Abstract
CYP2E1 encodes an enzyme that participates in the activation of several carcinogenic substances. Thus, numerous studies have investigated the association between CYP2E1 polymorphisms and colorectal cancer (CRC) risk, but inconclusive results have been obtained. We performed a meta-analysis to precisely evaluate the relationship of CYP2E1 rs2031920, rs3813867, and rs6413432 polymorphisms with the susceptibility to CRC. Scopus, Web of Science and PubMed databases were searched to identify eligible studies, and the association between the polymorphisms and CRC risk was then quantitatively synthesized using different genetic models. Eighteen studies with 23,598 subjects were selected for inclusion into the analysis. Significant association between rs2031920 and an increased CRC risk was observed in homozygous (OR = 1.496, 95% CI 1.177–1.901, P = 0.001), recessive (OR = 1.467, 95% CI 1.160–1.857, P = 0.001) and allele (OR = 1.162, 95% CI 1.001–1.349, P = 0.048) models. Significant association was not found for rs3813867 and rs6413432 (P > 0.05). In conclusion, our results suggest that rs2031920, but not rs3813867 and rs6413432, is associated with the risk of CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide and is associated with significant morbidity and mortality1. In 2020 alone, nearly two million new CRC cases and one million CRC-related deaths were reported2. Age is a well-established risk factor for CRC along with other environmental risk factors such as physical inactivity, obesity, high intake of red, low intake of fiber, tobacco smoking, and alcohol consumption3,4. In addition, genetics has unequivocally been implicated as a key determinant in the development of CRC5. Polymorphisms in cancer-related genes may therefore influence interindividual susceptibility to CRC6.
CYP2E1 encodes cytochrome P450 2E1 (CYP2E1), a phase I enzyme involved in the process of xenobiotic metabolism. CYP2E1 plays a key role in the conversion of xenobiotics into several highly reactive intermediate metabolites prior to their elimination by phase II enzymes7. For example, it is known to activate low-molecular-weight procarcinogens such as nitrosamines into active carcinogens directly involved in digestive tract oncogenesis. Consequently, higher CYP2E1 activity has been associated with higher rates of cancer progression8. The CYP2E1 gene contains more than ten well-characterized single nucleotide polymorphisms (SNPs) that may influence the activity of the enzyme and thus cancer risk9. The most commonly studied polymorphisms in CRC include rs3813867 (conventionally known as PstI) and rs2031920 (conventionally known as RsaI) in the 5’-flanking regions of the gene, as well as rs6413432 (conventionally known as DraI) in intron 6. These polymorphisms are known to have functional effects on cells. In particular, rs2031920 and rs3813867 have been associated with increased transcriptional and enzymatic activity of the gene. In addition, increased transcriptional activity of the CYP2E1 gene has been discovered in association with rs6413432, which is also associated with DNA single-strand breaks known to lead to cancer10. This is one of the reasons why we focused on these three polymorphisms in this study.
In addition, although these polymorphisms are frequently studied, the association between these polymorphisms and the risk of CRC remains inconclusive. For example, a study in the Hungarian population showed that the variant allele of rs3813867 was positively associated with an increased risk of CRC11. However, there were also studies showing a lack of association between the same polymorphisms and the risk of CRC8,12,13. Likewise, for rs2031920, while Silva et al.14 showed that individuals carrying the variant allele had an increased risk of developing CRC, Kury et al.15 found no association between the polymorphism and the risk of CRC. Similar discrepancies were also noted for rs641343216,17,18,19. These inconsistencies between the studies may be attributed to the small sample size of each study and the different genetic backgrounds and lifestyles of study participants from different populations. Meta-analysis can be used to resolve these inconsistencies. However, the last meta-analyses focusing on these three polymorphisms was published almost 9 years ago20,21 and new studies in this area have recently been added, which may lead to a different study result. This is another reason why we focus on these three polymorphisms. Therefore, in this work, a meta-analysis was conducted to obtain a more precise estimate of the association between CYP2E1 rs3813867, rs2031920, and rs6413432 polymorphisms and CRC risk.
Results
Study selection and characteristics
The flow diagram of the study selection process is shown in Fig. 1. A total of 152 records were identified in the PubMed, Scopus and Web of Science databases. Of these, 44 records were identified as duplicates and removed. After screening the titles and abstracts of the remaining studies, 81 studies were identified as potentially relevant. When the full-texts were reviewed, 63 studies were excluded for the following reasons: (i) they reported data on other CYP2E1 polymorphisms, were case-only studies, or did not contain useful information (N = 34); (ii) they were review articles (N = 23); (iii) the cases included patients with benign tumors such as adenomas (N = 3); (iv) they did not contain individual data for the rs3813867 and rs2031920 polymorphisms (N = 2); (v) study participants overlapped in more than one study (N = 1). Finally, 18 studies comprising a total of 10,302 cases and 13,296 controls were included in this meta-analysis.
