Abstract
Gold nanoparticles (AuNPs) have different unique properties and a wide range of applications in different fields. Thereby, there is a growing urgency for the production of AuNPs using a safe and an economic method. In this study, optimization of fermentation conditions by Bacillus subtilis NRC1 for extracellular AuNPs synthesis using response surface methodology was achieved. The data obtained from Plackett–Burman design followed by Box–Behnken design indicated the accuracy and reliability of the model and it could be used to navigate the design space with a reasonable accuracy. Numerical optimization of Bacillus subtilis NRC1 active extracellular filtrate production, showed the optimum conditions of 0.74% (w/v) casein hydrolysate, 3.99% (w/v) dextrin, 47 × 106 CFU/ml inoculum size at pH 7.76 and 25 \(^\circ\)C to give the maximum AuNPs biosynthesis. The model was highly valid and the obtained data had a confidence factor of 98.48%. Statistical optimization resulted in a 2.6-fold increase in AuNPs production compared with that of the non-optimized medium.
Similar content being viewed by others
Introduction
In the recent years, nanoparticles (NPs) have attracted a great attention in different applications in medicine, agriculture, electronics, optics, catalysis and in biosensors. It is very well known that NPs (within the size less than 100 nm) are effectively used for most applications due to similar size to biomolecules and easier penetration1.
Gold and its compounds have long been used as medicinal agents throughout the history of civilization with its earliest record dating back to 5000 years ago in Egypt2. A wide range of AuNPs have been developed by several physical, chemical and biological routes3. Green or biological synthesis of NPs has received an interesting attention as it avoids the use of harmful chemicals and hazardous substances. In addition, the synthesis of NPs is at mild conditions of pressure, temperature and pH and does not require the addition of external reducing, capping and stabilizing agents which minimizes their toxicity and expands their biomedical applications4,5,6.
Many biological candidates such as fungi, bacteria and plants are the excellent choice for an eco-friendly green synthesis of AuNPs7. Microbes are regarded as potent green nanofactories. They have been used for the synthesis of NPs due to their ease of handling, growing in a low-cost medium, maintaining the safety levels, having the potential of adsorbing the metal ions and reducing them into NPs by the enzymes produced by metabolic processes2,8. The extracellular NPs synthesis, is preferred over the intracellular NPs synthesis as it is easier for the NPs extraction from the medium9.
There are several Bacilli species that have the ability to synthesize AuNPs as Bacillus niabensis, Bacillus subtilis, Bacillus licheniformis, Bacillus megaterium, Bacillus marisflavi, Bacillus cereus, Endophytic Bacillus cereus, Brevibacillus formosus, Lysinibacillus sp. and Lactobacillus sp.3,10,11,12,13,14.
It was reported that the synthesis of bacterial AuNPs is linked with bioactive molecules derived from bacteria15. AuNPs produced by diverse microorganisms perform numerous functions which are associated in many fields of applications in medicine as diagnosis and treatment of cancer, anti-angiogenesis, anti-arthritic, antimalarial agents and others8.
The traditional optimization of microbial fermentation has been carried out by monitoring the effect of one variable at a time (OVAT) on an experimental response. One parameter is changed with keeping other variables at constant levels in this technique. Therefore, the interactive effects among the different parameters and the complete influence of the parameter on the response could not be detected. Besides, a large number of experiments is necessary to conduct the research with this technique and subsequently more required time and expenses. Response surface methodology statistical analysis (RSM) is a well-known statistical analysis method which is applied in the optimization of medium constituents and other critical factors responsible for the production of microbial biomolecules16,17. Therefore, RSM could reduce time and costs with more precise results18. Also, RSM analyzes the interactions of all factors in a process and explains the prediction of optimum conditions19.
The cytotoxic activity of biogenic AuNPs against different types of carcinoma was reported in the literature. Virmani et al.20 compared the anticancer behavior of the biogenic AuNPs and chemically synthesized AuNPs towards the cancerous (HeLa, MCF-7, A549 and H1299) and normal (HEK293) cell lines. Cell viability assay suggested that the biologically synthesized particles significantly inhibited the growth of cancer cells in a dose dependent manner, with an IC50 value of about 200 µg/ml and were found to be non-toxic towards the normal cells. Barabadi et al.21,21 reviewed the anticancer activities of biologically synthesized AuNPs against different cancer cell lines and they concluded that biologically synthesized AuNPs had anticancer activities against most of the tested cancer cell lines. In addition, Zarepour et al.23 reported the importance of biogenic AuNPs for leukemia therapy. The proposed molecular mechanisms of biogenic AuNPs-induced cytotoxicity in all studies involved over generation of reactive oxygen species and activation of apoptotic pathways resulting in a cell death.
