Abstract
Egg production traits are economically important in laying ducks. Genetic molecular mechanisms and candidate genes underlying these traits remain unclear. In this study, whole genome variants were identified through whole-genome resequencing using three high-egg producing (HEN) and three low-egg producing (LEN) laying ducks. The gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genome (KEGG) pathways for the genes of common differential variants between HEN and LEN ducks were determined. Frizzled class receptor 6 (FZD6) was further genotyped using the Sequenom MassARRAY iPLEX platform. The association of FZD6 gene polymorphisms with 73 egg production and weight traits in 329 female ducks were estimated. A total of 65,535 single nucleotide polymorphisms (SNPs) and 4,702 indels were identified across the genome. Fourteen GO terms and 14 KEGG pathways were determined for the genes of common differential variants, including MAPK signaling, Wnt signaling, melanogenesis and calcium signaling pathways, which are key functional pathways for poultry egg production reported in previous reports. Further analysis showed that 27 SNPs of FZD6 were associated with three early egg production of duck and egg weight traits, including egg production at 17 weeks (EP17), 18 weeks (EP18) and 19 weeks (EP19) and egg weight at 59 weeks (EW59). The FZD6 should be considered a novel candidate gene for egg production traits in laying ducks.
Similar content being viewed by others
Introduction
In the modern poultry industry, egg production traits, including egg number (EN), egg weight (EW), and age at first egg (AFE), are important reproductive and economic factors1,2 and improved egg production traits are usually the goals of poultry breeding companies3. In chickens, egg production traits are quantitative traits whose genetic architecture has been uncovered using a quantitative trait locus (QTL) analysis in a previous genome-wide association study (GWAS)4. Over 659 QTLs for egg-production traits have been found5 and many promising genes and mutants for egg-production traits have been identified6. In contrast the genetic basis underlying the egg production traits in ducks is still not fully understood, because the heritability of egg production traits is low to intermediate, ranging from 0.13 to 0.437. Some candidate genes for egg production traits have been investigated, mainly focused on the genes associated with reproductive physiology, such as the follicle-stimulating hormone receptor gene8, melatonin receptor gene9 and the prolactin gene10. However, candidate gene method has limited ability to identify genetic basis of egg production traits in duck.
With the development of proteomic technologies, it is possible to investigate the genetic basis for key traits of livestock and poultry by whole-genome resequencing. In pigs, 13,955,609 SNPs and 2,666,366 indels have been found and the common differential SNPs and indels between five high- and five low-prolificacy Yorkshire sows using whole-genome resequencing technology and several differential variants within candidate genes for reproduction traits were confirmed by comparing selective regions and published quantitative trait locus (QTL) data11. In chickens, whole-genome resequencing from eight chickens with slow- and fast-feathering rates showed 54,984 SNPs and mutated genes were involved in the response to stimuli, growth and reproduction processes and two SNPs involved in feather development were identified in the exonic regions of the Wnt signaling gene12. In ducks, selection signatures were uncovered by genome-wide comparisons among Mallards, indigenous-breeds and Pekin ducks and two selective sweeps for two important economic traits of the Pekin duck were identified through further fine-mapping based on resequencing of more than 1,000 ducks from an F2 segregating population generated by wild crossed with domestic birds13. In geese, the KIT gene underlying white or gray plumage color in Chinese domestic geese was identified by resequencing the whole genome of 18 geese from six populations, including white and gray goose breeds14.
There are 250–300 million Shan-ma ducks (Anas Platyrhynchos) raised every year and as one of the main Chinese indigenous egg laying duck types, makes up about half laying duck population raised in China. The breed originated from Longmen town, in Longyan city and in January 2017, the Ministry of Agriculture and Rural Affairs of China officially approved the registration and protection of agricultural products based on the Longyan Shan-ma duck from this region.
In this study whole-genome resequencing was performed on three full-sib high- and three full-sib low-egg producing Shan-ma ducks. The study aimed to identify the mutations across the genome and the genetic basis underlying egg production traits in egg-laying ducks.