The characteristics of the included studies are shown in Table 1. Of the included studies, two were in Chinese22,23 and the rest were in English. Seven studies were conducted on Asians17,22,23,24,25,26,27, seven studies were conducted on Caucasians11,13,14,15,16,28,29, and the remaining four studies included participants of other descendants such as Brazilians, Saudi Arabians or mixed ethnicities8,18,19,30. The most commonly used method for genotyping the polymorphisms was polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP), while a few studies used Taqman or microarray-based approaches. For the rs6413432 polymorphism, the distribution of genotypes in controls was consistent with HWE in all the included studies. For rs2031920, however, the genotype distribution deviated significantly from HWE in three studies22,27,30. For rs3813867, only one study reported a significant deviation from the HWE22. All of the studies had high methodological quality (≥ 5 stars on the Newcastle–Ottawa Scale) except for two studies22,30. The star ratings of the included studies are shown in Table 2.
Meta-analysis results: rs2031920
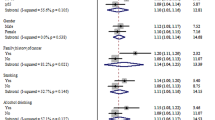
The results of the meta-analysis on the association of CYP2E1 rs2031920 polymorphism with CRC risk are shown in Table 3. Overall, pooled results from 10 studies (comprising 3,695 cases and 4,887 controls) revealed a significant association between the polymorphism and an increased CRC risk in the homozygous (OR = 1.496, 95% CI 1.177–1.901, P = 0.001), recessive (OR = 1.467, 95% CI 1.160–1.857, P = 0.001), and allele models (OR = 1.162, 95% CI 1.001–1.349, P = 0.048) (Fig. 2). Subgroup analysis by ethnicity revealed that there were significant associations between CYP2E1 rs2031920 polymorphism and CRC risk in Asians in homozygous (OR = 1.578, 95% CI 1.209–2.058, P = 0.001), recessive (OR = 1.526, 95% CI 1.176–1.980, P = 0.001), and allele models (OR = 1.231, 95% CI 1.031–1.469, P = 0.021), an observation consistent with the overall analysis (Table 3). In contrast, among Caucasians, only the homozygous and recessive models showed significant associations. Interestingly, the ORs of the associations were large in Caucasians (homozygous model, OR = 5.819, 95% CI 1.234–27.436; recessive model, OR = 5.720, 95% CI 1.214–26.954). No subgroup analysis was performed for other ethnicities as only one study was available for this subgroup30.
Similarly, subgroup analysis by study quality also revealed statistically significant results in the homozygous (OR = 1.899, 95% CI 1.220–2.954, P = 0.004), recessive (OR = 1.599, 95% CI 1.217–2.100, P = 0.001) and allele models (OR = 1.216, 95% CI 1.020–1.450, P = 0.030) in the high quality studies. On the other hand, no significant association between rs2031920 and the risk of CRC was observed in the low quality studies under all five genetic models (P > 0.05; Table 3).
Meta-analysis results: rs3813867
Pooled results from eight studies (comprising 4,055 cases and 4,801 controls) are shown in Table 4. The CYP2E1 rs3813867 polymorphism was not significantly associated with CRC risk in any of the genetic models studied (homozygous model, OR = 1.020, 95% CI 0.682–1.526, P = 0.923; heterozygous model, OR = 1.161, 95% CI 0.841–1.603, P = 0.366; dominant model, OR = 1.179, 95% CI 0.845–1.645, P = 0.333; recessive model, OR = 1.302, 95% CI 0.693–1.538, P = 0.876; allele model, OR = 1.175, 95% CI 0.862–1.602, P = 0.306) (Fig. 3). Subgroup analysis by ethnicity and study quality also revealed no significant association (Table 4).
Meta-analysis results: rs6413432
The association of the CYP2E1 rs6413432 polymorphism with susceptibility to CRC is shown in Table 5. The combined results from six case–control studies involving 2,552 cases and 3,608 controls showed that there was no significant association between the polymorphism and susceptibility to CRC in all genetic models studied. The combined ORs and their 95% CIs were as follows: homozygous model (OR = 1.307, 95% CI 0.673–2.540, P = 0.429); heterozygous model (OR = 1.142, 95% CI 0.790–1.650, P = 0.481); dominant model (OR = 1.172, 95% CI 0.811–1.694, P = 0.399); recessive model (OR = 1.146, 95% CI 0.745–1.762, P = 0.535); allele model (OR = 1.177, 95% CI 0.858–1.616, P = 0.313) (Table 5 and Fig. 4). No significant association was observed even after performing subgroup analysis by ethnicity. Subgroup analysis by study quality was not performed because all studies were of high quality.
