Abstract
Recently, populations of various species with very low genetic diversity have been discovered. Some of these persist in the long term, but others could face extinction due to accelerated loss of fitness. In this work, we characterize 45 individuals of one of these populations, belonging to the Iberian desman (Galemys pyrenaicus). For this, we used the ddRADseq technique, which generated 1421 SNPs. The heterozygosity values of the analyzed individuals were among the lowest recorded for mammals, ranging from 26 to 91 SNPs/Mb. Furthermore, the individuals from one of the localities, highly isolated due to strong barriers, presented extremely high inbreeding coefficients, with values above 0.7. Under this scenario of low genetic diversity and elevated inbreeding levels, some individuals appeared to be almost genetically identical. We used different methods and simulations to determine if genetic identification and parentage analysis were possible in this population. Only one of the methods, which does not assume population homogeneity, was able to identify all individuals correctly. Therefore, genetically impoverished populations pose a great methodological challenge for their genetic study. However, these populations are of primary scientific and conservation interest, so it is essential to characterize them genetically and improve genomic methodologies for their research.
Similar content being viewed by others
Introduction
Some of the main threats that can affect the viability of a population include habitat loss and fragmentation, which, often, lead to the isolation of small populations and, consequently, reduced genetic variation and inbreeding1,2. Loss of genetic diversity may have detrimental effects on both population fitness and viability in the long term by restricting evolutionary potential3. In addition, inbreeding, caused by mating between close relatives, can lead to inbreeding depression, reducing fitness and population growth in the short term4. Both of these factors significantly increase the probability of extinction of populations.
Indeed, a growing number of studies based on complete genomes have recently revealed very low genetic diversity in certain mammalian species of conservation concern. In some of these, heterozygosity is as low as ~ 100 SNPs/Mb (heterozygous sites or SNPs per mega-base), one order of magnitude below that of species with large populations. These include species such as the vaquita (Phocoena sinus), with 105 SNPs/Mb5, and the Iberian lynx (Lynx pardinus), with 102 SNPs/Mb6. In a few other species, the heterozygosity is yet another order of magnitude smaller, such as in the Island fox (Urocyon littoralis), with just 14 SNPs/Mb found in an individual from San Nicolas Island7. How these species have reached these extraordinarily low levels of genetic diversity and how this may be aggravated by consanguinity and inbreeding depression is still unclear2,8,9,10,11. To address these questions, genomic analyses of populations with extremely low genetic diversity are essential.
The Iberian desman (Galemys pyrenaicus), also known as the Pyrenean desman, is a small semi-aquatic mammal that inhabits clean rivers in mountains of the northern half of the Iberian Peninsula12,13. In recent years, the species has suffered significant population declines for reasons that are still being analyzed, but loss and fragmentation of the riparian habitat appears to be one of the most critical factors14. Previous double digest restriction site-associated DNA (ddRAD) and whole-genome sequencing studies of the species revealed that the Iberian desman has exceptionally low heterozygosity levels15,16. The lowest levels were recorded in the Pyrenees, where heterozygosity ranged from 12 to 116 SNPs/Mb, covering the two lowest orders of magnitude reported in mammals. Studies based on kinship networks revealed important connectivity problems for the species due to physical barriers, such as dams, which are leading to populations with very high inbreeding levels in the upper parts of rivers17,18. Ecological barriers, including the desiccation of rivers, the presence of predators such as the American mink, and contamination coming from urban areas14, can also isolate populations.
Here, we analyze a population of Iberian desman from the western Pyrenees, where it was previously shown, using relatively few individuals, that the species exhibits shallow genetic diversity throughout the area15,16. To obtain genomic sequences and SNPs for the analyses, we applied the ddRADseq (ddRAD sequencing) technique, which is popular in population genomic studies due to the ease of obtaining data from a large number of individuals19, and assembled the reads using the recently sequenced Iberian desman genome15. We determined the heterozygosity rate and the inbreeding coefficient for 45 individuals to describe the genetic diversity of this population in detail and to understand whether genetic diversity and inbreeding were homogeneous in this area or whether these values were more extreme in some particular localities. We also tested the resolution power of the SNPs obtained by ddRADseq to genotype individuals in these populations of extremely low genetic variability and showed that most of the methods tested were unable to perform a correct individualization. Solving these methodological problems is crucial to address critical conservation problems in populations of species with extremely low genetic diversity.