Results and discussion
Phenotype statistics and genome variants
Statistics results of egg number (EN) and egg weight (EW) traits between HEN and LEN duck number ducks at 71 weeks are shown in Table S1. Egg number and total egg weight in the HEN group were significantly increased to 22.97% (P = 0.007) and 27.38% (P = 0.011), respectively, compared with the LEN group. There was no significant difference in EW between the two groups.
A total of 82.69 Gb raw base reads were obtained from three HEN and LEN ducks, shown in Table S2. Clean data reads of each sample ranging from 12.42 to 15.13 Gb were acquired after the quality control. The sequencing quality was high with Q20 and Q30 ratio greater than 92%. Mapping rate, average genome depth and coverage were obtained through clean reads mapped to the duck reference genome in Table S3, with the mapping rate of each sample more than 94%. The average sequencing depth ranged from 9.02 to 10.76 times and four times coverage ranged from 91.80% to 94.89%. The sequencing quality, mapping rate and average sequencing depth met the requirements for detecting the genomic variations.
A total of 7,243,250 SNPs and 864,777 indels were identified in the Shan-ma duck genome in Table 1. The number of SNPs and indels in the Shan-ma duck detected in this study were less than other duck types15,16, because the trial birds were full sibling individuals of one breed. In this study, the number of SNPs in intergenic, intronic and exonic regions including exonic stop gain, exonic stop loss, exonic synonymous, and exonic non-synonymous were 3,531,432 (52.21%), 3,232,152 (47.79%) and 114,824 (1.70%), respectively.
In the exonic region, 79,997 (69.67%) of the SNPs were synonymous mutations, 34,357 (29.92%) were non-synonymous mutations, 421 (0.37%) were stop gain and 49 (0.04%) were stop loss. The transition to transversion ratio (Ts/Tv) was 2.429, which indicated the SNP quality was comparable to human and animal Ts/Tv ranges of 2.0 to 2.4 (Li et al., 2020). For the indels, 365,133 (42.22%) were insertion and 499,644 (57.78%) were deletion variants. In the exonic region, indels consisted of 2135 frameshift, 959 non-frameshift, 54 stop gain and nine stop-loss variants.
Common differential variations, annotation, and function enrichment analyses
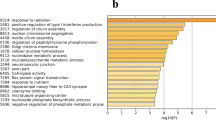
A total of 65,535 common differential SNPs and 4,702 common differential indels were identified between the HEN and LEN groups. The detailed annotated information is shown in Tables S4 and S5. The common differential SNPs and indels participated in 2027 and 954 genes, respectively, including 2417 common genes. These 2417 genes with enriched gene ontology (GO) terms and Kyoto encyclopedia of genes and genome (KEGG) pathways were determined through annotation using the online analysis tool Database for Annotation, Visualization and Integrated Discovery (DAVID, version v2021q4) (https://david.ncifcrf.gov/)17. Fourteen significantly enriched GO terms with FDR corrected P value < 0.05 were identified, including two molecular function GO terms, including nucleic acid binding transcription factor activity and transcription factor activity sequence-specific DNA binding, two cell components GO terms, including plasma membrane and cell periphery and ten biological process GO terms, including homophilic cell adhesion via plasma membrane adhesion molecules, signaling, cell communication, regulation of signaling, single organism signaling, regulation of cell communication, regulation of signal transduction, signal transduction, cellular response to organic substance and cell–cell adhesion via plasma-membrane adhesion molecules in Table S6.
Fourteen KEGG pathways were significantly enriched (FDR corrected P value < 0.05) as shown in Table S7. Four of 14 pathways, including MAPK signaling, Wnt signaling, melanogenesis and calcium signaling pathways were associated with poultry egg production in previous studies18,19,20. Because nine SNPs including exon 3: c.C855T:p.L285L, c.T852C:p.I284I, c.C741T:p.N247N, c.G645C:p.P215P, c.A435G:p.R145R, c.A405G:p.T135T, c.T372C:p.N124N, c.A329G:p.N110S and exon 4: c.A1437G:p.K479K located at two exons of FZD6 were found using whole-genome resequencing and enriched in both Wnt signaling and melanogenesis signaling pathways, it was selected for further study.
