytogeographic domain and 2n chromosome number reported for the species are presented in Table 1. The number of individuals of each species for each analysis is presented in Supplementary Table S1. The localization of occurrence of each access, individual identification and families are presented in Supplementary Table S2.
Nuclear 2C value and GC%
Young leaves from each germplasm (Supplementary Table S2) were used for nuclear 2C value and GC% measurements. Solanum lycopersicum L., 1753, ‘Stupické’ was used as internal standard (2C = 2.00 pg)10. 2 cm2 leaf fragment from each Psidium germplasm and from the S. lycopersicum were simultaneously chopped43 for about 30 s in a Petri dish containing 0.5 mL OTTO-I 44 modified for species of the Myrtaceae family (0.1 M citric acid, 0.5% Tween 20, 50 µg mL-1 RNAse, 2 mM dithiothreitol, and 7% polyethylene glycol 2000 – PEG)37.After adding 0.5 mL of the same buffer, the resulted suspensions were incubated for 3 min, filtered on a 30 μm diameter nylon filter (Partec) in a 2.0 mL microtube, and centrifuged at 100 xg for 5 min. The supernatant was discarded and 100 μL of the same buffer was added to the pellet, which was homogenized in vortex and incubated for 10 min. Subsequently, 0.5 mL of modified OTTO-II staining buffer (400 mM Na2HPO4H2O, 2 mM dithiothreitol, 50 µg mL-1 RNAse, and 75 µg mL-1 propidium iodide (PI, excitation/emission wavelengths: 480–575/550–740 nm) was added to the10,44. The suspensions were filtered through 20 µm nylon mesh (Partec) into tubes (Partec) and kept for 30 min in the dark. Then, the suspensions were analyzed in a flow cytometer (BD Accuri C6 flow cytometer, Accuri cytometers, Belgium) equipped with a 488 nm laser source to promote emissions at FL2 (615—670 nm) and FL3 (> 670 nm). The fluorescence peaks of the G0/G1 nuclei of each access and the standard were identified in the histograms using BD Accuri™ C6 software. G0/G1 peaks with coefficient of variation (CV) less than 5% were considered for nuclear 2C value measurement in pg by the formula: nuclear 2C value of the access (pg) = [(mean G0/G1 peak channel of the access)*2.00 pg S. lycopersicum]/(mean G0/G1 peak channel of S. lycopersicum).
For GC%, nuclear suspensions were generated following the procedure adopted to measure the nuclear 2C value with some modifications: (a) the OTTO I and II buffers were not supplemented with RNAse, and (b) the OTTO II buffer was supplemented with 1.5 μM of 4’,6-diamidino-2-phenylindole (DAPI, excitation/emission wavelengths: 320–385/400–580 nm). The suspensions were analyzed with a Partec PAS flow cytometer (Partec GmbH, Munster, Germany), equipped with an 388 nm UV mercury arc lamp and a GG 435–500 nm band-pass filter. AT% was measured using the formula45%ATsample = %ATstandard*[(RDAPI/RPI)1/r], in which: %ATS. lycopersicum = 64.50% 11,46; R = ratio of the fluorescence intensity of the access/standard; r = 3 for DAPI46. From the AT%, the GC% was calculated by the following formula: GC% = 100—AT%. The data corresponding to the nuclear 2C value and GC% of Psidium accesses were submitted to clustering by the Toucher method optimized by Euclidean distance, in which the variables were separately evaluated (nuclear 2C value and GC%). The analyses were conducted in the Genes computer program45.
Percentage of methylated cytosines (5-mC%) in the genome
The 5-mC% data of P. guajava accesses were revisited from our previous study47. For P. cattleyanum, P. guineense, P. myrtoides, P. gaudichaudianum, P. friedrichsthalianum and Psidium sp. the unpublished 5-mC% was measured based on the methodology used for P. guajava47.
