Abstract
Akkermansia muciniphila, a commensal bacterium commonly found in healthy gut microbiota, is widely considered a next-generation beneficial bacterium candidate to improve metabolic and inflammatory disorders. Recently the EFSA’s Panel on Nutrition, Novel food, and Food Allergens has declared that pasteurized A. muciniphila DSM 22959T (also MucT, ATCC BAA-835) can be considered safe as a novel food, opening the door to its commercialization as a food supplement. Despite its recognized health benefits, there is still little information regarding the antimicrobial susceptibility of this species and reference cut-off values to distinguish strains with intrinsic or acquired resistance from susceptible strains. In this study, we combined a genomic approach with the evaluation of the antibiotic susceptibility in five human A. muciniphila isolates. Genomic mining for antimicrobial resistance genes and MICs determinations revealed that only one strain harboring tetW gene showed resistance to tetracycline, whereas all A. muciniphila strains showed low sensitivity to ciprofloxacin and aminoglycosides with no genotypic correlation. Although all strains harbor the gene adeF, encoding for a subunit of the resistance-nodulation-cell division efflux pump system, potentially involved in ciprofloxacin resistance, the susceptibility towards ciprofloxacin determined in presence of efflux pump inhibitors was not affected. Overall, our outcomes revealed the importance to extend the antibiotic susceptibility test to a larger number of new isolates of A. muciniphila to better assess the safety aspects of this species.
Similar content being viewed by others
Introduction
Akkermansia muciniphila is a Gram-negative anaerobic bacterium abundantly present in the human intestine where it uses mucin as carbon and nitrogen source1,2. Several studies demonstrated that this species is beneficial to host health and its abundance correlated inversely with people suffering from metabolic syndrome (e.g.: obesity, diabetes, cardiometabolic disease) and inflammatory bowel disease3,4,5. In particular, the type strain, A. muciniphila DSM 22959T (also MucT, ATCC BAA-835), has been extensively studied and is considered a next-generation beneficial bacterium due to its protective effect against obesity and metabolic disorders6,7,8. The first intervention study targeting overweight/obese insulin-resistant humans showed that supplementation with A. muciniphila DSM 22959T is safe, well-tolerated and improves metabolic parameters9. Nevertheless, the use of A. muciniphila in food or pharmaceutical formulations depends on the demonstration of its efficacy and safety within regulatory frameworks. In 2020, EFSA's Panel on Biological Hazards (BIOHAZ) did not recommend this species for the Qualified Presumption of Safety (QPS) list due to safety concerns10. Although they recognized the safe and well-tolerated administration of live or pasteurized cells of A. muciniphila DSM 22959T in humans and mice, they stressed the possible involvement of the ability to degrade mucin with pathological processes and a possible association of this species with neurological diseases. Moreover, EFSA’s panel highlighted the presence of several antimicrobial resistance genes (ARGs) in the genomes of this species. A. muciniphila together with other anaerobic gut commensals associated with human health (Bacteroides spp., Clostridium butyricum, Faecalibacterium prausnitzii) are gaining significant attention for their potential application in food supplements and pharmaceutical formulations, however these genera and microbial species do not have a history of safe use yet11,12. An important aspect to consider, when evaluating their safety, is the potential to carry ARGs that could be acquired from harmful bacteria. The gut microbiome, the main source of these health-promoting microbes, is considered a reservoir of ARGs and the anaerobic commensal species represent the main contributors to the human intestinal resistome13,14. Specifically, A. muciniphila is thought to be particularly plastic and prone to gaining antimicrobial resistance (AMR)15,16. In Europe, information on AMR for bacteria deliberately introduced into the food chain is of paramount importance to declaring a microorganism safe for human and animal consumption. For this purpose, phenotypic testing based on the determination of a minimum inhibitory concentration (MIC) for a selected group of antimicrobials, together with the complementary search of the whole genome sequences for the presence of known ARGs, should be performed17. Nevertheless, while genome mining for ARGs is routinely performed, standardized methods for the MIC evaluation have not yet been defined for this microorganisms. Furthermore, there is a lack of specific microbiological cut-off values to be used to distinguish strains with intrinsic or acquired resistance from susceptible strains18,19, mainly due to the limited number of available cultivable strains. Recently, EFSA Panel on Nutrition, Novel food, and Food Allergens (NDA) expressed a positive opinion on the safety of pasteurized A. muciniphila, opening the door for its use in food supplements and in food for special medical purposes20. With the introduction of A. muciniphila in the food chain, the evaluation of the antimicrobial susceptibility of this bacterium becomes fundamental to meet the safety recommendations of EFSA. To the best of our knowledge, only few studies have focused on the antibiotic resistance profile of A. muciniphila strains through phenotypic tests21,22,23, while most of the available information derives from genome data analysis15,23,24,25,26. Furthermore, as most of the phenotypic studies concern the type strain of this species, the antimicrobial susceptibility variation in A. muciniphila strains remains largely unexplored. As such, the aim of the present study is to characterize the AMR profile of novel human isolated strains of A. muciniphila including the type strain DSM 22959T, by integrating phenotypic and in silico approaches, to provide new insight into the safety of this promising species.