Association study
To further confirm the association between FZD6 and egg production traits in the Shan-ma duck, FZD6 in 329 female ducks was genotyped by Sequenom MassARRAY technology. The primers of FZD6 genotyped used in Sequenom MassARRAY platform are shown in Table S8. Fifty-three SNPs were found and 35 retained for further association study after sixteen SNPs were removed because the HWE P value was less than or equal to 4.86 × 10–5 and two SNPs excluded because MAF was less than 0.05 in Table 2. A total of 64 associations were identified between 29 SNPs of FZD6 and four traits, EP17, EP18, EP19 and EW59 in Table 3. Twenty-one SNPs associated with EP17 were identified, with the minor allele of 14 SNPs including A1546176G, A1546196T, C1546217T, A1546400G, G1546616A, A1547351G, A1547887T, C1548016T, C1548761T, G1564507A, C1567012T, A1567491G, T1570377C, and G1571845C which increased EP17 and the minor allele of seven SNPs T1551159C, G1553772A, A1553775G, C1553982G, T1554192C, T1554298C and C1558458T decreased EP17. Ten SNPs associated with EP18 were found, including the minor allele of four SNPs A1546176G, A1546196T, C1546217T and C1548016T, which increased EP18 and the minor allele of six SNPs T1551159C, C1553982G, T1554298C, C1558458T, A1562579G and A1571291G, which decreased EP18. Seventeen SNPs associated with EP19 were identified, including the minor allele of four SNPs A1546176G, A1546196T, C1546217T, and C1548016T, which increased EP19 and the minor allele of 13 SNPs, including T1551159C, G1553772A, A1553775G, C1553982G, T1554192C, T1554298C, C1558458T, C1558820T, A1562579G, G1567392C, C1570687T, A1571291G and G1572493C, which decreased EP19. Sixteen SNPs associated with EW59 were identified, including the minor allele of one SNP, G1571845C, which increased EW59 and 15 SNPs, including C1558458T, T1551159C, C1553982G, A1562579G, A1571291G, G1553772A, T1554192C, A1553775G, C1570687T, C1558820T, A1570698G, T1554298C, G1558943A, G1572493C and G1567392C, which deceased EW59.
The gene FZD6 is a member of the frizzled gene family, which encodes frizzled class receptor 6, which is a WNT signaling protein receptor. The WNT-frizzled selectivity plays a significant role in developmental biology, stem cell regulation oncogenesis and human disease21. In human disease, FZD6 is a core gene that is strongly linked to human ovarian cancer22. In animal reproduction, secreted frizzled-related protein completes the preparation for the next reproduction process during the transition from the young to the egg-laying phase23 and WNT-FZD6 interaction regulates the follicle-stimulating hormone selection for dominant follicle24. Based on above-mentioned information, it was believed that FZD6 could be considered a promising candidate gene for the egg production traits, which needs to be validated in large duck populations.
In Summary, whole genome variants, including 7,243,250 SNPs and 864,777 indels from between high- and low-egg number ducks were identified using high-throughput sequencing technology, with 65,535 common differential SNPs and 4,702 common differential indels also identified by comparing sequences from the two groups of ducks, which participated in 2,027 and 954 genes, respectively, including 2,417 common genes. These genes were significantly enriched 14 GO terms and 14 KEGG pathways. Four of 14 signaling pathways, including the MAPK, Wnt, melanogenesis and calcium signaling pathways have been shown to be related to poultry egg production in previous studies. An association of FZD6 with egg production traits has been verified as 29 SNPs were associated with four traits, EP17, EP18, EP19, and EW59. Importantly, pathways and candidate genes identified in this study will not only provide a new insight into the genetic basis underlying egg production traits in ducks but also improve better understanding the genetic architecture and molecular mechanisms of these traits in poultry.
Methods
Birds and samples
High-yield line Longyan Shan-ma ducks were raised in cages under the same environmental and nutritional conditions at the Longyan Shan-ma Duck Original Breeding Farm. Three high-egg (HEN) and three low-egg producing (LEN) female ducks from one full-sib family were used in this study. Blood was sampled from the brachial vein using citrate-based anticoagulant syringes before the ducks were euthanized by electrical stunning and exsanguination. The blood samples were snap-frozen in liquid nitrogen then held at − 80 °C until used.