Materials and methods
Samples of the Pyrenean desman
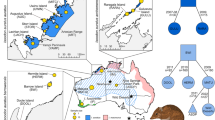
The population studied is located in the provinces of Gipuzkoa and Navarra (Spain), in the north of the Iberian Peninsula (Fig. 1). We used 45 tissue samples from Iberian desmans (a small piece from the tail tip) captured between 1997 and 2011 during monitoring works of the species promoted by the environmental authorities Diputación Foral de Gipuzkoa and Gobierno de Navarra (Supplementary Table S1 of the Supporting Information and Fig. 1). From these, 13 samples had already been used in previous works15,16,20.
(A) Map of the Iberian Peninsula showing the Iberian desman distribution range in grey. The study area is indicated with a red square. (B) Enlarged map showing the localities of the Iberian desmans used in this work. Each sampled individual is represented with different symbols for males (squares) and females (circles). The numbers indicate the localities (1: Aiaiturrieta-Ataun; 2: Amundarain-Zaldibia; 3: Leitzaran-Berastegi; 4: Erasote-Leitza; 5: Urumea; 6: Elama-Artikutza; 7: Ameztia-Labaien; 8: Ezpelura-Urrotz; 9: Sasoaran-Eugi; 10: Olazar-Eugi; 11: Aritzakun; and 12: Urrobi-Auritz). Red lines delimit the three main river basins: Cantabrian at the northwest, Adur at the northeast, and Ebro at the south. Populations 1 and 2 are possibly extinct. Human population nuclei with more than 500 inhabitants are indicated in dark gray. The map was constructed using QGIS 2.1453. The distribution range of Galemys pyrenaicus was generated using information from different sources13,54. The coastline shapefile was downloaded from Natural Earth (https://www.naturalearthdata.com). The shapefiles of the shaded relief map, rivers, main river basins, and human population nuclei were downloaded from Instituto Geográfico Nacional (https://www.ign.es). The river shapefile for France was obtained from DIVA-GIS (https://www.diva-gis.org).
Ethics statement
No animal was specifically captured for this study, as they had been captured for previous works to monitor these populations. Therefore, this study did not require ethics approval by a specific committee.
Construction of ddRAD libraries and sequence processing
DNA was extracted using the DNEasy Blood and Tissue Kit (QIAGEN). Genomic libraries were constructed following the ddRADseq protocol19 with some modifications. Each library was made in groups of 24 samples, and samples with low sequence yield were repeated in subsequent experiments, as indicated in Supplementary Table S1. First, the DNA was digested using the restriction enzymes EcoRI and MspI. The digested DNA was ligated to adapters P1 and P2, which bind to the EcoRI and MspI overhangs, respectively. Adapter P1 contains a different 5-nucleotide barcode for each sample so that they can be identified in the library sequences. All samples were then pooled and a fraction between 300 and 400 bp was selected in an E-Gel EX 2% agarose gel (Invitrogen). From this fraction, 16-cycle PCR amplifications were carried out using primers that anneal to the adapters and allow the generation of standard Illumina libraries. To minimize sequencing bias, 6 PCRs were performed for each sample. PCR products were concentrated in 20 μl using the MinElute PCR Purification Kit (QIAGEN). Finally, the library concentration was estimated using a Nanodrop, 400 ng were run in an EX 2% agarose gel, and the library was extracted in 30 μl using the QIAquick Gel Extraction Kit (QIAGEN). The libraries were sequenced using the NextSeq Sequencing System (Illumina) in the Genomics Core Facility at Pompeu Fabra University with the 150-cycle Mid Output kit and single-read sequencing.
The sequences obtained were filtered and assembled using the Stacks 2.60 package21. First, the process_radtags program was used to separate reads belonging to different individuals according to the barcodes using the recovery option (-r) and to filter out reads with a quality score limit (-s) of 10. After this step, reads from samples sequenced in different libraries were combined. This set of filtered reads is available at Dryad (see Data Availability section). Then, the reads were mapped to the reference genome of the Iberian desman15 with BWA v0.7.1722 using the mem algorithm. Subsequently, SAMtools v1.923 was used to produce BAM alignments where reads with a minimum mapping quality (-q) of 20 were kept. The mapped reads were processed using Gstacks from the Stacks package with the SNP calling model snp and both alpha thresholds for discovering SNPs and for calling genotypes of 0.01. Using the populations program of the same package, the sequences were saved in FASTA format after selecting loci with a minimum proportion of called individuals (r) of 0.9 (i.e., loci present in at least 90% of the individuals) and a minor allele frequency (MAF) of 0. Using the FASTA sequences of the loci, heterozygosity was estimated for each specimen as the number of heterozygous positions divided by the total sequenced length of the loci. The SNP dataset of all samples containing the first SNP from each locus was obtained with r = 0.9 and MAF = 0.025 and it was filtered for linkage disequilibrium (r2 > 0.8) with PLINK v1.90b6.2224. These SNPs were saved in PLINK and VCF formats for further analyses.