Sensitivity analysis
Sensitivity analysis was performed by sequentially omitting individual studies to assess the stability of the results. For rs2031920, the results of the homozygous, recessive, and dominant models were not altered with the omission of any individual study (Supplementary Information online). However, for the heterozygous model, omitting13,14,17,22,27 changed the results from non-significant to significant. However, this change was not unexpected, as the combined results of rs2031920 under the heterozygous model were at the borderline OR value (OR = 1.004; Table 3). A similar observation was noted in the allele model, where removal of several studies changed the results from non-significant to significant. Similar to the heterozygous model, the instability of the results for the allele model was not unexpected, as the lower limit of the 95% CI was 1.001, which is also a borderline value.
For rs3813867, the result for the homozygous model was also unstable for the same reason. However, for the heterozygous and recessive models, the results appeared to be largely driven by Fernandes et al.18and Kiss et al.11, respectively (Supplementary Information online). Indeed, the omission of Fernandes et al.18 also significantly altered the results of the homozygous and recessive models of rs6413432 (Supplementary Information online). For all other genetic models, the results did not change when any of the studies was omitted.
Publication bias diagnosis
The presence of publication bias was examined using Begg’s and Egger’s tests and visually verified using funnel plots (Figs. 5, 6, 7). For rs2031920, no significant publication bias was detected in the allele model (P > 0.05 for both Begg’s and Egger’s tests). However, both tests revealed a significant publication bias in the heterozygous (Begg's test P = 0.032, Egger's test P = 0.012) and dominant models (Begg's test P = 0.032, Egger's test P = 0.018). Apart from this, no publication bias was observed in the homozygous and recessive models in Begg’s test but appeared to be significant in Egger’s test (homozygous, Begg’s test P = 0.251, Egger’s test P = 0.035; recessive, Begg’s test P = 0.175, Egger’s test P = 0.037). ‘Trim and fill’ analysis was performed for all four genetic models that showed significant publication bias in at least one of the tests. The homozygous, heterozygous, dominant, and recessive models were found to have four, three, five, and four missing studies, respectively. However, imputation of these missing studies did not significantly change the results (homozygous, P = 0.052; heterozygous, P = 0.583; dominant, P = 0.341; recessive, P = 0.079).
For the rs3813867 polymorphism, no significant publication bias was detected by both the Begg’s and Egger’s tests (homozygous, Begg’s test P = 0.624, Egger’s test P = 0.141; heterozygous, Begg’s test P = 0.083, Egger’s test P = 0.541; dominant, Begg’s test P = 0.138, Egger’s test P = 0.306). Similarly, no publication bias was detected for rs6413432 (homozygous, Begg’s test P = 0.497, Egger’s test P = 0.113; heterozygous, Begg’s test P = 0.851, Egger’s test P = 0.957; dominant, Begg’s test P = 0.348, Egger’s test P = 0.963; recessive, Begg’s test P = 0.174, Egger’ test P = 0.128; allele, Begg’s test P = 0.348, Egger’s test P = 0.912).
Discussion
CYP2E1, located on chromosome 10q26.3, encodes the CYP2E1 enzyme that is mainly localized in the liver. CYP2E1 belongs to the phase I group of drug-metabolizing enzymes that are involved in the metabolism of several small molecules such as ethanol, acetaminophen and procarcinogens like nitrosamines and azo compounds31. The enzyme has been extensively studied as it is directly involved in the metabolic activation of more than 85 xenobiotics to hepatotoxic or carcinogenic metabolites32. In addition, CYP2E1 is known to be the most active CYP450 isoenzyme because of its ability to reduce molecular oxygen to highly reactive oxygen species (ROS) even in the absence of a substrate33. Excessive levels of the ROS accelerate cancer development by acting on messengers in intracellular signaling pathways, leading to activation of lipid peroxidation, DNA damage, and carcinogenesis34. For these reasons, CYP2E1 is one of the most intensively studied cytochrome genes in cancer35.
Over the decades, several studies have focused on a few important polymorphisms of CYP2E1 that can potentially affect the function of the gene. These include the rs2031920 and rs3813867 polymorphisms, which are located in the 5'-regulatory region of CYP2E1, as well as the rs6413432 polymorphism, which is located in intron 6 of the gene36. The rs2031920 polymorphism has been associated with higher transcriptional and enzymatic activity due to the replacement of cytosine with thymine at position 1,019 of the gene37. Meanwhile, the rs3813867 polymorphism of CYP2E1 results from the substitution of guanine with cytosine at the 1259th position, whereas rs6413432 involves a substitution of thymine with adenine at the 7678th position of the gene38. These substitutions may lead to altered binding affinity of transcription factors and other regulatory elements, causing changes in the amount of protein product and subsequently the risk of cancer39.
