Abstract
Migration of methane-rich fluids at submarine cold seeps drives intense microbial activity and precipitation of authigenic carbonates. In this study, we analyzed microbially derived authigenic carbonate samples recently recovered from active gas hydrate mounds on the southwestern slope of the Chukchi Borderlands (CB), western Arctic Ocean. Our main aim was to characterize the distribution patterns of trace elements in carbonate-hosted lipid fractions to assess metalloenzyme requirements of microbes involved in anaerobic oxidation of methane (AOM). We measured stable isotopes, trace elements, lipid biomarkers, and genomic DNA, and results indicate the dominance of AOM-related lipid biomarkers in studied carbonate samples, as well as a predominant occurrence of the anaerobic methanotrophic archaea (ANME)-1. We also report evidence for significant preferential enrichments of various trace elements (Li, Ni, Co, Cu, Zn, and Mo) in the total lipid fractions of CB carbonates, relative to elemental compositions determined for corresponding carbonate fractions, which differ from those previously reported for other seep sites. We hypothesize that trace element enrichments in carbonate-hosted lipid fractions could vary depending on the type of AOM microbial assemblage. Additional work is required to further investigate the mechanisms of lipid-bound trace elements in cold seep carbonates as potential metalloenzymes in AOM.
Similar content being viewed by others
Introduction
Cold seeps occur at ocean margins worldwide1,2, corresponding to methane-rich fluid emanations from the seabed into the water column3,4. The seepage rates of reduced fluids can vary substantially at the seafloor5, leading to a large spectrum of redox conditions in subsurface sediments at cold seeps6. A key biogeochemical process in submarine methane seeps is the anaerobic oxidation of methane (AOM), which efficiently consumes a substantial fraction of methane released into the overlying water column7,8. During AOM, alkalinity levels strongly increase in the surrounding pore waters because of the production of biocarbonate (HCO3−), promoting carbonate supersaturation and precipitation2,9. Accordingly, authigenic carbonate precipitation is typically encountered in cold seeps10,11, which can serve as an archive for investigating methane release events in both modern and ancient seeps12,13.
AOM is typically mediated by a consortium of anaerobic methanotrophic archaea (ANME) and sulfate-reducing bacteria (SRB)14,15. However, recent studies have also reported that ANMEs can also use other electron acceptors, such as manganese (Mn)- and iron (Fe)-rich oxyhydroxides, to promote methane oxidation in cold seeps16. Notably, other trace metals such as nickel (Ni), cobalt (Co), molybdenum (Mo), tungsten (W), and zinc (Zn) can also be involved in AOM, acting as enzymatic co-factors17,18. Most of these findings have been obtained from culture experiments17,19. A recent study characterized, for the first time, the trace element geochemistry of the total lipid fractions preserved in authigenic carbonates from various seeps worldwide, such as the Congo, Nile deep-sea, and Niger fans, and the Gulf of Mexico20, suggesting that such an approach could be used to identify trace metals essential to microbial activity in cold seeps. A previous study reported marked enrichment of Ni, Co, Mo, and W in carbonate-hosted lipid fractions, which were linked to previously identified enzymatic pathways involved in AOM20. Nonetheless, information about how AOM may be influenced by changes in trace metal bioavailability in cold seeps remains scarce.
Recently, gas hydrate mound structures have been discovered in the Chukchi Borderlands (CB) of the western Arctic Ocean21. Geochemical information on the origin of the emitted gas and pore fluid properties was obtained within this mound structure during the R/V ARAON expedition ARA09C in 201822. However, thus far, biogeochemical signatures preserved in authigenic carbonates have not been investigated in these newly discovered gas hydrate mounds. In this study, we provide a detailed geochemical characterization of a series of authigenic carbonate samples using oxygen and carbon stable isotopes and mineralogy, as well as trace element abundances, lipid biomarkers, and nucleic acids. The main objectives of this study were to i) evaluate key environmental factors controlling carbonate precipitation in the CB mounds, ii) characterize geochemical and microbial community characteristics in the CB mounds, and iii) assess the relation of trace metals in microbial activity associated with AOM by comparing our new data for CB geochemical and microbial signatures with those previously obtained at other seeps worldwide20. We aimed to test the hypothesis that specific trace element enrichments in the total lipid fractions extracted from authigenic carbonates could be indicative of preferential trace metal utilization by different ANME communities. Our study sheds new light on the metalloenzyme requirements of microbes involved in AOM.
Results
Mineralogy and stable isotopic compositions
The authigenic carbonate samples collected in this study (Fig. 1, see also Fig. S1 in the Supplementary Information) were dominated by calcite, with a minor contribution from dolomite (Table 1). The δ13C values of carbonates (δ13Ccarbonate) were depleted, varying between − 37.0 and − 32.8‰. The δ18O values of carbonates (δ18Ocarbonate) ranged from 3.9 to 5.6‰, with more enriched values occurring in samples from the deeper sediment layers (Table 1). The measured δ13Ccarbonate and δ18Ocarbonate values of bulk CB carbonates partially overlapped with those determined for a series of seep carbonate samples previously investigated20 (Table 1 and Fig. S210,23,24,25,26,27).