Whole-genome resequencing and quality control
The genomic DNA (gDNA) of blood samples was extracted using the phenol–chloroform method. The integrity of the DNA was estimated using electrophoresis in a 1% agarose gel and the purity of DNA was assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher, Foster City, CA, USA). The concentration of DNA was measured using an Invitrogen Qubit 2.0 fluorimeter (Thermo Fisher Scientific, Foster City, CA, USA). The OD260/OD280 ratios were between 1.8 and 2.0 and the concentration above 1.5 μg of each DNA sample were used to construct the DNA libraries. The DNA samples were randomly interrupted using a Covaris crusher (S220; Covaris, LLC, Woburn, MA, USA) to create fragments of 350 bp in length. The DNA Libraries were constructed using a TruSeq Library Construction Kit (Illumina Inc., San Diego, CA, USA) following the manufacturer's instructions. The DNA library was sequenced on the HiSeq 2500 high-throughput sequencing platform (Illumina Inc., San Diego, CA, USA).
Raw data reads were obtained by sequencing and clean data reads were acquired following the quality control (QC) procedure to remove unusable reads. Usable reads contained the Illumina library construction adapters, more than 10% unknown N bases and one end of the read had to contain more than 50% of low-quality bases, with a sequencing quality value less than or equal to five.
Detection and annotation of genomic variants
All clean reads were aligned to the reference duck genome (BGI_duck_1.0) using Burrows–Wheeler aligner (BWA) software (version 0.7.8-r455) with default parameters25. The SNPs and indels were detected using the SAMtools (version 0.1.19)26 with the parameters as “-q 1 -C 50 -m 2 -F 0.002 -d 1000” and the filtering criteria “the mapping quality > 20 and the depth of the variate position > 4”. The functional annotation of these variants was carried out using ANNOVAR (version 2013Aug23)27 and known genes and region annotations were determined using the UCSC genome browser database28.
Differential variants, annotation, and function enrichment analyses
To identify differential variants across genomes between high- and low-egg producing ducks, the SNPs with the same genotype were distinguished in each group first, then differential variants and then the above-mentioned differential or “common differential variants”29 were determined between the two groups. Genes located within 500 kb with these common differential variants were annotated.
The GO enrichment analyses for the genes of the common differential variants were conducted using the DAVID v6.8 online server (https://david.ncifcrf.gov/home.jsp)17. The KEGG pathway function enrichment analyses for the genes of the common differential variants were performed in web-based software Kobas 3.0 (http://kobas.cbi.pku.edu.cn/)30,31. The copyright permission (No. 221304) to publish the corresponding KEGG pathways was officially granted by Kanehisa Laboratories32,33,34. False discovery rates (FDR) with a corrected P value < 0.05 were considered significant for GO terms and pathways.
Association study of FZD6 with egg number traits
Based on the reproductive physiological function of genes in the pathways, FZD6 was used for the further association study with 73 egg production traits in 329 female ducks, as seen in Table 4. The region 1,546,176 to 1,572,493 bp on chromosome 2 of FZD6 was genotyped by the Sequenom MassARRAY platform (Sequenom, San Diego, CA, USA), according to the manufacturer’s instructions. Quality control of SNP genotyping data were estimated by Haploview 4.1 software35 and any with a minimum allele frequency (MAF) of less than 0.05 and a Hardy–Weinberg equilibrium (HWE) test P value < 1.0 × 10–4 were excluded. Associations between SNPs and egg production traits were analyzed with the general linear model (GLM) in PLINK v1.9036 using the following model:
where Y is the trait value, µ is the overall mean, F is the family effect, BW is the body weight effect, G is the fixed effect of genotype and e is the random error.