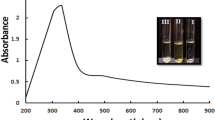
In a previous study the authors synthesized AuNPs using extracellular filtrate of isolated Bacillus subtilis NRC1, characterized the produced AuNPs using different physical methods, and studied their biological activities with their application as liver anti- fibrosis agent (under publication). Bacillus subtilis AuNPs were characterized using UV–visible spectroscopy which showed a surface plasmon resonance (SPR) spectrum at 540–550 nm. Energy dispersive X-ray spectroscopy (EDX) peak of AuNPs was shown at 2 keV, which confirmed the presence of elemental Au. High resolution transmission electron microscopy (HRTEM) figures of AuNPs showed well dispersed particles with spherical, hexagonal and oval shapes, with the size of 4.7–25 nm. Selected area electron diffraction pattern (SAED) for a single particle showed ring pattern with very fine spots indicating the polycrystallinity structure of the tested AuNPs. Dynamic light scattering spectroscopy (DLS) study revealed, the average diameter size of the particles was 23 nm. AuNPs were negatively charged as determined by zeta potential measurement and the charges were −25 mV. Fourier transform infrared spectroscopy (FTIR) spectra of AuNPs revealed that they were capped with reducing biomolecules secreted by Bacillus subtilis. The spectra strongly indicated the presence of proteins covering them which might play a role in the synthesis and stabilization of NPs.
In the current study, optimization of the growth variables of Bacillus subtilis NRC1 for production of AuNPs active extracellular filtrate was studied. Plackett–Burman design followed by Box–Behnken design were applied in this study. Also, the optimum cultural and nutritional conditions for maximum AuNPs production were obtained and validated.
Material and methods
Microorganism
The bacterial culture NRC1 was isolated from a golden earring and molecularly identified using 16S rRNA gene sequence as Bacillus subtilis NRC1 and it was deposited in NCBI GenBank with an accession number of MK33315224. It was used for the green synthesis of AuNPs.
Biosynthesis of AuNPs
The extracellular bacterial filtrate at an appropriate dilution and 2 mM HAuCl4 (1:1) were incubated for 24 h at 30 °C under static and illuminated conditions. The change in solution colour from pale yellow to purple was indicated, as positive AuNPs synthesis.
UV–vis investigation
The effect of different parameters on the biosynthesis of AgNPs were characterized by absorption spectral scan using double beam UV–vis spectrophotometer (Carry 100 Ultraviolet–visible spectrophotometer, Agilent, USA) at a resolution of 1 nm in a wave length range between 300 and 700 nm to determine their surface plasmon resonance peaks. Under the experimental conditions, both the size and concentrations of nanoparticles may vary and both of these affect the UV–vis spectrum. Therefore, the absorbance intensity was taken as an indication of an increase or decrease in the nanoparticles synthesis.
Optimization of cultural and nutritional variables for improving AuNPs synthesis using response surface methodology
Different cultural and nutritional independent variables were screened with special focusing on the enhancement of AuNPs synthesis. Primarily, the effects of nitrogen sources, carbon sources and metal ions were screened using one variable at a time (OVAT) method, where each of the variables was amended while other variables being constant, then response surface statistical methodology was used to determine the significance of these variables and interactions with other variables.
Effect of carbon sources, nitrogen sources and metal ions using OVAT method
Effect of nitrogen sources
This experiment was designed to find out the effect of certain nitrogen sources in the medium on the efficiency of bacterial filtrates for AuNPs synthesis25. Different nitrogen sources were added individually to 25 ml nutrient broth in 250 ml Erlenmeyer flasks, at a concentration that is equivalent to the nitrogen content in 0.2% of sodium nitrate. These nitrogen sources were sodium nitrite, potassium nitrate, casein hydrolysate, urea, ammonium phosphate, ammonium sulphate, peptone and yeast. Then the media were inoculated with Bacillus subtilis NRC1 and incubated for 3 days at 30 \(^\circ\)C under shaking at 150 rpm.
Effect of carbon sources
In this experiment attempts were made to find out the effects of some carbon sources on the growth and biomolecules production for efficient AuNPs synthesis25. Individually supplemented nutrient broth medium with 15 carbon sources representing mono, di and poly-saccharides at 1% were added to 25 ml nutrient broth in 250 ml Erlenmeyer flasks. Carbon sources were starch, dextrin, lactose, maltose, sucrose, glucose, fructose, xylose, arabinose, raffinose, mannose, galactose, glycerol, sodium acetate and tripotassium citrate. These flasks were inoculated with Bacillus subtilis NRC1 and incubated for 3 days at 30 \(^\circ\)C under shaking at 150 rpm.
Effect of mineral salts
Metal ions were tested for their ability to increase the biomolecules in the bacterial filtrates which were used in AuNPs synthesis26. Different mineral salts were added individually at the reported concentrations in the literature to 25 ml nutrient broth in 250 ml Erlenmeyer flasks (calcium sulphate 10−4 M, magnesium sulphate 10−4 M, sodium molybdate 10−6 M, manganese sulphate 10−4 M and zinc sulphate 10−4 M). These flasks were inoculated with Bacillus subtilis NRC1 and incubated for 3 days at 30 \(^\circ\)C under shaking at 150 rpm.