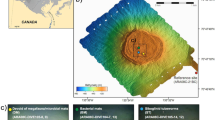
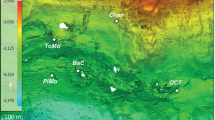
Map showing (a) the mound structures discovered in the Chukchi Borderlands (CB) with authigenic carbonates investigated in this study and (b) four comparison study sites: Congo fan (CF), Nile deep-sea fan (NDSF), Niger fan (NF), and Gulf of Mexico (GoM). The bathymetric map was generated with the International Bathymetric Chart of the Arctic Ocean Version 3.0 (https://www.ngdc.noaa.gov/mgg/bathymetry/arctic). The sub-bottom profile (SBP) image was acquired using a SBP120 equipment (frequency; 12 kHz, wide-beam angle; 90–120°, Kongsberg). The global ocean map was generated with the Ocean Data View version 5.1.4 (GlobalHR, https://odv.awi.de).
Trace element compositions
Trace element data for carbonate (1 M AA leachates), sulfide (3 M HNO3 leachates), detrital silicate (HF-HCl digestion), and lipid fractions are reported in Tables S1–S4 in the Supplementary Information. Compared to JLs-1 (carbonate reference material; Triassic limestone), the carbonate fractions investigated in this study were generally characterized by much higher trace element abundances (Fig. 2 and Fig. S3), many of which (lithium (Li), transition metals, strontium (Sr), REE, lead (Pb), and thorium (Th)) were enriched up to a few hundred times. In contrast, three elements (titanium (Ti), barium (Ba), and W) showed relatively lower concentrations (up to tenfold depletion compared to JLs-1).
Enrichment factors of (a) carbonate fraction (1 M AA leachates) data normalized to values for the JLs-1 carbonate reference material, (b) total lipid fraction data normalized to values for the JLs-1 carbonate reference material, and (c) total lipid fraction data normalized to values for the corresponding carbonate fractions (1 M AA leachates).
Trace element data for the extracted sulfide-rich solid phases (i.e., 3 M HNO3 leachates) were normalized to the data for corresponding carbonate fractions, indicating that some elements (Ti, copper (Cu), zirconium (Zr), and Th) in the carbonate and sulfide fractions were enriched as much as 10 to 100 times (Fig. S3). Trace element abundances in the detrital silicate fractions were also normalized to the Post Archean Australian Shale (PAAS) reference values28. Some elements, such as Li, Rb (rubidium), Ba, W, Pb, Th, and U, were relatively enriched in the detrital silicate fractions relative to PAAS (Fig. S3). For the lipid fractions, several trace elements displayed a large range of abundance values (Li: 2.8–29.9 ppm, transition metals: 0.5–300 ppm, Rb: 2.2–4.1 ppm, Sr: 12.0–45.0 ppm, Ba: 13.0–22.0 ppm, Tables S1–S4). In contrast, REEs displayed very low abundances (< 1.34 ppm) in the lipid fractions. Notably, the lipid fraction data normalized to JLs-1 and the carbonate fractions of the CB carbonates clearly showed marked enrichment of Ti, Ni, Cu, Zn, and Pb (as much as 10–1000 times; Fig. 2, see also Fig. S4).
Lipid biomarker characteristics
Concentrations of archaeal and bacterial lipids showed large variations in the CB authigenic carbonates (Fig. 3, see also Table S5). Among isoprenoid hydrocarbons, the irregular tail-to-tail isoprenoids, such as crocetane, are often coeluted with phytane. Considering that the isotopic compositions of the crocetane/phytane peaks in CB carbonates were with − 28.5 to − 25.4‰ (Table S5), i.e., not 13C-depleted, it appears that crocetane was nearly absent in our samples, but phytane derived from the degradation of chlorophyll and other pigments was present with the values of 0.01–0.05 μg g− 1 dry weight (dw). The C25 compound 2,6,10,15,19-pentamethylicosane (PMI) was detected in the range of 0.02–0.04 μg g− 1 dw. Moreover, isoprenoid dialkyl glycerol diethers (isoprenoid DGDs), such as archaeol and sn-2-hydroxyarchaeol, were the most predominant archaeal lipids in the samples, displaying concentration ranges of 0.02–0.18 μg g− 1 dw and 0.02–0.32 μg g− 1 dw, respectively. For bacterial lipids, non-isoprenoid DGDs (i.e., If, IIa, and IId) with anteiso pentadecyl moieties or cyclopropyl groups attached at both the sn-1 and sn-2 positions were identified, ranging between 0.01 and 0.11 μg g− 1 dw. With respect to other DGDs (series I, II, and III), we could not fully determine their presence due to the detection limit. Among fatty acids (FAs) detected, saturated FAs (e.g., C16:0 and C18:0) were the most predominant, ranging between 0.04 and 0.62 μg g− 1 dw (Table S5). Other FAs (e.g., i-C15:0, ai-C15:0, C16:1ω7, C18:1ω9, C18:1ω7, C20:0, and C22:0) were approximately 2 to 10 times lower in concentration (0.01–0.23 μg g− 1 dw) compared with the predominant saturated FAs (Table S5). The δ13C values of PMI ranged from − 88.2 to − 61.5‰ (Fig. 3, see also Table S5). The δ13C values of archaeol and sn-2-hydroxyarchaeol varied between − 102.9 and − 82.9‰ and between − 108.0 and − 87.5‰, respectively (Fig. 3, see also Table S5). Finally, the δ13C values of non-isoprenoid DGDs ranged from − 88.6 to − 50.6‰ (Fig. 3, see also Table S5).