The sex of all individuals was determined bioinformatically by mapping the reads against a Y-chromosome loci database as in previous work16.
Population structure analysis
Population structure was analyzed with STRUCTURE 2.3.425 using the admixture model, correlated allele frequencies, and a number of populations (K) ranging from 1 to 6. A total of 1,000,000 iterations were run with a burn-in of 100,000. For each K value, 10 independent runs were performed and summarized with CLUMPP26. The optimal value of K was estimated with the method of Evanno et al.27, as implemented in STRUCTURE HARVESTER28.
The principal component analysis (PCA) was performed with the KING v.2.2.5 program29.
Relatedness and inbreeding coefficients
We calculated the kinship coefficients between pairs of Iberian desmans with the KING program29. KING uses a method to infer the kinship coefficient between pairs of individuals which does not require allele frequency information. Specifically, we used the KING-robust method, which is not affected by population structure, with the kinship option. Negative kinship values and pairs with a flag error of 0 or 0.5, corresponding to unrelated individuals, were not used. The kinship coefficients were doubled to convert them to relatedness coefficients.
The relatedness coefficients were also calculated using the RELATED program30,31. We used the dyadml estimator32, which was previously shown to be the most adequate for SNPs derived from ddRADseq data18, and the full nine states identity-by-descent (IBD) model, which takes inbreeding into account. Confidence intervals (95%) were calculated with 100 bootstraps, and only relatedness values with confidence intervals that did not overlap 0 were used. Using the same model, we estimated the inbreeding coefficient for each Iberian desman. The inbreeding coefficient was also estimated with PLINK24, which estimates this value based on the observed versus expected number of homozygous genotypes. It should be taken into account that these two estimates of the inbreeding coefficient, based on SNP frequencies without genomic position information, measure relatively recent inbreeding events and do not reflect past inbreeding, as do measures based on runs of homozygosity estimated from whole genomes15.
Test of individualization using duplicate samples
As the relatedness coefficients were obtained from specimens with very low genetic diversity and very high inbreeding levels, we tested the accuracy of the estimates in this special context. For this, we used the reads of duplicate samples from the same individual that were repeated in different runs to increase their coverage (Supplementary Table S1). Only samples with a minimum of 200,000 reads, which were found to be sufficient for this purpose in preliminary analysis, were compared in a new assembly. In it, duplicate samples were included, without combining them, together with all the other samples in the study, giving rise to the comparison of 23 replicated samples. In addition, we recorded how these programs behaved when comparing three individuals from Amundarain-Zaldibia, with extreme inbreeding levels and which were probably related to one another. We had two samples for one of these individuals, leading to five comparisons between highly inbred and related individuals. For this analysis, we generated the SNP dataset with the same parameters as above. In addition, we generated three extra datasets with higher MAF filter values (0.1, 0.2, and 0.3, respectively) to test the effect of this parameter.
The samples were individually identified using different methods. The KING program and the RELATED program with the three-states IBD model (with no inbreeding) were used to estimate relatedness coefficients and the individualization was performed considering that duplicate samples have a relatedness coefficient of 133. We also used the duplicate option in KING to detect duplicates automatically. Finally, we tested other programs for detecting duplicates and kinship relationships: the clustering method implemented in PLINK using the "–cluster –matrix" option24; COLONY v2.0.6.5 with allelic dropout and false allele rates of 0.001 each33,34, as they proved to be the best error rates in preliminary analyses; and the pedigree-reconstruction methods PRIMUS v1.9.035 and VCF2LR36.