Results and discussion
Comparative genome sequence analyses
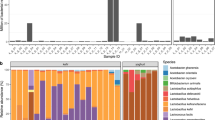
In our study, we isolated A. muciniphila strains from feces of healthy volunteers. Species-specific qPCR assay2 along with the analysis of the melt curve allowed to select fecal samples positive for the presence of A. muciniphila to be used for a further enrichment step and isolation procedure. Although 9 donors out of 16 (= 56%) tested positive, a total of 10 isolates of A. muciniphila were obtained from only 5 subjects, highlighting that difficulties in culturing A. muciniphila from fecal samples could limit the isolation and characterization of new strains25,27. The A. muciniphila isolates, preliminarily identified via partial 16S rRNA gene sequencing, were differentiated into 5 subtypes, different from the type strain, by molecular typing (Supplementary Table S1 and Figure S1). Each subtype belonged to a different donor, indicating that different subjects could be colonized by different strains. Then, one isolate for each subtype was selected for whole-genome sequencing. General genomic features of the A. muciniphila genomes are summarized in Table 1 and in Supplementary Figure S1. De novo assembly of the genomic data, after contigs cleaning (cut-off 500 bp), revealed genome sizes ranging from 2.65 to 3.42 Mbp according to previous observations (average: 3.01 Mbp; median: 2.97 Mbp, completeness of assemblies 98%, contaminations detected ranged from 1.02 to 9.77%)15. The G + C contents of the genomes ranged from 54.9 to 55.8%. The presence of a high value of marker genes duplicated in strain Amap1 justifies the high number of very small contigs and consequently the extra length of the genome. However, this does not lead to a significant increase in terms of the number of unique predicted genes. Previous large-scale genomic-based analyses15,26, focusing on Akkermansia spp diversity, have shown that A. muciniphila is not the only species of the genus Akkermansia. These studies highlighted the presence of at least 4 new candidate species, which are characterized by remarkable whole-genome divergence despite similar 16S rRNA gene sequences. Therefore, the 16S rRNA analysis alone is not sufficiently discriminatory for the identification of the Akkermansia spp.. To unequivocally identify the new strains at the species level, the whole-genome Average Nucleotide Identity (ANI) values were calculated using fastANI between the newly sequenced genomes and the A. muciniphila type strain genome28. Compared to the wild type, all five strains exhibited ANI scores ranging between 97.4 and 99% and were therefore confirmed as A. muciniphila according to the ANI species limit definition29. We also performed a pan-genome analysis on the 5 isolates, along with 188 publicly available Akkermansia genomes (NCBI). The comparative genomic analysis of the newly isolated and publicly available (n = 188) Akkermansia genomes identified a pan-genome of 19,738 genes, including 212 core genes (shared between the 99 and 100% of the strains), 34 soft-core genes (between 95 and 99%), 4618 shell genes (between 15 and 94%), and 14,874 cloud genes (less than 15% of the strains) (Fig. 1). The large pan-genome and the small core genes show great genetic diversity within Akkermansia spp. The coding sequence alignment of the 193 genomes performed for the pan-genome analyses, with the Roary matrix built on the presence-absence of shell genes, highlighted 3 clusters (Fig. 2). According to Karcher et al.26, two clusters referred to the new putative species, namely SGB9228 and SGB9223, the third to A. muciniphila species. The analysis revealed that the new five strains were all identified as A. muciniphila species.
Phylogenetic tree of the 193 Akkermansia sp. genomes. The shell genes were used for the clusterization based on gene presence (dark blue) or absence (white). The genomes are clusterized into three groups corresponding to Akkermansia sp. SGB9228 (11 genomes), SGB9223(69 genomes), A. muciniphila (113 genomes). The yellow zone of the tree highlights the position of the newly isolated A. muciniphila strains characterized in this study.
Antimicrobial resistance genes and mobile DNA elements
EFSA-FEEDAP guidelines encourage the use of whole-genome sequences to characterize new candidate bacterial strains regarding their potential functional traits of concern, such as the presence of genes encoding for, or contributing to resistance to antimicrobials relevant to their use in humans and animals17. To investigate the presence of ARGs in A. muciniphila strains, a homology-based search was performed using Resistance Gene Identifier (RGI) on Comprehensive Antibiotic Resistance Database (CARD)30. To date, most of the available information on the antibiotic resistance profile of A. muciniphila derives from genome data analysis15,23,24,25,26. When Van Passel and colleagues24 analyzed the genome of A. muciniphila DSM 22959T, they found two potential β-lactamase genes and one gene coding for a 5-nitroimidazole antibiotic resistance protein, commonly found in other intestinal anaerobic bacteria. However, Gòmez-Gallego3 reported that the recent inspection of the genome annotation of A. muciniphila DSM 22959T did not reveal any ARGs of concern. This was further confirmed by Machado et al.23 in a recent publication focused on the antimicrobial resistance profile of A. muciniphila DSM 22959T. According to the previous studies15,26, the most frequently ARGs found in A. muciniphila genomes are adeF, aph(6)-Id, sul2, InuC and aph(3″)-Ib. ARGs found in the genomes of the new A. muciniphila stains are listed in Table 2. The gene aph(6)-Id, associated with resistance to aminoglycosides, involved in streptomycin inactivation31, and the gene sul2, one of the most prevalent determinants of sulfonamide resistance32, were specifically found in the genomes of A. muciniphila Vtp7 and Amap1. The latter also carries the tetW gene, which confers resistance to tetracycline33. All strains were also characterized by the presence of the gene adeF. This gene encodes a membrane fusion protein belonging to the resistance-nodulation-cell division (RND) efflux pump system AdeFGH34, potentially involved in resistance to fluoroquinolones and tetracycline. However, according to our findings, the A. muciniphila strains characterized in this work, harbor only the adeF gene, while the other components of the AdeFGH operon were not detected, thus limiting its potential activity in fluoroquinolones—and tetracycline-resistance mechanism. Nevertheless, it is important to underline that Karcher et al.26, in their study on genomic diversity and ecology of human-associated Akkermansia species, found the gene adeF consistently in most of the A. muciniphila genomes, but never present in the other Akkermansia spp. genomes.