Despite being extensively investigated, the association between CYP2E1 polymorphisms and CRC risk remains inconclusive, as conflicting results have been reported in different studies. These conflicting results can be attributed to numerous factors, including the sample size of individual studies, ethnicity of study participants, geographical variations, as well as environmental factors such as dietary habits5. To address these discrepancies, we conducted a systematic review and meta-analysis to combine the results of previous studies, in order to yield a more accurate estimation on the association between CYP2E1 polymorphisms and CRC risk. In contrast to a pooled analysis, a meta-analysis considers the characteristics of individual studies and weighs them appropriately based on well-accepted statistical parameters, such as sample size, before combining them, thereby reducing the potential for erroneous conclusions40. We demonstrated a statistically significant association of the CYP2E1 rs2031920 polymorphism with CRC risk under the homozygous (OR = 1.496, 95% CI 1.177–1.901, P = 0.001), recessive (OR = 1.467, 95% CI 1.160–1.857, P = 0.001) and allele (OR = 1.162, 95% CI 1.001–1.349, P = 0.048) models. This observation may be attributed to the location of the polymorphism, which falls within the transcriptional regulatory region of CYP2E1. Therefore, the nucleotide substitution of this polymorphism could affect the binding of transcription factors to the 5'-flanking region of CYP2E1, thereby altering its mRNA expression levels41. The positive association between rs2031920 polymorphism and cancer susceptibility has also been studied in tumor types other than CRC, including cancers of the head and neck42,43, esophagus41, lung44, stomach45, urological organs46, and urinary tract47. However, there were also a few studies suggesting the opposite association, whereby the rs2031920 polymorphism may serve as a protective factor as in the case of nasopharyngeal cancer in the Tunisia populations48 and bladder cancer in Asians49. These observations suggest that the distribution of allele or genotype frequency varies in different populations and association may be different between cancer types. Therefore, we stratified our meta-analysis by ethnicity to gain better insight into the impact of ethnic diversity on the association of these polymorphisms with the risk of CRC. That being said, subgroup analyses revealed slight differences in risk association between Asians and Caucasians. This could be explained by the higher allele frequency of the rs2031920 c2 allele in the Asian population compared to the Caucasian population, which is consistent with the observation of Wang et al.9 that different ethnic groups generally have not only differences in the living environment, dietary habits, and genetic backgrounds, but also in the frequency distribution of CYP2E1 genotypes.
The results of this meta-analysis suggested that there was no significant association of the rs3813867 polymorphism with susceptibility to CRC under all genetic models examined. Our finding was in agreement with previous studies that also found no association with CRC risk in study samples from Australia12, Spain13 and Brazil8. In contrast, a significant association was found in the study by Kiss et al.11, whereas other studies by Kury et al.15 and Kim et al.25 found a positive association between the polymorphism and CRC risk only in individuals who regularly consumed red meat. These discrepancies highlight the possible existence of gene–gene or gene-environment interactions in influencing the effects of genetic polymorphisms on CRC risk50,51.
Similarly, no significant association was observed between rs6413432 polymorphism and CRC risk in our study, which is consistent with studies in other populations, such as in Lebanese by Darazy et al.29, in Saudi Arabians by Saeed et al.19, and in Malaysians by Chong et al.17. In other cancers, several previous studies also supported our findings, showing a non-significant association between the rs6413432 polymorphism and susceptibility to urinary cancer47 and gastric cancer52. In contrast to the aforementioned rs2031920 polymorphism, the functional effect of rs6413432 has not been conclusively proven, but it is thought to enhance transcriptional activity and affect CYP2E1 expression and the catalytic activity of the encoded enzyme53. Nevertheless, further studies are needed to increase statistical power to detect and confirm even the slightest effect of the rs6413432 polymorphism on the risk of CRC54.
The results of our meta-analysis were consistent with previous meta-analyses by Peng et al.20 and Jiang et al.21. However, because more studies were used (and more participants – a total of 23,598 subjects – were included) for analysis in the present work, our study power is higher and the risk estimate is therefore more reliable. In addition, the meta-analysis by Jiang et al.21 was limited to participants from Western populations only, and no study represented the Asian population. Thus, the results of our study are more representative of the global population. This, together with the inclusion of recent studies in this area of research, allows us to present the most up-to-date summary and assessment of the associations between the three CYP2E1 polymorphisms and the risk of CRC. Apart from that, unlike the previous meta-analyses by Jiang et al.21 and Peng et al.20, the current meta-analysis used Scopus as one of the databases to search for relevant articles. In general, Scopus includes a wider range of journals and also contains more articles than the Web of Science55, allowing more relevant studies to be identified and included in the analysis. We also performed an additional stratified analysis based on study quality using the Newcastle–Ottawa scale to provide a comprehensive picture of the evidence based on all included studies. These are the strengths of the present work.