Response surface methodology statistical analysis
Optimization of fermentation conditions experiments for AuNPs biosynthesis by Bacillus subtilis NRC1 extracellular filtrate was carried out using two designs: Firstly, Plackett–Burman (PBD) followed by Box–Behnken (BBD). RSM designs were constructed using Design – Expert software (Stat-Ease Inc., Minneapolis, MN, USA, ver. 7.0). All experiments were done with 25 ml nutrient broth in 250 ml Erlenmeyer flasks. The flasks were inoculated with Bacillus subtilis NRC1 and incubated for 3 days under shaking at 150 rpm and 30 \(^\circ\)C. The responses in these experiments were absorbance intensities at the maximum peaks that are specific for the biosynthesis of AuNPs. All the experiments were carried out in triplicate and the average values were recorded.
Plackett Burman design (PBD)
Plackett–Burman experiments were carried out for screening the most significant variables affecting AuNPs synthesis. The PBD was based on six independent variables; (A): casein hydrolysate concentration (%), (B): dextrin concentration (%), (C): initial pH, (D): incubation period (days), (E): inoculum size (CFU × 106/ml) and (F): temperature (°C). This design contained 15 runs in 3 levels with 3 center points as shown in Tables 1 and 2. The responses in these experiments were the absorbance intensities at the maximum peaks that are specific for the biosynthesis of AuNPs.
Plackett Burman design is based on the first order linear equation:
where Y is the predicted response (production of AuNPs), β0 is the model intercept, βi is the variables estimates and Xi represents the coded independent variable which were coded as A, B , C,…. in this study.
The results of PBD followed by BBD were used for determining the significant variables that were effective for the AuNPs synthesis by the produced filtrate of Bacillus subtilis NRC1.
Box–Behnken design (BBD)
Response surface modeling and optimization of AuNPs biosynthesis conditions were accomplished using a quadratic BBD. Through this design, five variables; (A): casein hydrolysate concentration (%), (B): dextrin concentration (%), (C): initial pH, (D): inoculum size (CFU × 106/ml) and (E): temperature (°C), each with three levels comprising forty six runs (Tables 3 and 4). The responses in all experiments were expressed in terms of absorbance intensities of the biosynthesized AuNPs.
The second degree polynomial equation, which includes all interaction terms, was used to calculate the predicted response:
where Yresponse represents the predicted response (biosynthesis of gold nanoparticles), β0 is the intercept, βi is the linear coefficient, βii signifies the quadratic coefficients while βij is the cross product coefficients and Xi , X2i , Xj represent the coded independent variables. In the present study the independent variables were coded as A, B and C therefore, the second order polynomial equation was as follows:
Data obtained from both designs were analyzed by analysis of variance (ANOVA) and multiple regression analysis using Design Expert software, Version 7.0 (Stat-Ease Inc. Minneapolis, MN, USA). Value of "Prob > F" less than 0.05 was used as a parameter to indicate the significant model terms. The equations for the model were generated and the interpretation of the data was based on the signs (positive or negative effect on the response) and statistical significance of coefficients (P < 0.05). 3D surface graphs were designed by using Design Expert software to analyze the relationship between the different variables.
Model validation
In order to determine the precision of the statistical experimental model (BBD) and to obtain the optimum conditions for maximum AuNPs biosynthesis, the independent variables were numerically optimized according to the model using Design Expert software ver. 7.0 where the values of AuNPs biosynthesis were theoretically calculated. The optimum conditions were experimentally applied and the produced response was compared with the theoretical results.
Nitrate reductase assay
Nitrate reductase activity was tested according to27. Potassium nitrate was added to the extracellular bacterial filtrate (enzyme source) and incubated in the dark for 24 h. Then, diazo coupling reagent (1% sulfanilamide in 3 N HCl and 0.02% N-(1-naphthyl) ethylene diamine hydrochloride) was added to the reaction mixture and diluted 10 folds. After incubation for 30 min in dark at 30 °C, optical density was recorded at 540 nm. Nitrate reductase activity was expressed as micromoles of nitrite produced per ml of filtrate per min (µmol /ml/ min).
Results
Effect of carbon sources, nitrogen sources and metal ions using OVAT method
Effect of medium supplementation with different nitrogen sources on the synthesis of AuNPs
As shown in Table 5, casein hydrolysate gave the highest absorbance intensity (2.20) for AuNPs synthesis. While it was noted that AuNPs synthesis decreased significantly by addition of potassium nitrate, ammonium phosphate and ammonium sulphate in comparison with nutrient broth medium.
Effect of medium supplementation with different carbon sources on the synthesis of AuNPs
The obtained results in Table 5 showed that dextrin gave the highest absorbance intensity (2.39) for AuNPs. Also, maltose and arabinose showed a valuable enhancement for AuNPs synthesis. On the other hand, sucrose, glucose and anhydrous glycerol completely inhibited AuNPs synthesis.
Effect of medium supplementation with different metal ions on the synthesis of AuNPs
Table 5 showed that the supplementation of nutrient broth medium with the tested metal ions had a negative impact on the AuNPs synthesis.