ARGs harbored in microbial commensals can become a significant hazard if transferred and acquired by pathogens, considering that the gut provides ideal conditions for gene exchange being an environment rich in microbes14. According to Guo et al.15 A. muciniphila acquired genes during its evolution, from a wide range of taxa associated with human intestinal habitat, through horizontal gene transfer. Furthermore, de Nies et al.16 highlighted that A. muciniphila has a plastic genome, that is particularly prone to acquire ARGs under antibiotics selective pressure. In light of the above, we wanted also to screen the A. muciniphila strains for the presence of mobile genetic elements (MGEs), possibly involved in the ARGs transfer. The only identified trait of concern is related to the Amap1 strains, which has a putative transposon (Tn6205) in its genome associated to the AGRs aph(6)-Id and sul2, thus posing risks of gene transfer events35. One insertion sequence (IS), ISAmu1, has been identified in the genomes of A. muciniphila Sap1 and Amap1 as well as in A. muciniphila DSM 22959T. In general, ISs can move ARGs as part of a composite or compound transposon, that is a region bounded by two copies of the same IS that can move as a single unit. However, there are examples of a single IS mobilizing an adjacent region that may contain one or more ARGs. ISs can also affect antibiotic resistance by driving the expression of the adjacent genes36. However, in the newly isolated A. muciniphila strains the ISs sequences identified are not flanked by ARGs. Plasmids, which could be involved in ARGs mobilization, were found in none of the A. muciniphila strains (Supplementary Figure S1).
Antibiotic sensitivity
To fully address the antibiotic resistance profile of the A. muciniphila strains, we performed a phenotypic test based on MIC determination for a selected group of antibiotics. According to the EFSA guidelines17, for Gram-negative bacteria the antibiotics tested should be those for Enterobacteriaceae. For antibiotic susceptibility testing, the culture medium used must allow the growth of the strains under assessment and not interfere with antibiotics17. IsoSensitest (IST) is the nutrient medium recommended by the British Society for Antimicrobial Chemotherapy. However, for specific bacteria, other formulations may be required17,37. In our study, we used a modified version of IST (sIST) to allow the growth of A. muciniphila. Therefore, we first verified whether the additional medium ingredients could interfere with antibiotic sensitivity. To this aim, we used E. coli Nissle as a reference strain of the Enterobacteriaceae family. MICs determined in sIST were compared with those obtained in cation-adjusted Mueller Hinton Broth, a conventional susceptibility test medium38. MICs determined in the two media agreed (Table 3), as such, we can state that the sIST components did not interfere with the antibiotic sensitivity assay. Overall, the MICs obtained for the antimicrobials tested are similar between the new A. muciniphila isolates and the type strain, showing a similar level of sensitivity within the species (Table 3). All strains were sensitive to ampicillin, tetracycline, colistin, fosfomycin, and sulfamethoxazole. Only the strain Amap1 showed resistance towards tetracycline as expected due to the presence of tetW gene in its genome. Strains Amap1 and Vtp7, which were genotypically predicted to be resistant to sulfonamides, due to the presence of gene sul2, resulted sensitive to sulfamethoxazole comparably to all other strains. Indeed, the presence of ARGs does not always translate into a resistant phenotype, as already reported for bacterial isolates with silent ARGs39. The latter underlines the importance of combining genotypic and phenotypic tests to better define antimicrobials resistance. All A. muciniphila strains, including the type strain, showed low sensitivity to ciprofloxacin and aminoglycosides (gentamicin, kanamycin, streptomycin). Comparing the phenotypic susceptibilities with the genotypic AMR profiles, all strains harbor a gene potentially involved in ciprofloxacin resistance mediated by antimicrobial efflux (adeF). Concerning the low sensitivity to aminoglycosides, only the Amap1 and Vtp7 strains have a genetic determinant presumably involved in streptomycin resistance (aph (6) -Id)31, indicating that poor sensitivity to this class of antibiotics in all the strains tested could be due to a more general intrinsic mechanism. Indeed, intrinsic resistance to aminoglycoside is common in anaerobic bacteria40,41. Furthermore, it is known that some Gram-negative bacilli are resistant to aminoglycoside due to a transport defect or change in outer membrane permeability and this mechanism causes cross-reactivity to all aminoglycosides42,43. According to EFSA recommendations, the MIC values obtained should be compared with public data on the specific species under study. To the best of our knowledge, few published studies have addressed the antibiotic susceptibility profile of A. muciniphila through phenotypic tests21,22,23. Moreover, these studies tested different molecules using different methodologies, making difficult compare data. Dubourg et al.21 has highlighted the increased colonization of the human intestinal microbiota by the Verrucomicrobia phylum following a broad-spectrum antibiotic regimen. In this context, they evaluated the sensitivity of A. muciniphila DSM 22959T to different antibiotics, using the E-test method on Wilkins-Chalgren agar plates with 5% blood. They found that the type strain is susceptible to imipenem, piperacillin/tazobactam, and doxycycline, and resistant to vancomycin, metronidazole and penicillin G. Later, Cozzolino et al.22 tested the susceptibility of A. muciniphila DSM 22959T to antimicrobials recommended for Lacticaseibacillus rhamnosus, using the E-test strips on Brain Heart Infusion agar plate. The MIC values they obtained for ampicillin, tetracycline, and streptomycin (respectively 2, 0.75, 128 mg L−1) are in line with those obtained in this study, indeed, the MIC values for gentamicin and kanamycin (respectively 4, 12 mg L−1) were consistently lower compared to our data, so that they categorized this strain as susceptible to these aminoglycosides. This discrepancy can be partially explained by the different methods used for phenotypic testing (broth dilution in our work vs. E-test method in Cozzolino et al.22) or by the growth medium used (sIST in our work vs BHI Cozzolino et al.22). Disagreements caused by methodologies used for classifying microbial species into resistant or susceptible phenotype are present in the literature19,44. However, it is important to highlight that broth dilution method is widely accepted for phenotypic tests based on MIC determination. Recently, Machado et al.23 evaluated the sensitivity of A. muciniphila DSM 22959 T strain to the antimicrobials using a mucin-supplemented Pepton Yeast Glucose and comparing two methods (broth microdilution vs E-test method). Their results showed that DSM 22959T is resistant to gentamicin, kanamycin, streptomycin, and ciprofloxacin, consistently with our findings. Furthermore, we found our results in line with the EFSA 2021 document20 and Ouwerkerk et al.45, where authors reported that A. muciniphila BAA-835 (= DSM 22959T) presented high resistance levels to aminoglycosides, and ciprofloxacin similarly to other A. muciniphila strains.