Statistical analysis
Statistical analysis of egg production and weight traits between high- and low-egg production birds was performed with the independent-sample T test using SPSS 19.0 (IBM, Armonk, NY, USA) and the results are presented as the mean ± standard deviation (SD). For the traits used in the association study, abnormal values of these traits were excluded before analysis using Grubbs’ method and the descriptive statistics were analyzed by Minitab 17.0 Statistical Software (Minitab Inc., State College, PA, USA).
Ethics statement
This study protocol was approved by the Longyan University Ethics Committee. All animal studies were conducted in accordance with the Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China). This study is also reported in accordance with the ARRIVE guidelines (https://arriveguidelines.org).
Data availability
All the data is available on reasonable request from the corresponding author.
References
Wang, C. et al. Molecular cloning, expression profile, polymorphism and the genetic effects of the dopamine D1 receptor gene on duck reproductive traits. Mol. Biol. Rep. 39, 9239–9246. https://doi.org/10.1007/s11033-012-1797-3 (2012).
Yuan, J. et al. Identification of promising mutants associated with egg production traits revealed by genome-wide association study. PLoS ONE 10, e0140615. https://doi.org/10.1371/journal.pone.0140615 (2015).
Sitzenstock, F. et al. Efficiency of genomic selection in an established commercial layer breeding program. Genet. Sel. Evol. 45, 29. https://doi.org/10.1186/1297-9686-45-29 (2013).
Goto, T. & Tsudzuki, M. Genetic mapping of quantitative trait loci for egg production and egg quality traits in chickens: A review. J. Poult. Sci. 54, 1–12. https://doi.org/10.2141/jpsa.0160121 (2017).
Hu, Z. L., Park, C. A. & Reecy, J. M. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 47, D701–D710. https://doi.org/10.1093/nar/gky1084 (2019).
Liu, Z. et al. Genome-wide association analysis of egg production performance in chickens across the whole laying period. BMC Genet. 20, 67. https://doi.org/10.1186/s12863-019-0771-7 (2019).
Lin, R. L., Chen, H. P., Rouvier, R. & Marie-Etancelin, C. Genetic parameters of body weight, egg production, and shell quality traits in the Shan Ma laying duck (Anas platyrhynchos). Poult Sci. 95, 2514–2519. https://doi.org/10.3382/ps/pew222 (2016).
Xu, J. et al. Molecular characterization, expression profile of the FSHRgene and its association with egg production traits in muscovy duck. J. Genet. 96, 341–351. https://doi.org/10.1007/s12041-017-0783-x (2017).
Feng, P. et al. Polymorphisms of melatonin receptor genes and their associations with egg production traits in Shaoxing duck. Asian-Australas J. Anim. Sci. 31, 1535–1541. https://doi.org/10.5713/ajas.17.0828 (2018).
Purwantini, D. et al. Prolactin gene polymorphisms and associations with reproductive traits in Indonesian local ducks. Vet. World 13, 2301–2311. https://doi.org/10.14202/vetworld.2020.2301-2311 (2020).
Li, X. et al. Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics 112, 199–206. https://doi.org/10.1016/j.ygeno.2019.01.014 (2020).
Qiu, M. et al. Whole-genome resequencing reveals aberrant autosomal SNPs affect chicken feathering rate. Anim Biotechnol, 1–13. https://doi.org/10.1080/10495398.2020.1846545 (2020).
Zhou, Z. et al. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 9, 2648. https://doi.org/10.1038/s41467-018-04868-4 (2018).
Wen, J. et al. Genomic scan revealed KIT gene underlying white/gray plumage color in Chinese domestic geese. Anim. Genet https://doi.org/10.1111/age.13050 (2021).
Gu, L. et al. Genetic characteristics of Jiaji Duck by whole genome re-sequencing. PLoS ONE 15, e0228964. https://doi.org/10.1371/journal.pone.0228964 (2020).
Lee, D. et al. Population analysis of the Korean native duck using whole-genome sequencing data. BMC Genomics 21, 554. https://doi.org/10.1186/s12864-020-06933-z (2020).
Huang, D. W. et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35, W169-175. https://doi.org/10.1093/nar/gkm415 (2007).