Lipid biomarker signatures of authigenic carbonates from the CB: (a) concentrations of major lipid biomarkers and ratios of sn-2-hydroxyarchaeol relative to archaeol, and (b) carbon isotopic compositions of major lipid biomarkers. Note that the dashed line indicates the average value of δ13C of methane (δ13CCH4) while the shaded area shows the full range of δ13CCH4 values.
Microbial compositions
The archaeal phyla detected in the studied samples were Euryarchaeota, Lokiarchaeota, Bathyarchaeota, and Thaumarchaeota (Table S6), with Euryarchaeota having the highest abundance, accounting for > 85% of the total archaeal sequences. Among them, the relative abundance of archaea ASV032, which belongs to ANME-1, was dominant (up to 99.5%; Fig. 4). Another archaea, ASV095, which belongs to Methanosarcinales, comprised 0.5% to 64.4% of archaeal sequences. In the bacterial communities, Firmicutes, Proteobacteria, Atribacteria, Actinobacteria, Bacteroidetes, Planctomycetes, and WM88 were predominant (Table S6).
Discussion
Constraints on fluid sources and environmental conditions during carbonate precipitation
Magnesium (Mg) calcite was the most dominant mineral (23–46 wt. %) in all carbonate samples retrieved from the CB mounds (Table 1). Generally, the environmental conditions during which carbonate precipitation occurs at cold seeps can be inferred from the mineralogical composition of carbonates23,29. For instance, aragonite precipitation is believed to be favored at sub-oxic conditions characterized by high dissolved sulfate concentrations in pore waters (i.e., near the seafloor), while high-Mg calcite and dolomite form preferentially under more reducing conditions and low sulfate levels30. Mg-bearing carbonate minerals such as dolomite are often encountered in relatively deeply buried sulfate-depleted sediment layers31. By analogy, the Mg-calcite carbonates encountered at the CB mounds most likely indicate precipitation under reducing conditions within the sediment column32. Moreover, considering that dissolved sulfide can catalyze dehydration of Mg2+ ions in fluids, enabling more rapid precipitation of Mg-calcites as opposed to aragonite33, the CB mounds may be subjected to anoxic conditions over long periods of time, since sulfide-enriched microenvironments could have been shaped by AOM processes under anoxic conditions. Furthermore, preferential precipitation of calcite over aragonite may occur at relatively low temperatures34, given that the temperature in the bottom seawaters of the Chukchi Sea is typically below 1 °C21. Thus, these properties could potentially explain the predominance of calcite over aragonite in the carbonate samples.
The measured δ13Ccarb values (− 37.0 to − 32.8‰; Table 1 and Fig. S2) were significantly lower than the typical values (~ 0‰) of marine bicarbonate (i.e., dissolved inorganic carbon in seawater)35. In general, the δ13Ccarb values reflect the source of dissolved carbon incorporated during carbonate precipitation36. The potential contributions of these different carbon isotopic signatures include (1) methane (δ13CCH4 for biogenic origin; < -60‰, and δ13CCH4 for thermogenic origin; − 50 to − 30‰) and (2) organic matter (ca. − 25‰)37,38. Furthermore, the extent of 13C-depletion in carbonates can be an indicator of specific microbial processes39,40. Considering that the carbon isotopic composition of ascending methane (δ13CCH4; − 95.7 to − 44.1‰) investigated at the CB mounds reflects a mixture of biogenic and thermogenic sources22, the microbial oxidation using this ascending methane can produce 13C-depleted bicarbonate (HCO3−) in sediments. We could not determine the precise age of carbonate samples using the U-Th method, due to the large amounts of detrital Th in our samples. Nonetheless, considering the sampling depths of our carbonate samples (see Fig. S1), they might be weakly related to the current sulfate-methane transition zone (SMTZ). Thus, the isotopic depletion of all carbonates (δ13Ccarb < − 30‰) at the CB mounds suggests that a substantial fraction of the carbon involved in carbonate precipitation was derived from the ascending methane-rich fluids, likely related to ancient methane releases similar to those that occurred in the Barents Sea41. Pore fluids become 18O-enriched (δ18Ofluid) during clay mineral dehydration or gas hydrate dissociation, which results in high δ18Ocarb values30,42,43. At the studied site, gas hydrate layers have been identified at sediment depths below 3–4 m22. In this regard, the calcite-dominated carbonates (δ18Ocarb; 5.1 ± 0.6‰ VPDB (n = 7), Table 1 and Fig. S2) investigated at the CB mounds display 18O-enriched isotopic signatures. Therefore, these results may indicate that they precipitated from similarly enriched pore waters, resulting from gas hydrate dissociation.