Optimization of Bacillus subtilis NRC1 filtrate production for AuNPs synthesis using RSM
Screening of the significant variables using Plackett Burman design (PBD) for AuNPs synthesis
Fifteen runs with different combinations of the independent variables were tested and their influences on AuNPs synthesis were shown in Table 2. The data obtained for AuNPs production were statistically analyzed using analysis of variance (ANOVA) and were shown in Table 6. According to these data, the model F-value of 66.78 and p-value of 0.0027 referred to the significance of the model. In addition, A, C, D, F, AB, AC, AE and BC were significant model terms (p-value less than 0.05). F-value of 726.57 implied a significant model curvature. However, the "Lack of Fit F-value" of 0.54 implied that, the Lack of Fit was not significant relative to the pure error.
The determination coefficient (R2) of the model was 0.99, indicating that there was a very good fit between the observed and predicted values of AuNPs biosynthesis and implied that the model is reliable for this process. The determination coefficient (R2) value (is always between 0 and 1) provided a measure of how much variability in the observed response values. The closer the R2 value to 1, the stronger the model is and the better it predicts the response28. The model R2 was 0.99 and the adjusted-R2 was calculated to be 0.98, indicating a good agreement between the experimental and predicted values of AuNPs synthesis. The model adequate precision, which measured the signal to noise ratio, was 31.28 where it is preferable to be more than 49. A comparatively low rate of variation coefficient was 6.95% indicated a superior accuracy and reliability of the model experiments. Accordingly, this model could be used to navigate the design space with a reasonable accuracy.
Final equation in terms of coded variables
where A: casein hydrolysate; B: dextrin; C: initial pH; D: incubation period; E: inoculum size; F: temperature.
These positive and negative signs in the equation, where for the variable of a positive sign, AuNPs production increases with its increase and vice-versa. On the other hand, the variable of a negative sign, AuNPs production increases with its decrease and vice-versa29. Interactions between two variables could appear as a synergistic effect (positive coefficient) or as an antagonistic effect (negative coefficient)30.
Among the 6 variables, casein hydrolysate concentration, initial pH, inoculum size and incubation period showed positive signs of the effect on AuNPs synthesis; however other factors showed negative signs of the effect.
The contribution of interaction between casein hydrolysate and initial pH in AuNPs production represents the highest percentage in this model 10.92% followed by interaction between dextrin and initial pH then initial pH and casein hydrolysate with 8.32, 7.05 and 4.37, respectively, as shown in Table 7.
Casein hydrolysate concentration, dextrin concentration, initial pH, incubation period, inoculum size and temperature were chosen for further optimization using Box–Behnken design (BBD). They were found to exhibit a significant effect on AuNPs synthesis at the tested concentration levels.
Box–Behnken design (BBD) for optimization of AuNPs biosynthesis
The quadratic BBD experiments were carried out and average values were calculated and recorded. The experimental and predicted results according to BBD were shown in Table 4. Analysis of variance of BBD for AuNPs biosynthesis was shown in Table 8. Analysis of variance of the experimental data obtained by BBD showed that, the model F-value was 14.40 and p-value was < 0.0001 which implied a highly significant statistical model. The p-values of the model terms indicate significant parameters if they are < 0.05. The significant model terms were: A, C, AB, AC, AD, AE, BC, C2, A2B, A2D, AB2, AD2, B2E and BD2. The model "Lack of Fit F-value" of 2.02 implied that the Lack of Fit was not significant relative to the pure error. This could be shown by the relation between expermintal and predicted results (Fig. 1).
The model correlation coefficients; R2, adjusted- R2 and predicted- R2 values were 0.94, 0.87 and 0.69, respectively. The adjusted- R2 was in a reasonable agreement with the predicted-R2 which also indicated a strong relationship between experimental and predicted results. In the present model, the ratio of the model adequate precision which measures the signal to noise ratio was 15.62, indicating that the model generated an adequate signal to design space. The model coefficient of variation value was 16.58%. All the previously mentioned information indicated the accuracy and the reliability of the model and that it could be used to navigate the design space with a reasonable accuracy.
Final equation in terms of coded variables
where A: casein hydrolysate, B: dextrin, C: initial pH, D: inoculum size, E: temperature.
The 3D response surface plots (Fig. 2a-j) were used to demonstrate the relationship between AuNPs absorbance and the experimental levels of each independent variable. Each plot represents the interaction between two variables, whereas the other variables were kept at zero levels. Through simple scanning of the 3D-figures, we could determine the positive and negative effects of different variables.
3D response surface plot for the effect of different factors on AuNPs biosynthesis and factor-factor interactions between (a) casein hydrolysate and dextrin (b) casein hydrolysate and initial pH (c) casein hydrolysate and inoculum size (d) casein hydrolysate and temperature (e) dextrin and initial pH (f) dextrin and inoculum size (g) dextrin and temperature (h) initial pH and inoculum size (i) initial pH and temperature (j) inoculum size and temperature.