Yield and chemical composition of the essential oil
We revisited the data about yield and chemical composition of the essential oil previously published by our research group for P. guajava40,42, P. guineense48 and P. cattleyanum24. For the other accesses, the essential oil was extracted based on the methodology used for P. guajava41. The identification and semi-quantification of the leaf essential oil compounds were performed using gas chromatography with flame ionization detector (GC-FID QP2010SE, Shimadzu, Japan) and gas chromatography coupled to mass spectrometry (GC–MS QP2010SE, Shimadzu, Japan). For these analyses, the following conditions were adopted: the carrier gas used was He for both detectors with flow rate and linear velocity of 2.80 mL min− 1 and 50.80 cm sec− 1 (GC-FID) and 1.98 mL min− 1 and 50.90 cm sec− 1 (GC–MS), respectively; injector temperature was 220 °C at a split ratio of 1: 30; fused silica capillary column (30 m × 0.25 mm); Rtx-5MS stationary phase (0.25 μm film thickness); the oven temperature had the following programming: initial temperature of 40 °C, which remained for 3 min and then the temperature was gradually increased at 3 °C min− 1 until it reached 180 °C, remaining for ten minutes, with a total analysis time of 59.67 min; the temperatures used in the FID and MS detectors are 240 and 200 °C, respectively. The sample used was drawn from the vial in a volume of 1 μL of a 3% solution of essential oil dissolved in 95% hexane.
GC–MS analyses were performed in an electron impact equipment with an energy of 70 eV; scanning speed of 1000; scanning interval of 0.50 fragments.sec− 1 and detected fragments from 29 to 400 (m/z). GC-FID analyses were performed by a flame formed by H2 and atmospheric air with a temperature of 300 °C. Flow rates of 40 mL min− 1 and 400 mL min− 1 were used for H2 and air, respectively.
Identification of the essential oil compounds was performed by comparing the mass spectra in relation to available in the spectrophotometer database (Wiley 7, NIST 05 and NIST 05 s) and by the retention index (RI). For the RI calculation, a mixture of saturated C7-C40 alkanes (Supelco, USA) submitted under the same chromatographic conditions as the OE was used and the adjusted retention time of each compound was obtained using GC-FID. Then, the calculated values for each compound were compared with those in the literature49,50,51.
Correlation analysis
2C value, GC%, 5-mC%, yield and content of each compound present in the essential oil were subjected to Pearson’s correlation. The analysis was conducted in the R environment39 using the package “Agricolae” (https://CRAN.R-project.org/package=agricolae).
Terpene synthase gene (TPS) copy number in P. guajava and P. guineense
We showed the polyploidy influence in the copy number of the genes involved with essential oil synthesis, the terpene synthase genes (TPS). These genes encode enzymes that act in essential oil synthesis pathways27,39,52,53,54. For this, we used the sequence of genes functionally characterized and involved in the synthesis of terpenes (TPS genes), which have been described and available in database ID AB266390.1 and ID MK873024.1. Through the BLAST tool, the similarity of these sequences was evaluated in relation to the TPS genes from the P. guajava genome annotation (data of the research group). The alignments that presented a score of at least 80% were selected for the design of the primers. From this, the primers were designed in the conserved motives of these TPS genes, considering mainly exon regions. Primers were designed and evaluated using the OligoIDTAnalyzer program (IDT). We defined two pairs of primers: the first (F 5’-GGTGGGATGTCGATGCTAAA-3’ and R 5’-CTCTTCCTCCGTAACTCTGTATTG3’) specific to one predicted TPS gene orthologue with an amplicon 500 pb; and a general primer pair (F 5’-CGATTCCGGCTACTTAGACATC-3’ and R 5’-GTTCTTCCAGCGTCCCATATAC-3’) aligned to the conserved motifs in eight predicted TPS genes of P. guajava genome, corresponding to sequences from 415 to 502 pb.