Trace element enrichments in authigenic carbonates
Compared to marine bioskeletal carbonates (i.e., JLs-1 limestone), most trace elements in the carbonate fractions are enriched in the studied cold seep carbonates, except for Ti, Ba, and W (Fig. 2a). These enrichments are well explained as reflecting the geochemical composition of surrounding pore waters, inherited from various early diagenetic processes that typically result in selective trace element enrichment44. In cold seeps, authigenic carbonates dominated by calcite typically incorporate higher amounts of trace elements (including REEs) compared to aragonite-rich samples, which form near the seafloor27. Moreover, sustained anoxic conditions in cold seeps generally explain why many redox-sensitive trace elements (Mo, U, Ni, V, Co, and Zn) are enriched in corresponding authigenic carbonates, such as in the CB carbonate samples45. Enrichment of Mo typically relates to Fe–Mn oxyhydroxide cycling in subsurface sediments and/or sulfide mineral formation, resulting from the presence of hydrogen sulfide in pore waters released from AOM46. Similarly, the observed enrichments identified for Ni, Co, and Zn reflect the remineralization of organic matter in sub-surface sediments, which also results in enriched sedimentary pore waters47,48.
The total lipid extracts of CB carbonates displayed particular trace element enrichments (Li, Ti, Ni, Co, Cu, Zn, and Mo) compared to the corresponding carbonate fractions (Fig. 2c). In contrast to the enrichments observed in leached carbonate fractions, which are directly inherited from the chemical composition of the pore waters from which they have precipitated, the enrichment associated with the lipid fractions hosted by authigenic carbonates can provide information on the bio-essential elements related to AOM-driven microbial activity at seeps20. Thus far, Ni and Zn have been identified in microbial organism cells, especially in seep-related microbial assemblages, where they are bound to specific sites of proteins and enzymes49,50. Co is present in cobamides involved in methyl group transfer during methanogenesis and methanotrophic processes17. Mo is bound to a pterin cofactor to form molybdopterin, which catalyzes electron redox reactions50, while Zn occurs as a single structural atom in both enzymes50 (e.g., methanol-coenzyme M methyltransferase enzyme (Mta) and Heterodisulfide reductase (Hdr)). Considering that microbial metalloenzymes play a key role in catalyzing major biogeochemical reactions, in particular AOM51, these enzymes may require some trace elements (e.g., Co, Ni, Mo, and Zn) as co-factors for electron transport or as catalytic centers at active sites50. Although trace elements (especially Cu and Ni) may be enriched in the organic (i.e., lipid) fractions due to their chemicophysical affinities to organic matter45, trace elements, such as Co, Ni, Mo, and Zn, were enriched, not only in the lipid fractions (Fig. 2b) but also in the carbonate fractions (Fig. 2a). Hence, the enrichment of Co, Ni, Mo, and Zn identified in the total lipid fractions associated with CB carbonates is most likely to reflect their implication in specific metal-rich enzymatic pathways related to AOM-driven methanotrophic activity. Fe is primarily present as Fe-S clusters used for electron transport and/or catalysis, while Ni is either bound to Fe-S clusters or in the center of a porphyrin unique to methanogen/methanotrophs containing cofactor F43052. Thus, their enrichment in the total lipid extracts separated from CB carbonates could similarly reflect their potential role as a limiting factor for methanotrophic activities because of their high bioavailability as essential metalloenzymes during AOM53.
Concerning Cu, which was also significantly enriched in the analyzed lipid fractions, it is worth mentioning that Cu-dependent anaerobic methanotrophic enzymes have not yet been identified, although Cu availability is important for aerobic methanotrophs54. For instance, an increase in Cu concentration can cause up to a 55-fold expression of particulate methane monooxygenase (pMMO) as a membrane protein found in aerobic methanotrophs50. Similar enrichments have been recently documented in cold seeps for light rare earth elements (e.g., La, Ce), also reflecting their preferential utility in aerobic methanotroph activity55. In the CB carbonates, the total lipid fractions extracted did not show any particular enrichment in light REE, indicating a decoupling between Cu and light REE. Given that Mg-calcite carbonates were predominantly precipitated under anoxic conditions in the CB mounds, the observed Cu enrichments in the CB carbonates would not be associated with the aerobic methanotrophy. Hence, additional studies are required to better assess the potential utility of Cu in AOM treatment. Similarly, it still needs to be elucidated why Li and Ti were enriched in the studied lipid fractions in the CB carbonates, and whether the potential bioavailability of Li and Ti as metalloenzymes could represent a limiting factor in methanogenesis and methanotrophic activity under anoxic conditions. Notably, these elements have previously been identified in microorganism cells, especially in seep-related microbial assemblages, where they were known to be bound to specific sites of proteins and enzymes49,50. Thus, to some degree, we speculate that the observed Cu, Li, and Ti enrichments in the total lipids of CB carbonates might be indicative of alternative microbial processes for AOM as metalloenzymes. Hence, in many physiological and biogeochemical aspects, further research is required to investigate the potential availability of Cu, Li, and Ti in anaerobic methanogens/methanotrophs in pure cultures.