Figure 2a illustrates the interaction effects between casein hydrolysate and dextrin, whereas it was found that the change in casein hydrolysate concentration showed no marked effect on AuNPs yield. While the increase in dextrin concentration from1%- 4% showed a gradual increase in AuNPs biosynthesis. Figure 2b depicts the interaction between casein hydrolysate and initial pH of fermentation medium. As shown in the figure, the maximum AuNPs synthesis was at the lowest and the highest levels of casein hydrolysate but AuNPs biosynthesis increased with the gradual increase in initial pH reaching maximum at pH 8. Figure 2c showed that maximum AuNPs biosynthesis was at 0.16% casein hydrolysate (the lowest concentration). However, the maximum yield was at the highest and the lowest inoculum size. Figure 2d represents the interaction between casein hydrolysate and temperature, maximum AuNPs biosynthesis was at the highest and the lowest levels of casein hydrolysate. While the change in temperature didn’t affect markedly their yield. As shown in Fig. 2e, by the increase in dextrin concentration, AuNPs production increased. Whereas, the change in the medium initial pH didn’t affect muchly their production. The interaction between dextrin and inoculum size was demonstrated in Fig. 2f, the gradual increase in dextrin concentration led to an increase in AuNPs yield reaching its maximum value at 4% dextrin. While their maximum yield was at an inoculum size near the central point. On the other hand, the increase in both dextrin concentration and temperature led to an increase in AuNPs as shown in Fig. 2g. While in Fig. 2h, the gradual increase in the fermentation medium initial pH slightly affected the nanoparticles productivity. However, the increase in bacterial inoculum size led to a sharp increase in AuNPs biosynthesis. Figure 2i depicts the interaction between initial pH and temperature, the plot reveals that the increase in both factors remarkably increased AuNPs yield. Figure 2j represents the 3D plot as a function of inoculum size and temperature. According to the figure, the change in temperature didn’t affect AuNPs. However, the increase in inoculum size till the highest value (280 CFU × 106/ml) led to maximum AuNPs yield.
Model validation
The numerical optimization for AuNPs synthesis was attempted with Design expert, producing optimum conditions of 0.74% (w/v) casein hydrolysate, 3.99% (w/v) dextrin, pH 7.76, 47 × 106 CFU/ml inoculum size and 25 \(^\circ\)C temperature to give the maximum AuNPs biosynthesis of 3.30 absorbance intensity. To substantiate the optimum concentrations, an experiment with the above particularized conditions was carried out and the result was corresponded to 3.25 ± 0.015. The experimental result was found to be very close to the anticipated value (predicted value) which was 3.30. This result showed that the model was highly valid and the obtained data had a confidence factor of 98.48%. This high degree of precision affirmed the reliability of the model. Statistical optimization resulted in a 2.6-fold increase in AuNPs production compared with that of the non-optimized medium. This will be cost effective for synthesis of AuNPs due to the high price of AuNPs precursor.
Nitrate reductase activity
The nitrate reductase activity was estimated in the extracellular filtrate after the growth of Bacillus subtilis NRC1 in this optimized medium. Nitrate reductase was found to be 4.3 µmol/ml/min.
Discussion
AuNPs have unique optical, electrical and photothermal properties. They have been extensively employed in the biomedical field; in drug delivery, imaging and treatment of major life threatening diseases such as cancer, rheumatoid arthritis, chronic inflammation, pathological neo-vascularization and neoplastic disorders3,14. Also, they have many applications in microbiology, medicine, environmental sensors and biosensors31.
As a result of the high price of the AuNPs precursor, the productivity of AuNPs must be increased.
RSM is a collection of statistical and mathematical techniques which are very useful for developing, improving and optimizing processes. RSM is a known statistical method for the optimization of the medium components and the growth conditions for the production of NPs30. Statistical experimental design methodologies are considered as powerful tools used for the optimization of target bioactive metabolites production. It has the advantages of taking into account the interaction among the factors, less time consuming, limited number of experiments and avoids the erroneous interpretation occurring in one-variable-at-a-time optimization. There are many available experimental designs such as Plackett–Burman (PBD), central composite (CCD) and Box–Behnken (BBD)32.
In this study, Plackett–Burman design (PBD) was applied, followed by Box–Behnken design (BBD) for the maximum AuNPs synthesis by Bacillus subtilis NRC1.
At the first, PBD was applied to analyze and conclude the most significant factors affecting AuNPs synthesis. The determination coefficient (R2) of this model was 0.99, indicating that there was a very good fit between the observed and predicted values of AuNPs biosynthesis and implied that the model is reliable for this process.
Casein hydrolysate concentration, dextrin concentration, initial pH, incubation period, inoculum size and temperature were chosen for further optimization using Box–Behnken design (BBD). They were found to exhibit a significant effect on AuNPs synthesis at the tested concentration levels.
The second-order design BBD plays an important role in response surface methodology especially when the design economy and precise prediction are desired. BBD is a kind of rotatable design which uses just three levels of each factor18.