Investigation of the drug efflux as resistance mechanism towards ciprofloxacin
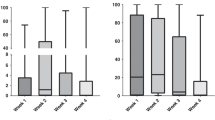
Ciprofloxacin is a fluoroquinolone antibiotic effective against Gram-negative and Gram-positive aerobic bacteria but with low potency against many anaerobic bacteria46. Reduced susceptibility to ciprofloxacin may be mediated by (1) alterations in the drug efflux; (2) mutations in the genes coding for drug targets (DNA gyrase and topoisomerase) or (3) change in outer membrane and/or porin permeability to the drug43,46. All A. muciniphila strains tested here showed scarce sensitivity to ciprofloxacin. Interestingly, we found that all the genomes harbored the gene adeF, encoding a membrane protein of a RND drug efflux complex putatively involved in fluoroquinolones resistance. According to previous observations26, this ARG is one of the most frequently found in the A. muciniphila genomes, therefore understanding its role within this species might be worthy of attention. To confirm the presence of active efflux pumps in the A. muciniphila strains, we measured the accumulation of ethidium bromide, a molecule known to be substrate of efflux pumps, in presence and absence of the efflux pump inhibitor (EPI) carbonyl cyanide 3-chlorophenylhydrazone (CCCP). CCCP is a protonophore cell membrane uncoupler that increases membrane permeability to protons, leading to a disruption of membrane potential, thus being a strong inhibitor of RND efflux pumps in Gram-negative bacteria47. The assumption of this test is that the higher is the concentration of substrate accumulated inside the bacterial cell, the lower is the efflux level and vice versa. The results showed that the intracellular accumulation of ethidium bromide is significantly (p < 0.01, T-test) increased with the addition of CCCP in all the strains, both in terms of total amount and kinetic of ethidium bromide accumulation (Fig. 3). This indirectly confirms that the lowest level of ethidium bromide accumulation, in the absence of CCCP, is determined by active efflux pumps in the A. muciniphila strains (Fig. 3). To investigate the involvement of efflux pumps in ciprofloxacin reduced susceptibility, the MICs were determined in absence and in presence of the EPIs CCCP and phenylalanine-arginine beta-naphthylamide (PaβN). PAβN is another broad-spectrum efflux pump inhibitor, capable of significantly reducing resistance to fluoroquinolones in P. aeruginosa48. It acts as a competitive inhibitor, preventing efflux of the antibiotics by binding the substrate-binding pocket of the efflux pumps and impairing the antibiotic bond to its affinity site48. In presence of efflux pump inhibitors, the MICs for ciprofloxacin were equal to or twofold lower than the MIC values determined without EPIs (Table 3). These slight variations in MICs represent the normal standard deviation of MIC dilution tests49. Since the addition of the EPIs did not result in a susceptible phenotype, it was excluded the active involvement of efflux pumps in the A. muciniphila reduced sensitivity to ciprofloxacin. Since genome analysis revealed no mutations in genes encoding ciprofloxacin targets, the low sensitivity to this molecule could be determined by its poor permeability to the outer membrane as reported for other Gram-negative43.
(A) Accumulation of ethidium bromide in A. muciniphila strains in absence (white bars) and in presence (black bars) of CCCP after 60 min of incubation. (B) Kinetics of ethidium bromide accumulation by A. muciniphila strains in absence (solid lines) and in presence (dashed lines) of CCCP. Data are shown as the average values of three replicates, with standard deviation. RFU, Relative Fluorescence Unit.
Conclusions
Our study provides new insights on the phenotypic and genotypic antibiotic resistance profile of A. muciniphila, showing a similar level of susceptibility among strains within this species. Between the antibiotics tested, all strains showed poor sensitivity to the fluoroquinolone ciprofloxacin and aminoglycosides, as expected for anaerobic bacteria40,42,43. The low sensitivity to these classes of antimicrobials, being widespread among all strains regardless of the presence of ARGs, appears to be caused by intrinsic mechanisms, such as change in the outer membrane and/or porin permeability to the drugs43. An ARG related to streptomycin inactivation (aph (6)-Id) was found in only two strains, with no effect on the antibiotic sensitivity compared to the other strains. We also investigated the involvement of efflux pump activity in reduced susceptibility to ciprofloxacin, since the adeF gene, which encodes for a component of the RND efflux pump system (AdeFGH), was found in all genomes. According to our results, no evidence of the role of an active drug efflux system related to ciprofloxacin reduced susceptibility was observed. However, the involvement of an active efflux against toxic compounds, such as ethidium bromide, was confirmed for all the strains. The ARG sul2 detected in two strains did not determine a resistant phenotype. Finally, only one of the strains showed traits of concern, harboring three ARGs, one conferring resistance to tetracycline (tetW) and two (aph (6)-Id, sul2) associated with an MGE (Tn6205).