However, there are also several limitations that need to be acknowledged in the present study. For instance, the modest number of included studies might still be insufficient to find a significant association between the rs3813867 and rs6413432 polymorphisms and the risk of CRC, although the power to detect a significant association was improved by this meta-analysis. In addition, there is a lack of ethnic diversity as there were no data on African populations in these eighteen studies, which focused mainly on Asians and Caucasians. Another limitation is the concern for the occurrence of publication bias, as only published studies were included. Nevertheless, imputation using the ‘trim-and-fill’ analysis showed that the results are unlikely to change even in the absence of publication bias. Finally, gene–gene or gene-environment interactions, which are known to also contribute to the risk of CRC, were not examined in the present work because of the lack of information in the included studies. However, the lack of gene–gene or gene-environment studies does not change the fact that a single gene can influence the risk of CRC, albeit modestly, as has been demonstrated in many other studies56,57,58,59,60,61,62. The reproducibility of the study results (both with individual studies and with previous meta-analyses) suggests that the study result was not likely due to chance alone. Although positive results from a single gene cannot usually be translated into clinical practice, knowledge of which low penetrance polymorphisms might influence the risk of CRC may shed light on which genetic pathways to focus on in designing a genetic screening panel in the future, which can undoubtedly contribute to a more individualized approach to medicine63.
In conclusion, the results of this meta-analysis suggest that the CYP2E1 rs2031920 polymorphism is associated with the risk of CRC. Although the rs3813867 and rs6413432 polymorphisms were not associated with the risk of CRC, subgroup analyses revealed some differences in the risk of association between Asians and Caucasians, and between high- and low-quality studies. Finally, in view of the limitations mentioned above, further studies with a better overall design are needed to verify the true association between CYP2E1 polymorphisms and CRC risk.
Methods
Literature search strategy and study selection
A literature search was performed in PubMed, Web of Science and Scopus databases up to February 24, 2022. The following keywords were used: “CYP2E1” AND “polymorphism” AND “colorectal cancer”. No language restriction was set. Studies were included if they met the following criteria: (1) examined the association between CYP2E1 gene polymorphisms and CRC risk; (2) case–control studies in design; and (3) contained sufficient data to estimate an odds ratio (OR) and its 95% confidence interval (CI). Non-research articles and studies conducted in non-human subjects were excluded. If more than one article was published by the same authors with the same or overlapping subjects, the study with the largest sample size or the most recent data was selected. References of eligible studies and relevant review articles were also screened to identify additional studies. The review was not prospectively registered.
Data extraction
The following information was extracted from each included study: first author’s name, year of publication, ethnicity (categorized as Asian, Caucasian, or Africans), country, total number of cases and controls, allele and genotype frequencies, genotyping methods, and deviation from Hardy–Weinberg equilibrium (HWE). When the HWE p-value was not reported, it was calculated using a Pearson’s χ2 test. All extracted information was recorded in an Excel spreadsheet.
Data synthesis
The quality of the included studies was assessed by two investigators using the Modified Newcastle–Ottawa Scale for Case–Control Studies of Genetic Association64. Studies that received ≥ 5 stars were considered to be of high quality. The strength of association between the CYP2E1 polymorphisms and CRC risk was assessed using the odds ratio (OR) and the corresponding 95% confidence interval (CI). Statistical significance of the pooled ORs was determined using the Z-test, and a P < 0.05 was considered statistically significant. In addition, heterogeneity among the studies was assessed using Cochran’s Q statistic test and the I2 statistic to quantify the proportion of total variation due to heterogeneity. An I2 value ≥ 50% was considered as having significant statistical heterogeneity, for which a random-effects model (the DerSimonian–Laird method) was used to calculate the pooled OR. Meanwhile, when heterogeneity was low, the fixed-effects model (Mantel–Haenszel method) was used to calculate the pooled OR. Sensitivity analysis was also performed to assess the robustness of the results. In addition, subgroup analyses were performed according to the ethnicity of the participants and the methodological quality of the studies. To assess the presence of publication bias among the included studies, Begg’s funnel plot and Egger’s linear regression tests were performed. If publication bias was identified, a ‘trim and fill’ analysis was performed to detect missing studies. All analyses were performed using STATA software, version 14.0 (StataCorp LP, College Station, TX, USA).
Data availability
The datasets supporting the conclusion of this article are included within the article (and the online Supplementary Information file).
References
Martinez-Romero, J., Bueno-Fortes, S., Martín-Merino, M., De Ramirez Molina, A. & de Lasrivas, J. Survival marker genes of colorectal cancer derived from consistent transcriptomic profiling. BMC Genom. 19, 45–60 (2018).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Barontini, J. et al. Association between polymorphisms of TAS2R16 and susceptibility to colorectal cancer. BMC Gastroenterol. 17, 1–7 (2017).