Test of relatedness, individualization and inbreeding estimates using simulated pedigrees
We also tested the accuracy of relatedness estimates and individualization using simulations of three artificial pedigrees, in which genotypes from our dataset were bioinformatically crossed using the script GetCrosses18. Each pedigree had founders from different areas (Supplementary Fig. S1). We also assessed the accuracy of the inbreeding coefficient calculations from RELATED using three pedigrees with additional crosses between relatives (Supplementary Fig. S2). In both cases, individuals with exceptional inbreeding levels were excluded as founders, leaving founders with inbreeding values that ranged from 0.0175 to 0.4783, to produce more generally applicable results. A total of 100 simulations were performed for each pedigree.
Results
Sequence assembly
After mapping the reads for each of the 45 Iberian desman to the reference genome (Supplementary Table S2), a total of 43,478 loci were assembled and a set of 1421 SNPs present in at least 90% of the individuals was generated. The genetic sexing led to the determination of 21 females and 24 males (Supplementary Table S1).
Population structure
Both the PCA (Fig. 2) and the STRUCTURE (Supplementary Fig. S3) analyses showed a certain degree of structure in the area. The optimal value of populations in STRUCTURE was K = 2, followed by a secondary peak at K = 4 (Supplementary Fig. S3B).
Principal component analysis of the Iberian desman individuals. Each locality is represented with a different color. The locality code, which can be found in the legend of Fig. 1, is also indicated.
Heterozygosity and inbreeding coefficients
Heterozygosity rates were very low for all the individuals, ranging from 26 to 91 SNPs/Mb (Supplementary Table S3 and Supplementary Fig. S4). As ddRADseq data is a subset of the genome, heterozygosity values calculated here may not be identical to those obtained from the whole genome sequence, but they are within the same order of magnitude. For example, for one desman from which the whole genome was obtained (IBE-C2769), heterozygosity was 116 SNPs/Mb when calculated from the whole genome15 and 80 SNPs/Mb from the ddRADseq data (Supplementary Table S3).
Individual inbreeding coefficients estimated with RELATED showed a mean value of 0.25 (Supplementary Table S3) and were highly variable among localities. The map showing color-coded values (Fig. 3) revealed exceptional inbreeding coefficients in the two most occidental localities, Amundarain-Zaldibia and Aiaiturrieta-Ataun, where most coefficients were higher than 0.7 and the four individuals presented a mean value of 0.76. In contrast, in two central localities within the analyzed area, Ezpelura-Urrotz and Ameztia-Labaien, the individuals had the lowest inbreeding values, with a mean of 0.11. Heterozygosity and inbreeding coefficient showed a strong negative correlation (R = − 0.93) when considering just this population of the Iberian desman.
Map of the Iberian desmans with their inbreeding coefficients, estimated with RELATED, represented using a color scale. The map was constructed as indicated in the legend to Fig. 1.
Individual inbreeding coefficients estimated with PLINK showed similar results, with a mean value of also 0.25 (Supplementary Table S3).
Pairwise relatedness and connectivity networks
We found 114 relationships between pairs of Iberian desmans determined using KING and 382 with RELATED. To visualize the connectivity patterns in the area, we represented the kinship networks on a map (Fig. 4 and Supplementary Fig. S5 for KING and RELATED relationships, respectively). As the relationships derived from RELATED were more abundant, these were subdivided into close and distant relationships according to a 0.2 threshold. Both relationships from KING and close relationship from RELATED predominantly showed connectivity between individuals from the same river or from neighboring localities. Only distant relationships determined using RELATED (Supplementary Fig. S5B) showed connectivity between individuals from different basins.
Maps plotting networks of related individuals detected by KING. Line thickness is proportional to the relatedness coefficient of the connected Iberian desmans. The map was constructed as indicated in the legend to Fig. 1.
Evaluation of the power to perform individualization and relatedness
We first tested the accuracy of the individualization by including 23 known pairs of replicates from different ddRAD libraries in a new assembly that rendered 688 SNPs. Theoretically, the relatedness coefficient between samples of the same individual should be 1. Both RELATED and KING showed values of this parameter close to 1 for all the pairs of replicates (Fig. 5; > 0.9 for RELATED and > 0.8 for KING), which, in principle, would be a good result in most situations as these values are much higher than the highest theoretical relatedness value between different individuals (0.5 for first-degree relationships). Thus, when applying a threshold of 0.8, which is the most favorable for both programs, all 23 replicates were detected (Table 1). As for the other options and programs used, the duplicate option from KING was able to detect only 21 out of 23 pairs of replicates whereas PLINK, COLONY, PRIMUS, and VCF2LR detected all the duplicate pairs.