Microbial signatures in authigenic carbonates
In this study, irregular hydrocarbons (e.g., PMI) and isoprenoid DGDs (e.g., archaeol and sn-2-hydroxyarchaeol) were identified as AOM-related lipids, showing the predominance of isoprenoid DGDs in most CB carbonate samples (Fig. 3). The δ13C composition of these lipids indicated relatively 13C-depleted signatures, compared to those reported for methane in the CB mounds (Fig. 3). In general, methane-derived microbial lipids are depleted in 13C in comparison to the methane source as a result of isotopic fractionation during methane assimilation56. The more 13C-depleted isoprenoid DGDs (i.e., archaeol and sn-2-hydroxyarchaeol) may indicate the occurrence of methanotrophs involved in AOM. Notably, CB-6 showed the most predominant 13C-depleted isoprenoid DGDs. This 13C-depleted pattern could be closely linked to active AOM taking place in sediment layers where the SMTZ currently occurs22 (90–120 cm sediment depths). On the other hand, because the depth of the SMTZ in methane seeps can fluctuate as a result of the upward methane fluxes57,58, low amounts of 13C-depleted lipids investigated in other CB carbonate samples could alternatively be explained as a result of the diffusive migration of methane-rich fluids59. In this context, the low amount of 13C-depleted microbial lipids in the CB carbonates below and above the present SMTZ may potentially reflect the weak AOM activities as a fossil signature during the change in methane fluxes60. Non-isoprenoid DGDs (If, IIa, and IId) derived from thermophilic and halophilic SRB61,62 were also detected in the studied CB carbonates (Fig. 3). Overall, the abundance of bacterial lipids in the CB carbonates was lower than that of AOM-related archaeal lipids. Considering previous studies in other cold seepages, such as the North Sea63 and the Eastern Mediterranean Sea64, the observed pattern between both lipid groups appear to reflect the process of AOM-coupled sulfate reduction. Furthermore, these bacterial lipids are more enriched in 13C compared to archaeal lipids (∆13C = 10–15‰), which reflects their involvement in AOM as syntrophic partners7,65. Based on these isotopic fractionations, the 13C-depleted non-isoprenoid DGDs in the CB carbonates (CB-1, -2, -6, and -7) may serve as evidence of syntrophic organisms involved in AOM. However, the δ13C values of the non-isoprenoid DGDs (− 53.9 ± 3.6‰, n = 5) in other CB carbonates (CB-3, -4, and -5) were slightly enriched in 13C relative to the ascending methane reported in the CB mounds. Therefore, these 13C-enriched lipids may indicate that these compounds originate from a mixed source mediating AOM as well as other processes. Our results should be further confirmed by determining quantitative and qualitative properties of source organisms obtained from different analytical approaches, such as potassium hydroxide (KOH) hydrolysis.
Based on the microbial lipids and nucleic acids, the chemotaxonomy of key microbes involved in AOM can be determined66. Three groups of ANME (ANME-1, ANME-2, and ANME-3) have often been reported in various cold seep environments, which are related to methanogens from the order Methanosarcinales and Methanomicrobiales58,67. The microbial communities dominated by ANME-1 biosynthesize archaeol and PMI with relatively low amounts of pentamethylicosenes (PMEs), whereas ANME-2 is characterized by the predominance of sn-2-hydroxyarchaeol (relative to archaeol) and abundant PMEs and crocetane/crocetenes66. Together with the presence of PMI in CB carbonates, the ratio of sn-2-hydroxyarchaeol to archaeol varied within the range 0.9–1.8 (Fig. 3), suggesting the dominant occurrence of ANME-1, which lived under anaerobically sulfate-depleted conditions68. These distinctive lipid patterns in most CB carbonates were also supported by higher proportions of ASV032, which belongs to ANME-1 (Fig. 4). Furthermore, the co-occurrence of syntrophic bacterial partners associated with ANME-1 can be indirectly inferred from the abundance of bacterial lipids, such as non-isoprenoid DGDs and fatty acids66. To date, 13C-depleted non-isoprenoid DGDs have been mainly found in prominent ANME-1 systems66. Thus, the less 13C-depleted non-isoprenoid DGDs in the CB carbonates appear to be associated with an unknown SRB linked to AOM69. Furthermore, the low abundance of branched fatty acids (i.e., i-C15:0 and ai-C15:0) in the CB carbonates may also indicate a negligible presence of typical SRBs (i.e., Desulfosarcina/Desulfococcus) associated with ANME-1. This inference may be partially supported by the incomplete sequence of Deltaproteobacteria. Hence, these lipid characteristics in CB carbonates, in combination with 16S rRNA gene pyrosequencing data, suggest that SRBs, not belonging to Deltaproteobacteria but possibly to other bacterial communities, would have been involved in AOM in the CB mounds. Notably, ANME-1 has been microscopically shown to occur as a single cell in cold seep environments70. Accordingly, ANME-1-related syntrophic relationships within the CB mounds appear to be more linked to other bacterial communities than typical SRB consortia.