The results of our study underline the urgent need for adequate microbiological breakpoints and standardized protocols to assess the antimicrobial susceptibility of A. muciniphila and to favor fair comparative analysis between different laboratories and institutes. Moreover, further studies involving a larger number of A. muciniphila strains are necessary to demonstrate the safety of this microbial species.
Materials and methods
Isolation of A. muciniphila strains
Informed consent was obtained from all subjects of the study. Ethical approval was not required for this study under local legislation and institutional requirements of the University of Milan. All methods were carried out in accordance with proper guidelines of the University of Milan which refer to the WHO Laboratory of Biosafety Manual. For the isolation of new A. muciniphila strains, fresh fecal samples were collected from 16 healthy adult human donors, aged 26 to 50 years (34 ± 8 years old) being part of this research group. To proceed with the isolation only with A. muciniphila positive-fecal sample, bacterial DNA was isolated from stool samples using DNeasy PowerLyzer PowerSoil Kit (QIAGEN), following the manufacturer’s instructions. Then, the molecular detection of A. muciniphila was carried out using species-specific real-time PCR analysis targeted to the variable regions of the 16S rRNA gene sequence of A. muciniphila2. Real-time PCR was performed using 50 ng of template DNA in a final reaction volume of 15 μl, using the EvaGreen™ kit (Bio-Rad) and following the manufacturer’s recommendations. PCRs were performed in duplicate on a CFX96 instrument (Bio-Rad). Data were recorded as threshold cycles (Ct) and analyzed using Bio-Rad CFX Manager™ software. Fecal samples positive for the presence of A. muciniphila (Ct < 30) were used for bacterial isolation following the method of Derrien et al. (2004). Briefly, serial dilutions of positive stool samples were inoculated in a mucin-containing medium for an enrichment step, before spread-plating on the same medium containing 1% (v/v) agar. Plates were incubated at 37 °C for 6 days in an anaerobic chamber (N2:H2:CO2 90:5:5). Colonies with different morphologies were picked up and grown in Brain Heart Infusion broth (Merck), supplemented with 0.5 g l−1 of L-cysteine hydrochloride and 0.25% (w/v) of mucin from porcine stomach (Merck). Isolation of DNA from bacterial cultures was performed using DNeasy UltraClean Microbial Kit (QIAGEN), following the manufacturer’s instructions. Isolates were identified as A. muciniphila by use of species-specific PCR2. All strains were also verified by partial 16S rRNA gene sequencing. The amplification was done using the universal primers P0 and P650. Finally, the new isolates of A. muciniphila were compared by BoxA1 PCR analysis according to van Belkum and Hermans51.
Bacterial strains and culture conditions
A. muciniphila DSM 22959T (also ATCC BAA-835; type strain) has been purchased by DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Germany). All strains of A. muciniphila were grown at 37 °C in an anaerobic chamber (N2:H2:CO2 90:5:5) for 48 h in yeast medium broth (YM). YM contained (l−1): 0.45 g KH2PO4; 0.45 g K2HPO4; 0.9 g (NH4)2SO4; 0.9 g NaCl; 0.1 g MgSO4; 0.1 g CaCl2; 4 g NaHCO3; 1.9 ml CH3COOH (35 mM); 1 g L-Cysteine HCl monohydrate; 2.5 g yeast peptone; 5 g yeast extract and 2.5 g glucose and, it has been further supplemented with 6 g L-threonine and 2.8 g N-acetyl-D-glucosamine52. Antibiotics sensitivity was determined using Iso-Sensitest broth (Oxoid) supplemented with (l−1): 0.5 g L-Cysteine HCl monohydrate, 0.01 g Hemin, 0.01 g Vitamin K1, 6 g L-threonine and 2.8 g N-acetyl-D-glucosamine, which were filter-sterilized and added to the autoclaved medium. Escherichia coli Nissle 1917 (serotype O6:K5:H1) was isolated from the probiotic product ECN (Cadigroup Farmaceutici) in Luria–Bertani (LB) agar (1,5%, w/v) plates. For the determination of the minimum inhibitory concentrations (MICs) of selected antimicrobials, E. coli Nissle 1917 was cultivated overnight in cation-adjusted Muller-Hilton broth38 or sIST broth, under aerobic conditions at 37 °C.