Distinguishing samples of different individuals is more problematic under high inbreeding scenarios because the relatedness values are much greater than the theoretical ones. When considering samples of different individuals, we evaluated whether these programs could discern three closely related individuals with very high inbreeding levels from Amundarain-Zaldibia. For one of these we had two samples, enabling five comparisons of related and highly inbred individuals. Indeed, RELATED showed relatedness values > 0.9 between samples known to belong to these different individuals, overlapping with the values found for replicate samples and therefore incorrectly individualizing the inbred desmans (Fig. 5 and Table 1). In the case of KING, all estimations between the different individuals, including the most inbred, were lower than 0.8, and thus lower than all the values between replicate samples, meaning that it correctly distinguished the inbred individuals (Fig. 5 and Table 1). PLINK, COLONY, PRIMUS, and VCF2LR also considered the highly inbred specimens to be replicates, giving erroneous individualization results (Table 1). Similar results for the identification of replicated and different individuals were obtained for all programs when different MAF filters were used (Supplementary Table S4), so it is probably better to use a MAF filter that does not remove too many SNPs.
We also tested the reliability of the relatedness coefficients estimated by simulating artificial pedigrees (Supplementary Fig. S6). We found a general overestimation of all the relationships tested in the three pedigrees, but the relatedness coefficients obtained using RELATED (Supplementary Table S5) were, in most cases, higher than those estimated with KING (Supplementary Table S6). Overestimation increased as more distant relationships were simulated. Only in the cases of parent–offspring and grandparent-grandchild, KING underestimated the relatedness coefficient (Table 2).
To test the performance of the inbreeding coefficient estimates with RELATED, we simulated different pedigrees with inbreeding (Supplementary Fig. S7). We found highly accurate estimates of the inbreeding coefficients in both the western and central areas, and only in pedigrees from the eastern area were some of the values obtained much higher than expected (Table 3 and Supplementary Table S7).
Discussion
Genetic analyses in populations with extremely low genetic diversity using ddRADseq
Identification of individuals from genetic marker data is important in many studies involving the monitoring of elusive species, for example, in estimating population density with capture-recapture, either using trapped specimens or non-invasive samples37,38. When there is ample marker information, individual identification is straightforward using various methods, including mismatch methods, pairwise relatedness analysis, or the COLONY program to handle multiple replicates33,39,40. Nevertheless, few studies have addressed the issue of differentiating them under conditions of both low genetic diversity and high levels of inbreeding, where individuals share a large proportion of their genotype41. In particular, exceptionally high inbreeding and low heterozygosity levels mean that some individuals appear to be almost clones at the genomic level. This is the case with the three highly inbred desman individuals from Amundarain-Zaldibia, which have an average inbreeding coefficient of 0.78 and an average heterozygosity of 27.3 SNPs/Mb, as well as a high degree of kinship between them, making it very difficult for the available individual identification programs to distinguish between replicates of the same individual and samples of different individuals (Fig. 5 and Table 1). In this study, we were able to use tissue samples of known origin and replicate libraries, making it a unique opportunity to address the methodological problems for detecting individuals in a context of low genetic diversity and high inbreeding levels. Of all the programs tested, those based on the pairwise relatedness between individuals (RELATED and KING) gave the best individualization results, as already shown in other studies39. In particular, KING is the only program that was able to differentiate between replicates of the same individual and samples of different individuals with high inbreeding levels (Table 1). The RELATED program was able to allocate all the known replicates, but failed to distinguish the highly inbred individuals as different samples. The different performance between RELATED and KING is probably due to the fact that the likelihood estimator implemented in RELATED uses the allele frequencies of the given set of samples to calculate the relatedness coefficient32, while the KING algorithm uses only SNP data from the pair of individuals tested each time, which makes the inference robust to the presence of population structure29. The main problem with algorithms that use allele frequencies, such as RELATED, is that they assume a homogeneous population structure and lead to inflated results among individuals of the same group when this condition is not fulfilled29; this is likely to happen in populations with certain population structure (Fig. 2 and Supplementary Fig. S3) and low levels of connectivity (Fig. 4 and Supplementary Fig. S5), such as those in this study.