The DNA sequences of the putative TPS were amplified from P. guajava and P. guineense genomic DNA using the primers. Amplification reaction consisted of 50 ng genomic DNA, 200 µM dNTPs, 0.5 µM each R and F primers, 1 U GoTaq enzyme (Promega), 1X GoTaq enzyme reaction buffer and 1.8 mM MgCl2. Amplification conditions were initial denaturation at 95 °C for 5 min, followed by 30 cycles of 95 °C for 1 min, 58 °C for 45 s, 72 °C for 1 min and a final extension at 72 °C for 5 min. The amplification products were evaluated on 1.5% agarose gel and NanoDrop. Then, DNA probes were generated for each putative gene by a second PCR reaction on the same conditions described above, differing by the labeling with Tetramethyl-rhodamine 5-dUTP (Roche) for the specific or ChromaTide Alexa Fluor 488–5-dUTP (Life Technologies) for the general. Fluorescent in situ hybridization (FISH) was performed in slides containing isolated and preserved nuclei to detect the number of hybridization signals corresponding to the TPS genes. Hybridization mix consisted of 50% formamide, 2X SSC and 200 ng of the probe. This mix was applied to the slide, which was covered with a coverslip, sealed with rubber cement and kept at 37 °C for 20 h. Post-hybridization wash was in 2X SSC at 42 °C for 20 min. Slides were counterstained with 4′,6-diamidino-2-phenylindole and analyzed on a photomicroscope Olympus BX60 equipped with epifluorescence and an immersion objective 100x/A.N. 1.4. At least 20 nuclei were scrambled for each species and for each gene using a 12-bit CCD digital video camera (Olympus) coupled to the photomicroscope and a computer with a digitizer plate. Captured images were processed by Image ProPlus 6.1 (Media Cybernetics).
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Govaerts, R., Dransfield, J., Zona, S., Hodel, D. R. & Henderson, A. World Checklist of Myrtaceae. Facilitated by the Royal Botanic Gardens, Kew. http://wcsp.science.kew.org/ (2022).
Proença, C. E. B. et al. Diversity, phylogeny and evolution of the rapidly evolving genus Psidium L. (Myrtaceae, Myrteae). Ann. Bot. https://doi.org/10.1093/aob/mcac005 (2022).
Proença, C. E. B., Costa, I. R. & Tuler, A. C. Psidium in Flora e Funga do Brasil. Jardim Botânico do Rio de Janeiro https://floradobrasil.jbrj.gov.br/FB10853 (2022).
Vitti, K. A., de Lima, L. M. & Filho, J. G. M. Agricultural and economic characterization of guava production in Brazil. Rev. Bras. Frutic. 42, 1–11 (2020).
Thakur, N., Upadhyay, S. & Preeti, S. Medicinal and traditional properties of Psidium guajava: a review. Octa J. Environ. Res. 8, 22–25 (2020).
Degla, L. H. et al. Pharmacognostical, biochemical activities and zootechnical applications of Psidium guajava (Myrtaceae), plant with high médicinal value in tropical and subtropical parts of the world: A review. J. Med. Plants 9, 14–18 (2021).
Bezerra, J. E. F., Lederman, I. E., Junior, J. F. da S. & Proença, C. E. B. In: Aracá. In: Vieira, R. F., Costa, T. S. A., Silva, D. B., Sano, S. M., Ferreira, F. R. Frutas Nativas da Região Centro-Oeste do Brasil. Frutas Nativas da Região Centro-Oste do Brasil. Brasília, DF: Embrapa. Informação Tecnológica: Embrapa Recursos Genéticos e Biotecnologia, p. 42–62 (2006).
Tuler, A. C. et al. Diversification and geographical distribution of Psidium (Myrtaceae) species with distinct ploidy levels. Trees Struct. Funct. https://doi.org/10.1007/s00468-019-01845-2 (2019).
Machado, R. M. & Forni-Martins, E. R. Psidium cattleyanum Sabine (Myrtaceae), a neotropical polyploid complex with wide geographic distribution: Insights from cytogenetic and DNA content analysis. Rev. Bras. Bot. 45, 943–955 (2022).
Praça-Fontes, M. M., Carvalho, C. R., Clarindo, W. R. & Cruz, C. D. Revisiting the DNA C-values of the genome size-standards used in plant flow cytometry to choose the “best primary standards”. Plant Cell Rep. 30, 1183–1191 (2011).
Doležel, J., Sgorbati, S. & Lucretti, S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 85, 625–631 (1992).
Feng, C. et al. A chromosome-level genome assembly provides insights into ascorbic acid accumulation and fruit softening in guava (Psidium guajava). Plant Biotechnol. J. 19, 717–730 (2021).
Vetö, N. M. et al. Transcriptomics analysis of Psidium cattleyanum Sabine (Myrtaceae) unveil potential genes involved in fruit pigmentation. Genet. Mol. Biol. 43, 1–11 (2020).
Wendel, J. F., Lisch, D., Hu, G. & Mason, A. S. The long and short of doubling down: Polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 49, 1–7 (2018).