Folaniyi, B. S. et al. Hypothalamic and ovarian transcriptome profiling reveals potential candidate genes in low and high egg production of White Muscovy Ducks (Cairina Moschata). Poultry Sci. 101310 (2021).
Zhang, T. et al. Transcriptome analysis of ovary in relatively greater and lesser egg producing Jinghai Yellow Chicken. Anim. Reprod. Sci. 208, 106114. https://doi.org/10.1016/j.anireprosci.2019.106114 (2019).
Tao, Z. et al. Comparative transcriptomic analysis of high and low egg-producing duck ovaries. Poult. Sci. 96, 4378–4388. https://doi.org/10.3382/ps/pex229 (2017).
Kilander, M. B., Dahlstrom, J. & Schulte, G. Assessment of Frizzled 6 membrane mobility by FRAP supports G protein coupling and reveals WNT-Frizzled selectivity. Cell Signal 26, 1943–1949. https://doi.org/10.1016/j.cellsig.2014.05.012 (2014).
Kumar, S. U., Kumar, D. T., Siva, R., Doss, C. G. P. & Zayed, H. Integrative bioinformatics approaches to map potential novel genes and pathways involved in ovarian cancer. Front. Bioeng. Biotechnol. 7, 391. https://doi.org/10.3389/fbioe.2019.00391 (2019).
Ren, J. et al. Divergently expressed gene identification and interaction prediction of long noncoding RNA and mRNA involved in duck reproduction. Anim. Reprod. Sci. 185, 8–17. https://doi.org/10.1016/j.anireprosci.2017.07.012 (2017).
Gupta, P. S. et al. Regulation and regulatory role of WNT signaling in potentiating FSH action during bovine dominant follicle selection. PLoS ONE 9, e100201. https://doi.org/10.1371/journal.pone.0100201 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. https://doi.org/10.1093/bioinformatics/btp324 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. https://doi.org/10.1093/bioinformatics/btp352 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164. https://doi.org/10.1093/nar/gkq603 (2010).
Navarro Gonzalez, J. et al. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res 49, D1046–D1057. https://doi.org/10.1093/nar/gkaa1070 (2021).
Jiang, J. et al. Whole-genome resequencing of holstein bulls for indel discovery and identification of genes associated with milk composition traits in dairy cattle. PLoS ONE 11, e0168946. https://doi.org/10.1371/journal.pone.0168946 (2016).
Wu, J., Mao, X., Cai, T., Luo, J. & Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34, W720-724. https://doi.org/10.1093/nar/gkl167 (2006).
Xie, C. et al. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–322. https://doi.org/10.1093/nar/gkr483 (2011).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551. https://doi.org/10.1093/nar/gkaa970 (2021).
Barrett, J. C. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. pdb ip71. https://doi.org/10.1101/pdb.ip71 (2009).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. https://doi.org/10.1086/519795 (2007).
Acknowledgements
This study was financed by the Key Research and Breeding Projects and Industrialization Foundations of Agricultural Improved Breeding in Breed Industry Innovation and Industrialization Project (2021-2025) of Fujian Province (zycxny20211014), Foreign Cooperation Projects of Fujian Provincial Science and Technology Department (2021I0045), Science and Technology Department Leading Project of Fujian Province (2022N0027), Fujian Province Undergraduate Innovation and Entrepreneurship Training Program (202111312016) and the Teacher Training Project of Longyan University. We thank International Science Editing (https://www.internationalscienceediting.cn) for language editing this manuscript.
Author information
Authors and Affiliations
Contributions
Y.L. and X.J. conceived and jointly supervised the study. Y.S. and Y.Z. contributed to the study design, performed statistical analyses and wrote the manuscript. Q.W., M.Z., J.L., E.X., M.L., Y.C., F.D., and H.P. contributed to analysis results and result interpretation. R.L., H.C. and W.Y. contributed to duck breeding. All authors contributed to the manuscript revision. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, Y., Zhang, Y., Wu, Q. et al. Whole-genome sequencing identifies potential candidate genes for egg production traits in laying ducks (Anas platyrhynchos). Sci Rep 13, 1821 (2023). https://doi.org/10.1038/s41598-022-21237-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-21237-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.