Potential metalloenzymes involved in methanotrophic activity
To further investigate the distribution patterns of trace elements (W, Li, Ti, Ni, Co, Cu, Zn, and Mo) presumably acting as potential metalloenzymes involved in AOM at cold seeps, principal component analysis (PCA) was performed using the data for total lipid extracts normalized to JLs-1 (Fig. 5a). We used these data to compare authigenic carbonate samples collected from different regions worldwide instead of those normalized to associated carbonate fractions (Fig. S5). Although W was not detected in our total lipid samples of the CB, this element is included because pronounced lipid-bound W anomalies are reported at other seep sites worldwide20. PCA was also performed using the archaeal and bacterial lipid data to compare the CB signatures with those from other cold seeps20 (Fig. 5b–c). The PCA results show that the combined distribution of trace elements and bacterial lipids in CB carbonates differs from those reported for other methane seeps worldwide (e.g., Congo fan; CF, Nile deep-sea fan; NDSF, Niger fan; NF, and Gulf of Mexico; GoM). This finding suggests that the enrichment of Ni, Co, Zn, Cu, Ti, and Mo in carbonate-hosted lipid fractions could potentially be related to different methanotrophic communities and their metalloenzymes. This hypothesis is plausible because ANME-1 is predominant in CB carbonates, while ANME-1 and Methanosarcinales (potential ANME-2) co-exist at other sites20 (Figs. 6, 7). Ni-containing methyl-coenzyme MCR is the key enzyme in AOM19. Two different MCR homologues, designated as proteins I and II71,72 have been isolated from cold seep areas: protein I contains (172S)-172-methylthio-F430, whereas protein II contains F43018,73. Considering that protein I is predominantly detected in ANME-174, we may infer that particular environmental conditions (e.g., temperature and partial pressure of methane) in the CB mound area specifically promote the synthesis of protein I for AOM75 compared to what can be observed at other seep sites. Future studies targeting deep gas-fluid sources and subsurface lithology are needed to unravel the crucial factors for determining discriminative ANME activities involved in the stimulated response under permafrost dissociation. The impact of lipid/enzyme degradation on the correlations between AOM-related lipids and trace elements should also be tested by comparing their relationships using data from modern and ancient carbonate samples. Previous studies have shown that there is a significant trace element enrichment difference between intra- and inter-crystalline lipid extractions in seep carbonates48. Hence, future studies should also explore the effect of the two-step extraction procedure on trace element distributions.
Comparison of lipid biomarker signatures of authigenic carbonates from the CB, Congo fan (CF), Nile deep-sea fan (NDSF), Niger fan (NF), and Gulf of Mexico (GoM): (a) concentrations of major lipid biomarkers and ratios of sn-2-hydroxyarchaeol relative to archaeol, and (b) carbon isotopic compositions of major lipid biomarkers with those of carbonates.
Conclusions
We analyzed a series of authigenic carbonate samples collected at methane seeps from the southwestern slope of the CB using various inorganic and organic proxies of fluid sources and biogeochemical processes (stable isotopes, mineralogy, trace elements, lipid biomarkers, and nucleic acids). The absence of crocetane, the abundance of PMI, and the relatively low ratio of sn-2-hydroxyarchaeol to archaeol in the CB carbonates indicated the dominant occurrence of ANME-1, as confirmed by 16S rRNA gene pyrosequencing. Specific trace elements (Ni, Co, Zn, Cu, Ti, and Mo) appeared to be enriched in the total lipid fractions extracted from CB carbonates, confirming recent findings of their utility as metalloenzymes in microbial processes related to AOM at cold seeps. The patterns of trace element enrichment and microbial lipid assemblages in CB carbonates reveal distinct specific characteristics compared to those from other seeps worldwide (e.g., Congo fan, Nile deep-sea fan, Niger fan, and Gulf of Mexico). This finding suggests that trace element enrichment in carbonate-hosted lipid fractions at cold seeps could vary depending on the type of AOM microbial assemblages among the ANME groups. Further work is required to elucidate the role of trace elements, such as Co, Zn, Cu, Ti, and Mo, as potential metalloenzymes in AOM during N2 fixation or aerobic methane oxidation coupled with denitrification. In this regard, pure-culture experiments are necessary to understand their bioavailability for metalloenzymes involved in AOM.
Material and methods
Sampling
Previous geophysical surveys (e.g., multibeam and sub-bottom profiling) conducted at the southwestern slope of the CB21 led to the discovery of gas hydrate mound structures characterized by acoustic blanking below the crest at certain depth intervals (Fig. 1). During the ARA09C expedition in 2018, sediment cores (ARA09C-ST07, -ST14, and -ST04/06) were retrieved from three mound structures, together with various authigenic carbonate samples recovered at different sediment depths between 38 and 415 cm (Fig. 1). The recovered carbonate samples were cleaned with ultrapure Milli-Q (MQ) water and air-dried prior to being crushed into powder using an agate and mortar.