The numerical optimization for AuNPs synthesis was attempted with Design expert, producing optimum conditions of 0.74% (w/v) casein hydrolysate, 3.99% (w/v) dextrin, pH 7.76, 47 × 106 CFU/ml inoculum size and 25 \(^\circ\)C temperature to give the maximum AuNPs biosynthesis of 3.30 absorbance intensity.
To our knowledge, there are few reports about using RSM for optimization of bacterial fermentation conditions for the biosynthesis of AuNPs.
Manivasagan et al.33 studied optimization of fermentation medium composition for maximum amylase production by using Streptomyces sp. MBRC-82 for the production of AuNPs using RSM. The variables chosen for PBD were soluble starch, yeast extract, peptone, K2HPO4, MgSO4.7H2O and NaCl. The significant factors resulted from the design were introduced in BBD for further optimization studies. They concluded the optimized medium components which were soluble starch (5.84 g/l), peptone (3.51 g/l) and NaCl (0.38 g/l) for maximum amylase production used in synthesis of extracellular AuNPs. While Manivasagan and Oh34 used PBD for screening of the significant variables for the production of fucoidanase by green synthesis using Streptomyces sp.. The enzyme is then used in the biosynthesis of AuNPs. Among the variables studied, the most significant variables identified as influencing the fucoidanase production were wheat bran, kelp powder and NaCl. They were selected and their optimal levels were identified using BBD. Maximum fucoidanase production could be achieved when the wheat bran, kelp powder and NaCl were at 3.34 g/l, 0.70 g/l, and 0.88 g/l, respectively. This optimization led to an enhancement of fucoidanase production from 3.71 U/ml to 16.74 U/ml, which was further used in AuNPs biosynthesis.
It was reported that Rane et al.35 carried out optimization of the production of biosurfactant by Bacillus subtilis ANR88 for application in AgNPs and AuNPs synthesis, using PBD. They selected 11 variables and it was found that only 4 variables significantly affected the culture growth which were ammonium ferric citrate, molasses, pH and inoculum size for biosurfactant production for AgNPs and AuNPs synthesis. Also, it was found that ammonium ferric citrate, molasses and pH showed a positive correlation with the biosurfactant production, while the inoculum size was found to have a negative effect on biosurfactant production. Also, the maximum biosurfactant yield was in the medium containing 4% molasses, 0.25% ammonium ferric citrate and pH 7. The produced biosurfactant efficiently used for synthesis almost uniform (in size and shape) AgNPs and AuNPs. Recently, Singh et al.14 used CCD to increase nitrate reductase activity through optimization of Bacillus licheniformis growth for using it in AuNPs biosynthesis. Maximum AuNPs biosynthesis was obtained using the optimized medium variables which contained 2.1 g/l glucose, 14.05 g/l peptone, 4.14 g/l yeast extract and 3.91 g/l KNO3.
Honary et al.36 studied the biosynthesis and mathematical optimization process for iron oxide NPs produced by Penicillium waksmaniis useing Box–Behnken experimental design to investigate the effects of different factors such as pH, temperature and FeCl3 concentration. The results showed R2 value of 0.9992 which indicated the accuracy and ability of the polynomial model. In the study of Barabadi et al.37 for biosynthesis optimization of AgNPs produced by Penicillium citrinum using BBD, the effective factors were AgNO3 (mM) concentration, solution pH, incubation temperature (°C), shaking rate (rpm) and incubation time (hours). They found that R2 value of the obtained model was 0.8894, indicating a good correlation between both observed and predicted values. Thus, they concluded the optimum conditions for the biosynthesis of AgNPs by Penicillium citrinum with a low cost and time.
In this study the nitrate reductase activity in extracellular filtrate of Bacillus subtilis NRC1 was found to be 4.3 µmol/ml/min. It was reported the important role of this enzyme in reduction of HAuCl4 into nanogold and their stabilization3,6,25,38.
Data availability
All data generated or analysed during this study are included in this published article.
References
Ghosh, S., Ahmad, R., Zeyaullah, M. & Khare, S. K. Microbial nano-factories: synthesis and biomedical applications. Front. Chem. 9, 1–19 (2021).
Li, X., Xu, H., Chen, Z.-S & Chen, G. Biosynthesis of nanoparticles by microorganisms and their applications. J. Nanomater. Review Article ID 270974, 16 (2011).
Shedbalkar, U., Singh, R., Wadhwani, S., Gaidhani, S. & Chopade, B. A. Microbial synthesis of gold nanoparticles: Current status and future prospects. Adv. Colloid Interface Sci. 209, 40–48. https://doi.org/10.1016/j.cis.2013.12.011 (2014).
Deepak, V., Kalishwaralal, K., Pandian, S. R. K. & Gurunathan, S. An insight into the bacterial biogenesis of silver nanoparticles, industrial production and scale-up. In Rai M. and Duran N. (Ed.) Metal Nanoparticles in Microbiology (pp 17–35). Berlin: Springer-Verlag. (2011).