This difference between programs in the power to estimate the relatedness coefficient can also be observed in the simulations with artificial pedigrees, where the estimates produced using RELATED were systematically inflated for most of the tested kinship categories compared to those obtained with KING (Table 2 and Supplementary Fig. S6). In any case, the simulations proved that, despite not being able to categorize the specific kinship categories, it is possible to detect a certain level of relatedness between the samples analyzed and construct relatedness networks to assess the level of connectivity in the area.
Estimations of the inbreeding coefficient of the artificial pedigrees using RELATED were generally more accurate than the relatedness coefficient (Table 3 and Supplementary Fig. S7), as observed in previous simulation works of populations with a higher genetic diversity and lower inbreeding17,18. This indicates that inbreeding estimations are more robust to the presence of population stratification and work well with both low and high levels of inbreeding. This is an important result because it allows the assessment of genetic health in inbred and low-diversity populations. The fact that the PLINK program, whose estimates are based on a different principle, gives inbreeding coefficients very similar to those obtained by RELATED, is another point that gives confidence to these results.
Extremely low genetic diversity and high inbreeding levels in the Iberian desman
Heterozygosity was unusually low in all specimens (Supplementary Table S3), with values ranging from 26 to 91 SNPs/Mb. These values are among the lowest found for the Iberian desman across its entire range15,16. In fact, they are among the smallest found for any mammal so far, only comparable to those found for the endangered and completely isolated Channel Island fox7, as estimated from its whole genome. It should be noted that heterozygosity values estimated from ddRADseq are not exactly the same as those estimated from whole genome data, but they are within the same order of magnitude15. Many other species have proven to prevail for long periods of time with low levels of genetic diversity5,6,7,42,43,44. Low genetic diversity diminishes the evolutionary potential of populations3, making it difficult to understand how these populations are able to survive with these exceedingly low levels of genetic diversity. Recent work based on genomic data suggests that these populations may have a low mutational load (i.e., a small proportion of deleterious recessive mutations that could become homozygous under inbreeding), as a consequence of purging of recessive strongly deleterious mutations in the past9,45,46. However, inbreeding should also be taken into account in these populations because it can have a negative impact when weakly and mildly deleterious variants, very difficult to purge, become homozygous4,47. Consequently, inbreeding may be a critical factor for predicting the fate of these low-diversity populations.
Indeed, the inbreeding coefficients determined for the Iberian desmans in this area were of great interest as they varied widely among the different localities (Fig. 3). In the central populations, where the density of the species is higher, inbreeding was relatively low, with many individuals having values < 0.1 and thus of an acceptable level for wild animals3. In contrast, there are some populations with extremely high values. In particular, the four individuals from Amundarain-Zaldibia and Aiaiturrieta-Ataun exhibited extremely high levels of inbreeding (Fig. 3, Supplementary Table S3). These values are typical of critically endangered species or populations, including the Attwater’s prairie-chicken (Tympanuchus cupido attwateri), with values of 0.65 in some individuals48; and grey wolves (Canis lupus) of the highly inbred population on Isle Royale, with values of 0.8149. The presence of highly industrialized areas and large human population nuclei, such as Ordizia and Beasain, downstream from the Amundarain-Zaldibia and Aiaiturrieta-Ataun desman populations, could have played a role in isolating these individuals from the other populations. Specifically, the channeling of the rivers and the destruction of the riparian habitat, together with water pollution around these urban areas, could create ecological barriers to dispersal and favor the inbreeding of the isolated populations. No close relationships were found between individuals from this area and the rest (Fig. 4 and Supplementary Fig. S5), so each population may have been isolated for several generations. Probably as a consequence of this, no desman was recorded in Aiaiturrieta-Ataun after 2001 and in Amundarain-Zaldibia after 2006 (the latest surveys having been carried out in 2018), suggesting that these populations could be extinct and are likely an example of the inbreeding-driven extinction vortex50,51,52. Early knowledge of this problem and an action aimed at promoting dispersal from genetically healthier nearby populations to these strongly inbred populations, whether natural or assisted, could have helped to reverse this situation.
There are other isolated desman populations of the Iberian desman with very low genetic diversity that could follow a similar extinction trajectory. Studies should be promoted to characterize not only their demographic status, but also the heterozygosity and inbreeding levels of these populations, in order to evaluate their conservation status and take the necessary measures to ensure their long-term viability.
Data availability
Filtered ddRADseq data is available in Dryad (https://doi.org/10.5061/dryad.brv15dvck). Additional data and figures may be found in Supporting Information.