Machado, R. M. et al. Population structure and intraspecific ecological niche differentiation point to lineage divergence promoted by polyploidization in Psidium cattleyanum (Myrtaceae ). Tree Genet. Genomes. https://doi.org/10.1007/s11295-022-01551-0 (2022).
Corneillie, S. et al. Polyploidy affects plant growth and alters cell wall composition. Plant Physiol. 179, 74–87 (2019).
Lynch, M. & Conery, J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000).
Wang, Y., Wang, X. & Paterson, A. H. Genome and gene duplications and gene expression divergence: A view from plants. Ann. N. Y. Acad. Sci. 1256, 1–14 (2012).
Wendel, J. F. Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249 (2000).
Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20, 116–122 (2004).
Myburg, A. A. et al. The genome of Eucalyptus grandis. Nature 510, 356–362 (2014).
Alonso, C., Pérez, R., Bazaga, P. & Herrera, C. M. Global DNA cytosine methylation as an evolving trait: Phylogenetic signal and correlated evolution with genome size in angiosperms. Front. Genet. 6, 1–9 (2015).
Alonso, C., Balao, F., Bazaga, P. & Perez, R. Epigenetic contribution to successful polyploidizations: Variation in global cytosine methylation along an extensive ploidy series in Dianthus broteri (Caryophyllaceae). New Phytol. https://doi.org/10.5465/AMBPP.2016.25 (2016).
Spadeto, M. S. et al. Intraspecific C-value variation and the outcomes in Psidium cattleyanum Sabine essential oil. Braz. J. Biol. 82, 1–8 (2022).
Pichersky, E. & Gershenzon, J. The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 5, 237–243 (2002).
Kulheim, C. et al. The Eucalyptus terpene synthase gene family. BMC Genomics 16, 450 (2015).
Padovan, A., Keszei, A., Külheim, C. & Foley, W. J. The evolution of foliar terpene diversity in Myrtaceae. Phytochem. Rev. 13, 695–716 (2014).
Ferreira Macedo, J. G. et al. Therapeutic indications, chemical composition and biological activity of native Brazilian species from Psidium genus (Myrtaceae): A review. J. Ethnopharmacol. 278, 114148 (2021).
De Souza, A. D. G., Resende, L. V., De Lima, I. P., Martins, L. S. S. & Techio, V. H. Chromosome number and nuclear DNA amount in Psidium spp. resistant and susceptible to Meloidogyne enterolobii and its relation with compatibility between rootstocks and commercial varieties of guava tree. Plant Syst. Evol. 301, 231–237 (2014).
Marques, A. M. et al. Refinement of the karyological aspects of Psidium guineense (Swartz, 1788): A comparison with Psidium guajava (Linnaeus, 1753). Comp. Cytogenet. 10, 117–128 (2016).
Da Costa, I. R. Estudos evolutivos em Myrtaceae: aspectos citotaxonômicos e filogenéticos em Myrteae, enfatizando Psidium e gêneros relacionados. Doctorate in Plant Biology – Universidade Estadual de Campinas, Instituto de Biologia. Campinas/SP (2009).
Mason, A. S. & Pires, J. C. Unreduced gametes: Meiotic mishap or evolutionary mechanism?. Trends Genet. 31, 5–10 (2015).
Sattler, M. C., Carvalho, C. R. & Clarindo, W. R. The polyploidy and its key role in plant breeding. Planta 243, 281–296 (2016).
Pereira, R. C., Davide, L. C., Techio, V. H. & Timbó, A. L. O. Duplicação cromossômica de gramíneas forrageiras: Uma alternativa para programas de melhoramento genético. Cienc. Rural 42, 1278–1285 (2012).
Arévalo-Marín, E. et al. The taming of Psidium guajava: Natural and cultural history of a neotropical fruit. Front. Plant Sci. 12, 1–15 (2021).
Machado, R. M. Distribuição geográfica e análise cariotípica dos citótipos de Psidium cattleianum Sabine (Myrtaceae). Dissertação (Mestrado em Biologia Vegetal) - Universidade Estadual de Campinas, Instituto de Biologia, Campinas, SP. (2016).
Carvalho, G. M. A., Carvalho, C. R. & Soares, F. A. F. Flow cytometry and cytogenetic tools in eucalypts: Genome size variation × karyotype stability. Tree Genet. Genomes 13, 1 (2017).