Salem, S. S. & Fouda, A. Green synthesis of metallic nanoparticles and their prospective biotechnological applications: an overview. Bio. Trace Elem. Res. https://doi.org/10.1007/s12011-020-02138-3.Online (2020).
Rónavári, A. et al. Green silver and gold nanoparticles: biological synthesis approaches and potentials for biomedical applications. Molecules 26, 1–39 (2021).
Ibrahim, S. et al. Optimization for biogenic microbial synthesis of silver nanoparticles through response surface methodology, characterization, their antimicrobial, antioxidant, and catalytic potential. Sci. Rep. 11, 770 (2021).
Menon, S., Rajeshkumar, S. & VenkatKumar, S. A review on biogenic synthesis of gold nanoparticles, characterization, and its applications. Resource-Efficient Technol. 3, 516–527 (2017).
Othman, A. M., Elsayed, M. A., Elshafei, A. M. & Hassan, M. M. Application of response surface methodology to optimize the extracellular fungal mediated nanosilver green synthesis. J. Genet. Eng. Biotechnol. 15, 497–504. https://doi.org/10.1016/j.jgeb.2017.08.003 (2017).
Li, Y. et al. Rapid biosynthesis of gold nanoparticles by the extracellular secretion of Bacillus niabensis 45: characterization and antibiofilm activity. J. Chem. 2, 1–7. https://doi.org/10.1155/2016/2781347 (2016).
San Diego, K. D. G., Alindayu, J. I. A. & Baculi, R. Q. Biosynthesis of gold nanoparticles by bacteria from hyperalkaline spring and evaluation of their inhibitory activity against pyocyanin production. J. Microbiol. Biotechnol. Food Sci. 8, 781–787. https://doi.org/10.15414/jmbfs.2018.8.2.781-787 (2016).
Srinath, B. S., Namratha, K. & Byrappa, K. Eco-friendly synthesis of gold nanoparticles by gold mine bacteria Brevibacillus formosus and their antibacterial and biocompatible studies. IOSR J. Pharm. 7, 53–60 (2017).
Pourali, P. et al. Biosynthesis of gold nanoparticles by two bacterial and fungal strains, Bacillus cereus and Fusarium oxysporum, and assessment and comparison of their nanotoxicity in vitro by direct and indirect assays. Electron. J. Biotechnol. 29, 86–93 (2017).
Singh, S., Dev, A., Gupta, A., Nigam, V. K. & Poluri, K. M. Nitrate Reductase mediated synthesis of surface passivated nanogold as broad-spectrum antibacterial agent. Gold Bull. 52, 197–216. https://doi.org/10.1007/s13404-019-00264-y (2019).
Qiu, R. et al. A biosynthesized gold nanoparticle from Staphylococcus aureus—as a functional factor in muscle tissue engineering. Appl. Mater. Today 22, 100905 (2021).
Shin, H. S. et al. Chemical and size effects of nanocomposites of silver and polyvinyl pyrrolidone determined by X-ray photoemission spectroscopy. Chem. Phys. Lett. 383, 418–422. https://doi.org/10.1016/j.cplett.2003.11.054 (2004).
Xiong, Y. H., Liu, J. Z., Song, H. Y. & Ji, L. N. Enhanced production of extracellular ribonuclease from Aspergillus niger by optimization of culture conditions using response surface methodology. Biochem. Eng. J. 21, 27–32 (2004).
Barabadi, H. et al. Microbial mediated preparation, characterization and optimization of gold nanoparticles. Braz. J. Microbiol. 45, 493–1501 (2014).
Könen-Adıgüzel, S. et al. Genotoxic, cytotoxic, antimicrobial and antioxidant properties of gold nanoparticles synthesized by Nocardia sp. GTS18 using response surface methodology. Mater. Res. Express. 5, 14. https://doi.org/10.1088/2053-1591/aadcc4 (2018).
Virmani, I. et al. Comparative anticancer potential of biologically and chemically synthesized gold nanoparticles. J. Clust. Sci. 31, 867–876 (2020).
Barabadi, H. et al. Emerging theranostic gold nanomaterials to combat colorectal cancer: A systematic review. J. Clust. Sci. 31, 651–658 (2020).
Barabadi, H. et al. Emerging antineoplastic gold nanomaterials for cervical cancer therapeutics: A systematic review. J. Clust. Sci. 31, 1173–1184 (2020).
Mostafavi, E. et al. Antineoplastic activity of biogenic silver and gold nanoparticles to combat leukemia: Beginning a new era in cancer theragnostic. Biotechnol. Rep. 34, e00714 (2022).
Gawdat, N.A. Biosynthesis of silver and gold nanoparticles by Bacillus spp. and their applications. Thesis for Degree of Doctor of Philosophy in Pharmaceutical Sciences (Microbiology and Immunology), Faculty of Pharmacy (Girls), Al-Azhar University, Egypt. (2021).
El-Bendary, M.A. Studies on the production and stability of Bacillus thuringiensis endotoxin. M.Sc thesis, Faculty of Science, Ain-Shams University, Egypt. (1994).