References
Nunney, L. & Campbell, K. A. Assessing minimum viable population size: Demography meets population genetics. Trends Ecol. Evol. 8, 234–239 (1993).
Keller, L. F. & Waller, D. M. Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 (2002).
Frankham, R. et al. Genetic Management of Fragmented Animal and Plant Populations. (Oxford University Press, 2017).
Charlesworth, D. & Willis, J. H. The genetics of inbreeding depression. Nat. Rev. Genet. 10, 783–796 (2009).
Morin, P. A. et al. Reference genome and demographic history of the most endangered marine mammal, the vaquita. Mol. Ecol. Resour. 21, 1008–1020 (2021).
Abascal, F. et al. Extreme genomic erosion after recurrent demographic bottlenecks in the highly endangered Iberian lynx. Genome Biol. 17, 251 (2016).
Robinson, J. A. et al. Genomic flatlining in the endangered island fox. Curr. Biol. 26, 1183–1189 (2016).
Hedrick, P. W. & Garcia-Dorado, A. Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol. Evol. 31, 940–952 (2016).
Teixeira, J. C. & Huber, C. D. The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. USA 118, e2015096118 (2021).
DeWoody, J. A., Harder, A. M., Mathur, S. & Willoughby, J. R. The long-standing significance of genetic diversity in conservation. Mol. Ecol. 30, 4147–4154 (2021).
García-Dorado, A. & Caballero, A. Neutral genetic diversity as a useful tool for conservation biology. Conserv. Genet. 22, 541–545 (2021).
Palmeirim, J. M. & Hoffmann, R. S. Galemys pyrenaicus. Mamm. Species 207, 1–5 (1983).
Kryštufek, B. & Motokawa, M. Species accounts of Talpidae. In Handbook of the Mammals of the World. Volume 8. Insectivores, Sloths and Colugos (eds R. A. Mittermeier & D. E. Wilson) 551–619 (Lynx Edicions, 2018).
Fernandes, M., Herrero, J., Aulagnier, S. & Amori, G. Galemys pyrenaicus. IUCN Red List of Threatened Species, e.T8826A12934876 (2008).
Escoda, L. & Castresana, J. The genome of the Pyrenean desman and the effects of bottlenecks and inbreeding on the genomic landscape of an endangered species. Evol. Appl. 14, 1898–1913 (2021).
Querejeta, M. et al. Genomic diversity and geographical structure of the Pyrenean desman. Conserv. Genet. 17, 1333–1344 (2016).
Escoda, L., Fernández-González, A. & Castresana, J. Quantitative analysis of connectivity in populations of a semi-aquatic mammal using kinship categories and network assortativity. Mol. Ecol. Resour. 19, 310–326 (2019).
Escoda, L., González-Esteban, J., Gómez, A. & Castresana, J. Using relatedness networks to infer contemporary dispersal: Application to the endangered mammal Galemys pyrenaicus. Mol. Ecol. 26, 3343–3357 (2017).
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S. & Hoekstra, H. E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7, e37135 (2012).
Igea, J. et al. Phylogeography and postglacial expansion of the endangered semi-aquatic mammal Galemys pyrenaicus. BMC Evol. Biol. 13, 115 (2013).
Rochette, N. C., Rivera-Colón, A. G. & Catchen, J. M. Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol. 28, 4737–4754 (2019).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Jakobsson, M. & Rosenberg, N. A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Earl, D. A. & vonHoldt, B. M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (2012).
Manichaikul, A. et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010).
Pew, J., Muir, P. H., Wang, J. & Frasier, T. R. related: An R package for analysing pairwise relatedness from codominant molecular markers. Mol. Ecol. Resour. 15, 557–561 (2015).
Wang, J. COANCESTRY: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 11, 141–145 (2011).
Milligan, B. G. Maximum-likelihood estimation of relatedness. Genetics 163, 1153–1167 (2003).
Wang, J. Individual identification from genetic marker data: Developments and accuracy comparisons of methods. Mol. Ecol. Resour. 16, 163–175 (2016).
Jones, O. R. & Wang, J. COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 10, 551–555 (2010).
Staples, J. et al. PRIMUS: Rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 95, 553–564 (2014).
Heinrich, V., Kamphans, T., Mundlos, S., Robinson, P. N. & Krawitz, P. M. A likelihood ratio-based method to predict exact pedigrees for complex families from next-generation sequencing data. Bioinformatics 33, 72–78 (2017).