Whole-genome sequencing and genomic analysis of A. muciniphila strains
The Whole-genome sequencing of the five A. muciniphila strains (Sap1, AmaP1, Vtp7, Rcp22, Amup9), made by Illumina NovaSeq 6000 platform, produced a total of 76′452′151 paired-end reads (Sap1: 10′542′938; Amap1: 10′766′503; Vtp7: 10′530′303; Rcp2: 26′681′187; Amup9: 17′931′220;) long 150 nt with GC content of 55%. The De novo assembling was performed by the assembly toolkit SPAdes 3.14.153. The software fastANI28 was used to compare by alignment-free computation of whole-genome Average Nucleotide Identity, the similarity between our draft genomes with 188 Akkermansia sp. complete genomes available on http://cmprod1.cibio.unitn.it/akkermansia_genomes/fna/. FASTQ data have been deposited in the European Nucleotide Archive (ENA) of the European Bioinformatics Institute under accession code PRJEB54610. The assembly has been upgraded by SPAdes using the genetic nearest complete genome found with fastANI as reference genome. The CheckM software was used for assessing the quality (completeness and contamination) of the assemblies54. The estimation of completeness and contamination is derived from collocated sets of genes that are ubiquitous and single-copy within a phylogenetic lineage. The fraction of marker genes that occur as duplicates is used to calculate the "Contamination" percentage. Instead, the "Completeness" value is obtained by the proportion of the missing markers to the total number of markers used. The number of CDS, RNAs and rRNA were determined using RAST tool kit55 associated with BAsic Rapid Ribosomal RNA Predictor (Barrnap) version 0.9 (https://github.com/tseemann/barrnap). The pangenome analyses was performed using Roary56 pan-genome pipeline which takes annotated assemblies in GFF3 format produced by Prokka57. Resistance Gene Identifier (RGI) on Comprehensive Antibiotic Resistance Database (CARD)30, updated April 2022, was used to predict the resistome of our strains based on homology and SNP models. RGI uses Prodigal, a protein-coding gene predictor for prokaryotic genome58. To include partial gene prediction, the Prodigal algorithm for small contigs (contigs < 20 kbp) was applied. “Perfect” and “Strict” algorithms were used to detect both perfect match, or previously unknown AMR genes variants, using detection models with CARD's curated similarity cut-offs to ensure the detected variant is likely a functional AMR gene. The Mobile Element Finder Tool35 was used to predict mobile genetic elements in our genomes by aligning the assembled contiguous sequences to reference sequences of previously known elements. The database used by Mobile Element Finder tool is the version 1.0.2 updated June 2020.
Plasmid DNA extraction and analysis
To verify the presence of plasmids in the isolated strains, extrachromosomal plasmid DNA was purified and subjected to agarose gel electrophoresis according to the method of Anderson and McKay59. Lactobacillus helveticus ATCC 15009, harboring three plasmids, was used as a positive control for plasmid DNA extraction60.
Total cell count and viability assessment by flow cytometry
Total cell counting and viability assessment of the bacterial cultures were performed by flow cytometry (FC), following the ISO 19344 procedure (2015)61, with some modifications. Briefly, bacterial samples were diluted (approximately 1 × 106 cells ml−1) in filtered phosphate-buffered saline (PBS) pH 7.4 to maintain an events rate in the flow lower than 2000 events s−1. The samples were stained with 0.1 µM SYTO™ 24 (Thermo Scientific) and 0.2 µM propidium iodide (PI; Sigma) and incubated in the dark at 37 °C for 15 min. FC analysis was performed with a C6 Plus flow cytometer (BD Biosciences, Milan Italy) with thresholds FSC-H 1000 and SSC-H 1000. All parameters were collected as logarithmic signals. Green (SYTO™ 24) and red (PI) fluorescence were detected in the FL1 (excitation 488 nm, emission filter 530/30) and FL3 (excitation 488 nm, emission filter 670 LP) channels, respectively. Electronic gates on the SYTO24/PI density plot were used to select and measure the total bacterial concentration (events ml−1), active fluorescent unit (AFU), and non-active fluorescent unit (nAFu), as described in ISO 19344 (2015)61.
Antimicrobial agents and MICs determination
According to EFSA-FEEDAP document 201817, the antibiotics included in the analysis were those tested for Enterobacteriaceae and other Gram-negative bacteria: ampicillin, gentamicin, kanamycin, streptomycin, tetracycline, ciprofloxacin, colistin, fosfomycin, and sulfamethoxazole (Merck). MICs for the antimicrobials listed above were determined by the standard macrodilution broth method38 in 24-wells plates, using serial two-fold dilutions of antimicrobials with a bacterial inoculum density of 5 × 105 AFU ml−1. To standardize the bacterial density in the inoculum suspensions, cell counting was performed by FC as described above. A bacterial suspension containing 1 × 108 AFU ml−1 was prepared by diluting an early stationary phase broth culture. The suspension was then diluted at 1:100 (1 × 106 AFU ml−1) in the suitable medium and 1 ml of this dilution was added as inoculum in a final volume of 2 ml. The growth controls contained only inoculated broth without antimicrobials, negative controls comprised media that was not inoculated. When ethanol and dimethylsulfoxide (DMSO) were used respectively as the diluents of tetracycline and sulfamethoxazole, their effect on cell growth was checked. The highest concentrations of ethanol and DMSO used were respectively: 0.63% (v/v) and 0.25% (v/v). In both cases, no inhibition of bacterial growth was seen. Incubation took place in an anaerobic chamber (N2:H2:CO2 90:5:5) at 37 °C for 48 h. E. coli Nissle was cultivated in aerobic conditions for 24 h at 37 °C. The MIC is the lowest concentration of antimicrobial where no visible growth is measured in the wells. MICs were determined at least in duplicates.