El-Bendary, M. A., Abo El-Ola, S., Moharam, M. & M.E.,. Enzymatic surface hydrolysis of polyamide fabric by protease enzyme and its production. Indian J. Fibre Text. Res. 37, 273–279 (2012).
Oza, G., Pandey, S., Shah, R. & Sharon, N. A mechanistic approach for biological fabrication of crystalline gold nanoparticles using marine algae, Sargassum wightii. Eur. J. Exp. Biol. 2, 505–512 (2012).
Kaushik, R., Saran, S., Isar, J. & Saxena, R. K. Statistical optimization of medium components and growth conditions by response surface methodology to enhance lipase production by Aspergillus carneus. J. Mol. Cat. B: Enzymatic. 40, 121–126 (2006).
Dhanya, G., Wetha, S., Madhavan, N. K., Rajeev, S. & Ashok, P. Response surface methodology for the optimization of alpha amylase production by Bacillus amyloliquefaciens. Bioresour. Technol. 99, 4597–4602 (2008).
El-Naggar, N. E. & Abdelwahed, N. A. M. Application of statistical experimental design for optimization of silver nanoparticles biosynthesis by a nanofactory Streptomyces viridochromogenes. J. Microbiol. 52, 53–63 (2014).
El-Domany, E. B., Essam, T. M., Ahmed, A. E. & Farghali, A. A. Biosynthesis physico-chemical optimization of gold nanoparticles as anti-cancer and synergetic antimicrobial activity using Pleurotus ostreatus fungus. J. Appl. Pharm. Sci. 8, 119–128 (2018).
El-Naggar, N.E-A., Mohamedin, A., Hamza, S.S. & Sherief, A-D. Extracellular biofabrication, characterization, and antimicrobial efficacy of silver nanoparticles loaded on cotton fabrics using newly isolated Streptomyces sp SSHH-1E. J. Nanomater. 2016, 17. https://doi.org/10.1155/2016/3257359 (2016).
Manivasagan, P., Venkatesan, J., Kang, K.-H., Sivakumar, K., Park, S.-J. & Kim, S.-K. Production of α-amylase for the biosynthesis of gold nanoparticles using Streptomyces sp. MBRC-82. Int. J. Biol. Macromol. 72, 71–78 (2015).
Manivasagan, P. & Oh, J. Production of a Novel Fucoidanase for the Green Synthesis of Gold Nanoparticles by Streptomyces sp. and Its Cytotoxic Effect on HeLa Cells. Mar. Drugs. 13, 6818–6837. https://doi.org/10.3390/md13116818 (2015).
Rane, A. N., Baikar, V. V., Kumar, V. R. & Deopurkar, R. L. Agro-industrial wastes for production of biosurfactant by Bacillus subtilis ANR 88 and its application in synthesis of silver and gold nanoparticles. Front. Microbiol. 16, 878–889. https://doi.org/10.3389/fmicb.2017.00492 (2017).
Honary, S., Barabadi, H., Ebrahimi, P., Naghibi, F. & Alizadeh, A. Development and optimization of biometal nanoparticles by using mathematical methodology: A microbial approach. J. Nano Res. 30, 106–115 (2015).
Barabadi, H. et al. Optimization of myco-synthesized silver nanoparticles by response surface methodology employing Box-Behnken design. Inorg. Nano-Met. Chem. 49, 33–43 (2019).
El-Bendary, M. A. et al. Myco-synthesis of gold nanoparticles using Aspergillus flavus: Characterization, optimization and cytotoxic activity. Curr Trends Microbiol. 12, 67–79 (2018).
Acknowledgements
The authors would like to thank and appreciate the National Research Centre of Egypt for the technical and financial supports of this work under the PhD project no. 13/7/11.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This work was supported by National Research Centre Grant number 13/7/11. Author Noha A. Gawdat has received the research support.
Author information
Authors and Affiliations
Contributions
M.A.E.-B.: Conceptualization; data curation; methodology; visualization; investigation; resources, drafted the work; revised the manuscript. S.S.A.: Conceptualization; data curation; methodology; resources; visualization; drafted the work; revised the manuscript. M.E.M.: Investigation; methodology, data curation, visualization; drafted the work; revised the manuscript. M.M.A.E.: made substantial contributions to the conception; design of the work; investigation; methodology; resources; software; visualization; acquisition, analysis, interpretation of data; writing the draft; revised the final manuscript. N.A.G.: Conceptualization; data curation; investigation; methodology; resources; software; visualization, drafted the work; revised the manuscript critically.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
El-Bendary, M.A., Afifi, S.S., Moharam, M.E. et al. Optimization of Bacillus subtilis NRC1 growth conditions using response surface methodology for sustainable biosynthesis of gold nanoparticles. Sci Rep 12, 20882 (2022). https://doi.org/10.1038/s41598-022-25324-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-25324-w
This article is cited by
-
Production of iron enriched Saccharomyces boulardii: impact of process variables
Scientific Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.