Royle, J. A., Fuller, A. K. & Sutherland, C. Unifying population and landscape ecology with spatial capture-recapture. Ecography 41, 444–456 (2017).
Carroll, E. L. et al. Genetic and genomic monitoring with minimally invasive sampling methods. Evol. Appl. 11, 1094–1119 (2018).
Ringler, E., Mangione, R. & Ringler, M. Where have all the tadpoles gone? Individual genetic tracking of amphibian larvae until adulthood. Mol. Ecol. Resour. 15, 737–746 (2015).
Wang, J. Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet. Res. 89, 135–153 (2007).
Taylor, H. R., Kardos, M. D., Ramstad, K. M. & Allendorf, F. W. Valid estimates of individual inbreeding coefficients from marker-based pedigrees are not feasible in wild populations with low allelic diversity. Conserv. Genet. 16, 901–913 (2015).
Benazzo, A. et al. Survival and divergence in a small group: The extraordinary genomic history of the endangered Apennine brown bear stragglers. Proc. Natl. Acad. Sci. USA 114, E9589–E9597 (2017).
Johnson, J. A. et al. Long-term survival despite low genetic diversity in the critically endangered Madagascar fish-eagle. Mol. Ecol. 18, 54–63 (2009).
Milot, E., Weimerskirch, H., Duchesne, P. & Bernatchez, L. Surviving with low genetic diversity: The case of albatrosses. Proc. R. Soc. B 274, 779–787 (2007).
Agrawal, A. F. & Whitlock, M. C. Mutation load: The fitness of individuals in populations where deleterious alleles are abundant. Annu. Rev. Ecol. Evol. Syst. 43, 115–135 (2012).
Kyriazis, C. C., Wayne, R. K. & Lohmueller, K. E. Strongly deleterious mutations are a primary determinant of extinction risk due to inbreeding depression. Evol. Lett. 5, 33–47 (2021).
Mathur, S. & DeWoody, J. A. Genetic load has potential in large populations but is realized in small inbred populations. Evol. Appl. 14, 1540–1557 (2021).
Hammerly, S. C., Morrow, M. E. & Johnson, J. A. A comparison of pedigree- and DNA-based measures for identifying inbreeding depression in the critically endangered Attwater’s Prairie-chicken. Mol. Ecol. 22, 5313–5328 (2013).
Adams, J. R., Vucetich, L. M., Hedrick, P. W., Peterson, R. O. & Vucetich, J. A. Genomic sweep and potential genetic rescue during limiting environmental conditions in an isolated wolf population. Proc. R. Soc. B 278, 3336–3344 (2011).
Blomqvist, D., Pauliny, A., Larsson, M. & Flodin, L. A. Trapped in the extinction vortex? Strong genetic effects in a declining vertebrate population. BMC Evol. Biol. 10, 33 (2010).
Fagan, W. F. & Holmes, E. E. Quantifying the extinction vortex. Ecol. Lett. 9, 51–60 (2006).
Palomares, F. et al. Possible extinction vortex for a population of iberian lynx on the verge of extirpation. Conserv. Biol. 26, 689–697 (2012).
QGIS_Development_Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project. http://qgis.osgeo.org. (2021).
Nores, C., Queiroz, A. I. & Gisbert, J. Galemys pyrenaicus. In Atlas y libro rojo de los mamíferos terrestres de España (eds L. J. Palomo, J. Gisbert, & J. C. Blanco) 92–98 (Dirección General para la Biodiversidad-SECEM-SECEMU, 2007).
Acknowledgements
This work was financially supported by research projects PID2020-113586GB-I00 and CGL2017-84799-P of MCIN/AEI/100.13039/501100011033, the latter also co-funded by "ERDF A way of making Europe", to J.C. O.H. was funded by DFG postdoctoral grant HA7255/1-1.
Author information
Authors and Affiliations
Contributions
L.E., O.H., J.G.E., and J.C. designed research, performed research and contributed samples. L.E., O.H., and J.C. analyzed data. L.E. and J.C. wrote the manuscript, with input from the other authors. All authors contributed to the interpretation of the results and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Escoda, L., Hawlitschek, O., González-Esteban, J. et al. Methodological challenges in the genomic analysis of an endangered mammal population with low genetic diversity. Sci Rep 12, 21390 (2022). https://doi.org/10.1038/s41598-022-25619-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-25619-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.