Mineral and stable isotope analysis
The bulk authigenic carbonate samples were ground to a fine powder (< 20 µm particle size) for mineral analysis using a Philips X-ray diffraction (XRD) system at the laboratories of the Crystallography research group (Central Laboratory for Crystallography and Applied Material Sciences, University of Bremen). The X-ray source was a Cu anode operated at 40 kV and 45 mA using CuKα radiation equipped with a diffracted beam graphite monochromator. Step scans were run from 5 to 65° (2θ) with a step size of 0.02° and a count time of 2 s per step (0.6°/min). Mineral identification was performed using the Philips X’ Pert HighScore software, which provides a semi-quantitative value for each identified mineral on the basis of relative intensity ratio values. The oxygen and carbon isotopic compositions (δ18O and δ13C) of the carbonate samples were measured at Beta Analytic (service sample number: 536510 to 536,516). In brief, the sample was placed into a clean vial and dissolved in H3PO4 to release CO2 into the headspace of the vial. The sample was placed in a temperature-controlled tray (72 °C), where CO2 equilibration took place for one hour. Finally, helium (He) was added to the sample and a mixture of He and CO2 was injected into a Finnegan MAT 251 mass spectrometer. All isotopic values were expressed using the δ-notation relative to the Vienna Pee Dee Belemnite (VPDB) standard and were reported in per mil (‰) with a standard deviation of less than 0.3‰ for both δ18O and δ13C values.
Trace element analysis
Homogenized carbonate samples (~ 5 g) were treated using the sequential extraction procedure previously described20 to selectively separate carbonate, sulfide, and detrital silicate fractions. We used the lipid fractions obtained from the solvent extraction method using a mixture of organic solvents (dichloromethane (DCM):methanol (MeOH), 2:1 v/v) to compare trace elements in lipid fractions with lipid biomarker data. The total lipid extracts (TLEs) were split into two aliquots for trace element analysis at IFREMER (Institut Français de Recherche pour l'Exploitation de la Mer; i.e., “French Research Institute for Exploitation of the Sea”) and lipid biomarker analysis at the Korea Polar Research Institute (KOPRI). All analyses were performed using an Element XR™ inductively coupled plasma-mass spectrometer (ICP-MS) at the Pôle Spectrométrie Océan (Brest, France). Polyatomic oxide and hydroxide interferences on the rare earth element (REE) were corrected using oxide formation rates determined by analyzing solutions of MQ–H2O, Ba + Ce, Pr + Nd, and Sm + Tb at the beginning of each measurement session and applied to all samples. Elemental concentrations were calculated using the Tm addition method76,77. The internal precision of all measurements was generally better than 5%. Repeated analyses of the JLs-1 reference material (Triassic limestone) were also performed, with a precision of < 10% for most trace elements, except for Li (11.8% relative standard deviation; RSD), Ti (61% RSD), and Zr (17% RSD). Note that the relatively poor RSD determined for Ti mostly reflects the very low abundance of this element in JLs-1 (4.1 µg/g). Significant Ti enrichments were measured in the lipid fractions, up to three orders of magnitude (Supplementary Tables); hence, much higher than the RSD determined for JLs-1. Therefore, we are confident that the measured Ti enrichments in the studied lipid extracts are meaningful, and thus, Ti data are still reported in this study. Because of the high Ba/REE ratios, several carbonate samples analyzed (including the carbonate reference material JLs-1) displayed anomalously high Eu (and to a lesser extent Gd) concentrations as a result of under-corrected interferences; hence, these two elements were not reported here.
Lipid biomarker analysis
Detailed procedures for lipid biomarker analyses have been previously described60. In short, one-half of the TLE was dried over anhydrous Na2SO4 and treated with tetrabutylammonium sulfite reagent to remove elemental sulfur. The TLE was chromatographically separated into apolar and polar fractions over an Al2O3 column (activated for 2 h at 150 °C). The apolar fraction was eluted using hexane:DCM (9:1), and 40 µL of 5α-androstane (10 µg mL− 1) was added as an internal standard. The polar fraction was recovered with DCM:MeOH (1:1 v/v) as an eluent and divided into two aliquots, to which either C22 7,16-diol (10 µg mL− 1) or C19 nonadecanoic acid (10 µg mL− 1) was added as an internal standard. Each aliquot was derivatized through silylation and methylation, prior to quantification by gas chromatography (GC) and identification by gas chromatography-mass spectrometry (GC–MS). GC and GC–MS conditions were as previously described60. Molecular compounds were determined by comparing their mass spectral fragmentation patterns and retention times with previously published data60. The δ13C values of lipid compounds were determined using an isotope ratio mass spectrometer (IRMS) connected to a GC via a combustion interface (glass tube packed with copper oxide, CuO, operated at 850 °C), as previously described60. Isotopic values were expressed as δ13C values per mil relative to VPDB. The δ13C values were corrected for the introduction of additional carbon atoms during silylation and methylation. The analytical errors were less than ± 0.4‰ for all lipid compounds.