Muhammad Nawawi, K. N. et al. Incidence and clinicopathological features of colorectal cancer among multi-ethnic patients in Kuala Lumpur, Malaysia: a hospital-based retrospective analysis over two decades. PeerJ 9, e12425 (2021).
Tan, S. C. Low penetrance genetic polymorphisms as potential biomarkers for colorectal cancer predisposition. J. Gene Med. 20, e3010 (2018).
Johani, F., Majid, M., Azme, M. & Nawi, A. Cytochrome P450 2A6 whole-gene deletion (CYP2A6*4 ) polymorphism reduces risk of lung cancer: a meta-analysis. Tobacco Induced Dis. https://doi.org/10.18332/tid/122465 (2020).
Guengerich, F. P. Cytochrome P450 2E1 and its roles in disease. Chem. Biol. Interact. 322, 109056 (2020).
Proenca, M. A. et al. A case-control study of CYP2E1 (PstI) and CYP1A1 (MspI) polymorphisms in colorectal cancer. Genet. Mol. Res. 14, 17856–17863 (2015).
Wang, L. et al. Genetic polymorphism analysis of cytochrome P4502E1 (CYP2E1) in a Chinese Tibetan population. Medicine 86, e8855 (2017).
Fang, Z. et al. Association between CYP2E1 genetic polymorphisms and urinary cancer risk: a meta-analysis. Oncotarget 8, 86853–86864 (2017).
Kiss, I. et al. Association between allelic polymorphisms of metabolizing enzymes (CYP 1A1, CYP 1A2, CYP 2E1, mEH) and occurrence of colorectal cancer in Hungary. Anticancer Res. 27, 2931–2937 (2007).
Butler, W. J., Ryan, P. & Roberts-Thomson, I. C. Metabolic genotypes and risk for colorectal cancer. J. Gastroenterol. Hepatol. 16, 631–635 (2001).
Landi, S. et al. A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet. Genom. 15(8), 535–546. https://doi.org/10.1097/01.fpc.0000165904.48994.3d (2005).
Silva, T. D., Felipe, A. V., Pimenta, C. A., Barão, K. & Forones, N. M. CYP2E1 RsaI and 96-bp insertion genetic polymorphisms associated with risk for colorectal cancer. Genet. Mol. Res. 11, 3138–3145 (2012).
Kury, S. et al. Combinations of cytochrome P450 gene polymorphisms enhancing the risk for sporadic colorectal cancer related to red meat consumption. Cancer Epidemiol. Biomarkers Prev. 16, 1460–1467 (2007).
Cotterchio, M. et al. Red meat intake, doneness, polymorphisms in genes that encode carcinogen-metabolizing enzymes and colorectal cancer risk. Cancer Epidemiol. Biomarkers Prev. 17, 3098–3107 (2008).
Chong, E. T. J., Lee, C. C., Chua, K. H., Chuah, J. A. & Lee, P.-C. RsaI but not DraI polymorphism in CYP2E1 gene increases the risk of gastrointestinal cancer in Malaysians: a case-control study. BMJ Open 4, e004109 (2014).
Fernandes, G. M. M. et al. CYP1A1, CYP2E1 and EPHX1 polymorphisms in sporadic colorectal neoplasms case control study. World J. Gastroenterol. 22, 9974–9983 (2016).
Saeed, H. M. et al. Cytochrome P450 1A1, 2E1 and GSTM1 gene polymorphisms and susceptibility to colorectal cancer in the saudi population. Asian Pac. J. Cancer Prev. 14, 3761–3768 (2013).
Peng, H., Xie, S. K., Huang, M. J. & Ren, D. L. Associations of CYP2E1 rs2031920 and rs3813867 polymorphisms with colorectal cancer risk: a systemic review and meta-analysis. Tumour Biol. 34, 2389–2395 (2013).
Jiang, O. et al. CYP2E1 polymorphisms and colorectal cancer risk: a HuGE systematic review and meta-analysis. Tumour Biol. 34, 1215–1224 (2013).
Chen, K. et al. A case-control study on the association between genetic polymorphisms of metabolic enzymes and the risk of colorectal cancer. Zhonghua Liu Xing Bing Xue Za Zhi 26, 659–664 (2005).
Chen, M. B. et al. Polymorphisms of CYP2E1 RsaI and susceptibility of colorectal cancer. Chin. J. Cancer Prev. Treat 14, 409–411 (2007).
Gao, C.-M. et al. CYP2E1 RsaI polymorphism impacts on risk of colorectal cancer association with smoking and alcohol drinking. World J. Gastroenterol. 13, 5725–5730 (2007).
Kim, N. H. et al. Red meat intake, CYP2E1 and PPARγ polymorphisms, and colorectal cancer risk. Eur. J. Cancer Prev. 28, 304–310 (2019).