Ciprofloxacin susceptibility assay and efflux pumps inhibitors activity
Susceptibility of A. muciniphila strains to ciprofloxacin was also evaluated in the presence of two efflux pump inhibitors (EPIs), carbonyl cyanide 3-chlorophenylhydrazone (CCCP; Merck) or phenylalanine-arginine beta-naphthylamide (PaβN; Merck), to determine the contribution of efflux pumps to ciprofloxacin sensitivity. The concentrations of the EPIs used (CCCP 80 µM; PaβN 10 µM) was decided based on the determination of their MICs. A lower concentration of CCCP (40 µM) was used only for the strains Rcp22 and Amup9, while a concentration of 5 µM of PaβN was used for A. muciniphila DSM 22959 T and Amup9. CCCP was soluble in the growth medium with DMSO (1% v/v). DMSO alone was used as a control, and it did not affect cells growth. MICs were determined as mentioned above.
The efflux pumps activity in A. muciniphila strains was indirectly assessed by measuring the accumulation of ethidium bromide in the presence and absence of the efflux pump inhibitor CCCP62. Briefly, the bacterial strains were cultured in sIST as previously described. The cells were washed in PBS (pH 7.4) and diluted in the same buffer to reach a concentration of 1 × 106 AFU ml−1. Two sets of samples were prepared for each bacterial strain. In the first set only ethidium bromide was added at the final concentration of 10 µM. In the second set a sub-inhibitory concentration of CCCP (80 µM and 40 µM) was added and samples were incubated at 37 °C for 10 min. After the incubation, ethidium bromide was added (10 µM). Accumulation of ethidium bromide in cells was measured immediately and recorded every 10 min for 1 h by FC. FC analysis was performed with a C6 Plus flow cytometer (BD Biosciences, Milan Italy) with thresholds FSC-H 1000 and SSC-H 1000. All parameters were collected as logarithmic signals. Ethidium bromide fluorescence was detected in the FL3 channel (excitation 488 nm, emission filter 670 LP). Electronic gates on density plot of the ethidium bromide fluorescence against forward scatter (FSC-H) were used to select and measure the fluorescence intensity to determine the amount of ethidium bromide inside the cells. Indeed, ethidium bromide is a DNA-intercalating agent that fluoresces when bound to DNA. Therefore, fluorescence is higher when intracellular. Samples with an equal concentration of cells but without ethidium bromide were used as blanks and their fluorescence was subtracted from the other measurements.
Data availability
The raw reads are deposited in the European Nucleotide Archive (ENA) of the European Bioinformatics Institute under accession code: PRJEB54610.
References
Derrien, M., Vaughan, E. E., Plugge, C. M. & de Vos, W. M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476 (2004).
Collado, M. C., Derrien, M., Isolauri, E., de Vos, W. M. & Salminen, S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 73, 7767–7770 (2007).
Gómez-Gallego, C., Pohl, S., Salminen, S., De Vos, W. M. & Kneifel, W. Akkermansia muciniphila: A novel functional microbe with probiotic properties. Benef. Microbes 7, 571–584 (2016).
Cani, P. D. & de Vos, W. M. Next-generation beneficial microbes: The case of Akkermansia muciniphila. Front. Microbiol. 8, 1765 (2017).
Cani, P. D., Depommier, C., Derrien, M., Everard, A. & de Vos, W. M. Akkermansia muciniphila: Paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 19, 625–637 (2022).
Everard, A. et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 110, 9066–9071 (2013).
Plovier, H. et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113 (2017).
Ottman, N. et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE 12, e0173004 (2017).
Depommier, C. et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019).
EFSA-BIOHAZ & Koutsoumanis, K. et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 12: Suitability of taxonomic units notified to EFSA until March 2020. EFSA J. 18, 6174 (2020).
Saarela, M. Safety aspects of next generation probiotics. Curr. Opin. Food Sci. 30, 8–13 (2019).
Kumari, M. et al. Fostering next-generation probiotics in human gut by targeted dietary modulation: An emerging perspective. Food Res. Int. 150, 110716 (2021).
van Schaik, W. The human gut resistome. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 370, 20140087 (2015).
Barron, M. The gut resistome and the spread of antimicrobial resistance. Research in this article was presented at ASM microbe, the annual meeting of the American Society for Microbiology, held June 9–13, in Washington, D.C. (2022)
Guo, X. et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diversity, and global distribution in mammalian gut microbiotas. BMC Genom. 18, 800 (2017).
de Nies, L. et al. Evolution of the murine gut resistome following broad-spectrum antibiotic treatment. Nat. Commun. 13, 2296 (2022).
EFSA-FEEDAP & Rychen, G. et al. Guidance on the characterization of microorganisms used as feed additives or as production organisms. EFSA J. 16, 5206 (2018).
Martín, R. et al. Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: A step forward in the use of F. prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 8, 1226 (2017).
Machado, D. et al. Revealing antimicrobial resistance profile of the novel probiotic candidate Faecalibacterium prausnitzii DSM 17677. Int. J. Food Microbiol. 363, 109501 (2021).
EFSA-NDA & Turck, D. et al. Safety of pasteurised Akkermansia muciniphila as a novel food pursuant to Regulation (EU) 2015/2283. EFSA J. 19, 6780 (2021).
Dubourg, G. et al. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int. J. Antimicrob. Agents 41, 149–155 (2013).
Cozzolino, A. et al. Preliminary evaluation of the safety and probiotic potential of Akkermansia muciniphila DSM 22959 in comparison with Lactobacillus rhamnosus GG. Microorganisms 8, 189 (2020).
Machado, D. et al. Insights into the antimicrobial resistance profile of a next generation probiotic Akkermansia muciniphila DSM 22959. Int. J. Environ. Res. Public Health 19, 9152 (2022).
van Passel, M. W. et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 6, e16876 (2011).
Caputo, A. et al. Whole-genome assembly of Akkermansia muciniphila sequenced directly from human stool. Biol. Direct 19, 10–15 (2015).