Iannicelli, J. et al. Effect of polyploidization in the production of essential oils in Lippia integrifolia. Ind. Crops Prod. 81, 20–29 (2016).
Jiang, S. Y., Jin, J., Sarojam, R. & Ramachandran, S. A comprehensive survey on the terpene synthase gene family provides new insight into its evolutionary patterns. Genome Biol. Evol. 11, 2078–2098 (2019).
De Souza, T. S. et al. Chemotype diversity of Psidium guajava L. Phytochemistry 153, 129–137 (2018).
Mendes, L. A. et al. Larvicidal effect of essential oils from Brazilian cultivars of guava on Aedes aegypti L. Ind. Crops Prod. 108, 684–689 (2017).
Mendes, L. A. et al. Spring alterations in the chromatographic profile of leaf essential oils of improved guava genotypes in Brazil. Sci. Hortic. (Amsterdam) 238, 295–302 (2018).
Galbraith, D. W. et al. Flow cytometric analysis of the cell cycle. Lab. Proced. their Appl. https://doi.org/10.1016/b978-0-12-715001-7.50090-1 (1983).
Otto, F. DAPI staining of fixed cells for high-resolution flow cytometly of nuclear DNA. Methods Cell Biol. 33, 105–110 (1990).
Cruz, C. D. GENES - Software para análise de dados em estatística experimental e em genética quantitativa. Acta Sci. Agron. 35, 271–276 (2013).
Meister, A. & Barow, M. DNA base composition of plant genomes. Flow Cytom. Plant Cells Anal. Genes Chromosom. Genomes https://doi.org/10.1002/9783527610921.ch8 (2007).
Alves, L. B., Noia, L. R., Canal, G. B., Ferreira, A. & Ferreira, M. F. da S. Epigenetic variation in guava (Psidium guajava) genotypes during the vegetative and reproductive phases of the production cycle. Genet. Mol. Res. 19, (2020).
Bernardes, C. de O. Diversidade genética, caracterização e atividade de óleos essenciais em Psidium spp. (Tese (Doutorado em Genética e Melhoramento) - Universidade Federal do Espírito Santo, Alegre-ES, 2017).
Adams, R. P. Identification of essential oil components by gas chromatography/mass spectrometry. J. Am. Soc. Mass Spectrom. 8 (2007).
Linstrom, P. J. & Mallard, W. G. NIST Chemistry WebBook. Natl Inti Stand. Technol. http://webbook.nist.gov/chemistry (2018).
El-Sayed, A. M. The Pherobase: Database of pheromones and semiochemicals. http://www.pherobase.com/ (2019).
Alicandri, E. et al. On the evolution and functional diversity of terpene synthases in the Pinus species: A Review. J. Mol. Evol. https://doi.org/10.1007/s00239-020-09930-8 (2020).
Gao, Y., Honzatko, R. B. & Peters, R. J. Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat. Prod. Rep. 29, 1153–1175 (2012).
Butler, J. B. et al. Annotation of the Corymbia terpene synthase gene family shows broad conservation but dynamic evolution of physical clusters relative to Eucalyptus. Heredity (Edinb). 121, 87–104 (2018).
Funding
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília – DF, Brazil; grants 443801/2014-2 and 308828/2015–1), Fundação de Amparo à Pesquisa do Espírito Santo (FAPES/VALE, Vitória – ES, Brazil; grant 75516586/16) and VALE. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) – Finance Code 001.
Author information
Authors and Affiliations
Contributions
M.A.S.: Investigation, writing – original draft, Writing – review & editing. F.A.F.S.: Investigation, Writing – original draft, Writing – review & editing. W.R.C.: Supervision, Conceptualization, Writing – original draft, Writing – review & editing. L.A.M.: Investigation. L.B.A.: Investigation. A.F.: Supervision and Statistical Analysis. M.F.S.F.: Supervision, Conceptualization, Writing – original draft, Writing – review & editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Silva, M.A., Soares, F.A.F., Clarindo, W.R. et al. Genomic and epigenomic variation in Psidium species and their outcome under the yield and composition of essential oils. Sci Rep 13, 1385 (2023). https://doi.org/10.1038/s41598-023-27912-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-27912-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.