Morita, M. et al. Genetic polymorphisms of CYP2E1 and risk of colorectal cancer: the Fukuoka colorectal cancer study. Cancer Epidemiol. Biomark. Prev. 18, 235–241 (2009).
Sameer, A. S. et al. Role of CYP2E1 genotypes in susceptibility to colorectal cancer in the Kashmiri population. Hum. Genom. 5, 530 (2011).
Cleary, S. P., Cotterchio, M., Shi, E., Gallinger, S. & Harper, P. Cigarette smoking, genetic variants in carcinogen-metabolizing enzymes, and colorectal cancer risk. Am. J. Epidemiol. 172, 1000–1014 (2010).
Darazy, M. et al. CYP1A1, CYP2E1, and GSTM1 Gene polymorphisms and susceptibility to colorectal and gastric cancer among Lebanese. Genet. Test Mol. Biomarkers 15, 423–429 (2011).
le Marchand, L., Donlon, T., Seifried, A. & Wilkens, L. R. Red meat intake, CYP2E1 genetic polymorphisms, and colorectal cancer risk. Cancer Epidemiol. Biomarkers Prev. 11, 1019–1024 (2002).
Gao, J. et al. High CYP2E1 activity correlates with hepatofibrogenesis induced by nitrosamines. Oncotarget 8, 112199–112210 (2017).
Chen, J. et al. A comprehensive review of cytochrome P450 2E1 for xenobiotic metabolism. Drug. Metab. Rev. 51, 178–195 (2019).
Massart, J., Begriche, K., Hartman, J. H. & Fromenty, B. Role of mitochondrial cytochrome P450 2E1 in healthy and diseased liver. Cells 11(2), 288. https://doi.org/10.3390/cells11020288 (2022).
Harjumäki, R., Pridgeon, C. S. & Ingelman-Sundberg, M. CYP2E1 in alcoholic and non-alcoholic liver injury. Roles of ros, reactive intermediates and lipid overload. Int. J. Mol. Sci. 22(15), 8221. https://doi.org/10.3390/ijms22158221 (2021).
Pan, S. T. et al. Computational Identification of the paralogs and orthologs of human cytochrome P450 superfamily and the implication in drug discovery. Int. J. Mol. Sci. 17, 1020 (2016).
Wang, F. J. et al. Update meta-analysis of the CYP2E1 RsaI/PstI and DraI polymorphisms and risk of antituberculosis drug-induced hepatotoxicity: evidence from 26 studies. J. Clin. Pharm. Ther. 41, 334–340 (2016).
Godoy, F. R. et al. Increased DNA damage is not associated to polymorphisms in OGGI DNA repair gene, CYP2E1 detoxification gene, and biochemical and hematological findings in soybeans farmers from Central Brazil. Environ. Sci. Pollut. Res. Int. 26, 26553–26562 (2019).
Karakoc, M. D., Kortunay, S., Kara, C. O. & Topuz, B. CYP2E1 and ALDH2 gene polymorphisms in squamous cell head and neck cancer in the Turkish population. Int. J. Hematol. Oncol. 29, 61–69 (2019).
Farhat, F., Daulay, E. R. & Chrestella, J. The role of CYP2E1 polymorphism in the activation of procarcinogen metabolism of nasopharyngeal carcinoma. Int. J. Nasopharyng. Carcinoma 1, 107–109 (2019).
Savitz, D. A. & Forastiere, F. Do pooled estimates from meta-analyses of observational epidemiology studies contribute to causal inference?. Occup. Environ. Med. 78, 621–622 (2021).
Zhao, F. et al. Association between polymorphisms in the CYP1A1, CYP2E1 and GSTM1 genes, and smoking, alcohol and upper digestive tract carcinomas in a high-incidence area of northern China. Oncol. Lett. 18, 1267–1277 (2019).
Lu, D., Yu, X. & Du, Y. Meta-analyses of the effect of cytochrome P450 2E1 gene polymorphism on the risk of head and neck cancer. Mol. Biol. Rep. 38, 2409–2416 (2011).
Tang, K. et al. The PstI/RsaI and DraI polymorphisms of CYP2E1 and head and neck cancer risk: a meta-analysis based on 21 case-control studies. BMC Cancer 10, 575 (2010).
Ye, X. H. et al. Association between the CYP2E1 polymorphisms and lung cancer risk: a meta-analysis. Mol. Genet. Genom. 290, 545–558 (2015).
Elingarami, S. et al. Polymorphisms in NEIL-2, APE-1, CYP2E1 and MDM2 genes are independent predictors of gastric cancer risk in a Northern Jiangsu population (China). J. Nanosci. Nanotechnol. 15, 4815–4828 (2015).