Karcher, N. et al. Genomic diversity and ecology of human-associated Akkermansia species in the gut microbiome revealed by extensive metagenomic assembly. Genome Biol. 22, 209 (2021).
Guo, X. et al. Different subtype strains of Akkermansia muciniphila abundantly colonize in southern China. J. Appl. Microbiol. 120, 452–459 (2016).
Jain, C. et al. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114 (2018).
Rosselló-Móra, R. & Amann, R. Past and future species definitions for Bacteria and Archaea. Syst. Appl. Microbiol. 38, 209–216 (2015).
Alcock, B. P. et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48(D1), D517–D525 (2020).
Ashenafi, M. et al. Purification and characterization of aminoglycoside phosphotransferase APH(6)-Id, a streptomycin-inactivating enzyme. Mol. Cell. Biochem. 387, 207–216 (2014).
Du, J. et al. Occurrence and abundance of tetracycline, sulfonamide resistance genes, and class 1 integron in five wastewater treatment plants. Environ. Sci. Pollut. Res. 21, 7276–7284 (2014).
Roberts, M. C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245, 195–203 (2015).
Leus, I. V. et al. Substrate specificities and efflux efficiencies of RND efflux pumps of Acinetobacter baumannii. J. Bacteriol. 200, e00049-e118 (2018).
Johansson, M. H. K. et al. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109 (2021).
Partridge, S. R., Kwong, S. M., Firth, N. & Jensen, S. O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e00088-e117 (2018).
Klare, I. et al. Evaluation of new broth media for microdilution antibiotic susceptibility testing of Lactobacilli, pediococci, lactococci, and bifidobacteria. Appl. Environ. Microbiol. 71, 8982–8986 (2005).
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 30 th ed. In:CLSI supplement M100. (Clinical and Laboratory Standards Institute, Wayne, PA, 2020).
Neuert, S. et al. Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Front. Microbiol. 27, 592 (2018).
Pence, M. A. Antimicrobial resistance in clinically important anaerobes. Clin. Microbiol. Newsletter. 41, 1–7 (2019).
Rasmussen, B. A., Bush, K. & Tally, F. P. Antimicrobial resistance in anaerobes. Clin. Infect. Dis. 24, S110–S120 (1997).
Mingeot-Leclercq, M. P., Glupczynski, Y. & Tulkens, P. M. Aminoglycosides: Activity and resistance. Antimicrob. Agents Chemother. 43, 727–737 (1999).
Delcour, A. H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta. 1794, 808–816 (2009).
Kulengowski, B., Ribes, J. A. & Burgess, D. S. Polymyxin B Etest® compared with gold-standard broth microdilution in carbapenem-resistant Enterobacteriaceae exhibiting a wide range of polymyxin B MICs. Clin. Microbiol. Infect. 25, 92–95 (2019).
Ouwerkerk, J. P. et al. Comparative genomics and physiology of Akkermansia muciniphila isolates from human intestine reveal specialized mucosal adaptation. Microorganisms 10, 1605 (2022).
Jacoby, G. A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41, S120–S126 (2005).
AlMatar, M., Albarri, O., Makky, E. A. & Köksal, F. Efflux pump inhibitors: New updates. Pharmacol. Rep. 73, 1–16 (2021).
Jamshidi, S., Sutton, J. M. & Rahman, K. M. Computational study reveals the molecular mechanism of the interaction between the efflux inhibitor PAβN and the AdeB Transporter from Acinetobacter baumannii. ACS Omega 2, 3002–3016 (2017).
ISO 10932 IDF 223. Milk and milk products—Determination of the minimal inhibitory concentration (MIC) of antibiotics applicable to bifidobacterial and non-enterococcal lactic acid bacteria (LAB) (2010).
Di Cello, F. & Fani, R. A molecular strategy for the study of natural bacterial communities by PCR-based techniques. Minerva Biotecnologica 8, 126–134 (1996).
van Belkum, A. & Hermans, P. W. BOX PCR Fingerprinting for molecular typing of Streptococcus pneumoniae. Methods Mol. Med. 48, 159–168 (2001).
Belzer and De Vos. Method of culturing Akkermansia. Patent WO2016177801A1 (2016).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Ramy, K. A. et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008).
Page, A. J. et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Seemann, T. et al. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Hyatt, D. et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 119 (2010).
Anderson, D. G. & McKay, L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46, 549–552 (1983).
Fortina, M. G., Parini, C., Rossi, P. & Manachini, P. L. Mapping of three plasmids from Lactobacillus helveticus ATCC 15009. Lett. Appl. Microbiol. 17, 303–306 (1993).
ISO 19344, IDF 232. Milk products—Starter cultures, probiotics and fermented products—Quantification of lactic acid bacteria by flow cytometry. (2015).
Blair, JM., Piddock, LJ. How to measure export via bacterial multidrug resistance efflux pumps. mBio 7, e00840–16 (2016).
Author information
Authors and Affiliations
Contributions
R.F., G.G., D.M., S.A. conceptualized and designed the work. R.F. performed the experiments and data analysis. G.G. performed the bioinformatics data acquisition and analysis. R.F. draft the manuscript. G.G., D.M., S.A. revised the manuscript critically. All the authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Filardi, R., Gargari, G., Mora, D. et al. Characterization of antibiotic-resistance traits in Akkermansia muciniphila strains of human origin. Sci Rep 12, 19426 (2022). https://doi.org/10.1038/s41598-022-23980-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23980-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.