Lin, Y.-C. et al. Cytochrome P450 2E1 RsaI/PstI polymorphism is associated with urologic cancer risk: evidence from a meta-analysis. Int. J. Clin. Exp. Med. 8, 8927 (2015).
Fang, Z., Wu, Y. & Zhang, N. Association between CYP2E1 genetic polymorphisms and urinary cancer risk: a meta-analysis. Oncotarget 8, 86853–86864 (2017).
Ben Chaaben, A. et al. Genetic polymorphism of cytochrome P450 2E1 and the risk of nasopharyngeal carcinoma. Bull. Cancer 102, 967–972 (2015).
Yin, X. et al. Association of CYP2E1 gene polymorphisms with bladder cancer risk: a systematic review and meta-analysis. Medicine 97, e11910 (2018).
Choi, J., Jia, G., Wen, W., Shu, X. O. & Zheng, W. Healthy lifestyles, genetic modifiers, and colorectal cancer risk: a prospective cohort study in the UK Biobank. Am. J. Clin. Nutr. 113, 810–820 (2021).
Najafi, S. et al. Gene regulation by antisense transcription: a focus on neurological and cancer diseases. Biomed. Pharmacother. 145, 112265 (2022).
Wu, M. S. et al. Genetic polymorphisms of cytochrome p450 2E1, glutathione S-transferase M1 and T1, and susceptibility to gastric carcinoma in Taiwan. Int. J. Colorectal. Dis. 17, 338–343 (2002).
Zhang, M.-X., Liu, K., Wang, F.-G., Wen, X.-W. & Song, X.-L. Association between CYP2E1 polymorphisms and risk of gastric cancer: an updated meta-analysis of 32 case-control studies. Mol. Clin. Oncol. 4, 1031 (2016).
Jackson, D. & Turner, R. Power analysis for random-effects meta-analysis. Res. Syn. Methods 8, 290–302 (2017).
Falagas, M. E., Pitsouni, E. I., Malietzis, G. A. & Pappas, G. Comparison of PubMed, Scopus, web of Science, and google scholar: strengths and weaknesses. FASEB J. 22, 338–342 (2008).
de Jong, M. M. et al. Low-penetrance genes and their involvement in colorectal cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 11, 1332–1352 (2002).
Donald, N., Malik, S., Mcguire, J. L. & Monahan, K. J. The association of low penetrance genetic risk modifiers with colorectal cancer in lynch syndrome patients: a systematic review and meta-analysis. Fam. Cancer 17, 43–52 (2018).
Houlston, R. S. & Tomlinson, I. P. M. Polymorphisms and colorectal tumor risk. Gastroenterology 121, 282–301 (2001).
Kury, S. et al. Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer 7, 326 (2008).
Tan, S. C. Low penetrance genetic polymorphisms as potential biomarkers for colorectal cancer predisposition. J. Gene. Med. 20, e3010 (2018).
Valle, L., Vilar, E., Tavtigian, S. V. & Stoffel, E. M. Genetic predisposition to colorectal cancer: syndromes, genes, classification of genetic variants and implications for precision medicine. J. Pathol. 247, 574–588 (2019).
Zheng, Y., Wang, J. J., Sun, L. & Li, H. L. Association between CYP1A1 polymorphism and colorectal cancer risk: a meta-analysis. Mol. Biol. Rep. 39, 3533–3540 (2012).
Aggarwal, N. et al. The association of low-penetrance variants in DNA repair genes with colorectal cancer: a systematic review and meta-analysis. Clin. Transl. Gastroenterol. 8, e109 (2017).
Tan, S. C. et al. Association between MIR499A rs3746444 polymorphism and breast cancer susceptibility: a meta-analysis. Sci. Rep. 10, 3508 (2020).
Acknowledgements
This work was supported by the Fundamental Research Grant Scheme of the Ministry of Higher Education, Malaysia (No. FRGS/1/2019/SKK08/UKM/02/9). Open Access funding enabled and organized by Projekt DEAL. The authors would like to thank Academic Proofreading (www.academicproofreading.uk) for English language editing.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.A.K.S. conducted the search, extracted data, performed the quality assessment appraisal, conducted statistical analysis, and drafted the manuscript. H.S. contacted authors for additional information not reported in published articles. N.A. provided statistical guidance on study power calculation. A.A. and M.P. contributed to the interpretation of the results. R.J. provided significant input and feedback on the draft manuscript. S.C.T. conceptualized the study, independently extracted the data, edited the draft manuscript, and provided supervision. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharzehan, M.A.K., Sito, H., Abdullah, N. et al. Association between CYP2E1 polymorphisms and colorectal cancer risk: a systematic review and meta-analysis. Sci Rep 12, 20149 (2022). https://doi.org/10.1038/s41598-022-24398-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-24398-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.