Nucleic acid analysis
Genomic DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, USA). DNA samples were subjected to polymerase chain reaction (PCR) amplification, library preparation, and paired-end Illumina MiSeq sequencing (2 × 300 bp) at the Integrated Microbiome Resource (IMR), Dalhousie University, Canada (http://cgeb-imr.ca). The primer pairs 515F/ 926R and 956F/ 1401R targeting the V4–V5 and V6–V8 regions were used to amplify bacterial and archaeal 16S rRNA genes, respectively78,79. Amplicons were sequenced using the paired-end (2 × 300 bp) Illumina MiSeq system (Illumina, USA) at the Integrated Microbiome Resource (IMR), Dalhousie University, Canada (http://cgeb-imr.ca). Samples that were not amplified were excluded from further analyses. The adapter and primer sequences were removed using Cutadapt version 2.1080, and the resultant sequences were processed using DADA2 version 0.9.581 to infer amplicon sequence variants (ASVs). For the quality trimming process, we applied a filtering option of maxN = 0, maxEE = c (2,2), and truncQ = 2. The low-quality tails of both reads were removed with truncLen = c (270,210). Denoising was performed after trimming based on the DADA2 error model. Sequences were dereplicated, and the core sample inference algorithm was applied to the dereplicated data. Paired reads were merged, and the chimeric sequences were removed. The following processes were performed after constructing a sequence table of ASVs to assign taxonomy using the software mothur82. Taxonomic assignments of representative ASV sequences were determined against the EzBiocloud database using sequence similarity searches83. After taxonomic assignment, archaea and unknown ASVs were removed for bacterial analysis, and bacteria and unknown ASVs were removed for archaeal analysis. All sequence data used in this study were deposited in the Sequence Read Archive at the National Center for Biotechnology Information under accession number PRJNA825649. Phylogenetic trees of major archaeal ASVs of Methanomicrobia with relative abundances greater than 3% were constructed using the maximum-likelihood algorithm84 with the general time reversible evolutionary model using MEGA X85. The robustness of the tree topologies was assessed using bootstrap analyses based on 1,000 replications of the sequences.
Statistical analysis
To provide a general view of the variability of trace element abundances and microbial lipid distributions in the carbonate samples, gaps in the data set were first filled as previously described86. When some components were not determined, a value of one-half of the minimum value detected for that variable in the whole data set was set as the limit of detection. These values were then replaced by a random number between zero and the limit of detection. Finally, the fractional abundances of trace elements and microbial lipids were obtained by normalizing each concentration to the summed concentrations of all trace elements and microbial lipids, respectively. Based on the fractional abundances, PCA was performed using R software version 3.4.2 (package information; FactoMineR, Vienna, Austria).
Data availability
All DNA sequence data generated for this study were deposited in the Sequence Read Archive at the National Center for Biotechnology Information under accession number PRJNA825649.
References
Campbell, K. A. Hydrocarbon seep and hydrothermal vent paleoenvironments and paleontology: Past developments and future research directions. Palaeogeogr. Palaeoclimatol. Palaeoecol. 232, 362–407 (2006).
Suess, E. Marine cold seeps and their manifestations: Geological control, biogeochemical criteria and environmental conditions. Int. J. Earth Sci. 103, 1889–1916 (2014).
Judd, A. & Hovland, M. Seabed Fluid Flow (Cambridge University Press, 2007).
Torres, M. E. et al. Fluid and chemical flux in and out of sediments hosting methane hydrate deposits on Hydrate Ridge, OR, I: Hydrological processes. Earth Planet. Sci. Lett. 201, 525–540 (2002).
Gründger, F. et al. Methane-fuelled biofilms predominantly composed of methanotrophic ANME-1 in Arctic gas hydrate-related sediments. Sci. Rep. 9, 9725 (2019).
Solomon, E. A., Kastner, M., Jannasch, H., Robertson, G. & Weinstein, Y. Dynamic fluid flow and chemical fluxes associated with a seafloor gas hydrate deposit on the northern gulf of mexico slope. Earth Planet. Sci. Lett. 270, 95–105 (2008).
Hinrichs, K. U. & Boetius, A. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry. in Ocean Margin Systems 457–477 (Berlin, Germany: Springer-Verlag, 2002).
Reeburgh, W. Oceanic methane biogeochemistry. Am. Chem. Soc. 107, 486–513 (2007).
Ussler, W. & Paull, C. K. Rates of anaerobic oxidation of methane and authigenic carbonate mineralization in methane-rich deep-sea sediments inferred from models and geochemical profiles. Earth Planet. Sci. Lett. 266, 271–287 (2008).
Feng, D. & Chen, D. Authigenic carbonates from methane seeps of the northern Congo fan : Microbial formation mechanism. Mar. Pet. Geol. 27, 748–756 (2010).
Mazzini, A., Ivanov, M. K., Parnell, J., Stadnitskaia, A. & Cronin, B. T. Methane-related authigenic carbonates from the black Sea: Geochemical characterisation and relation to seeping fluids. Mar. Geol. 212, 153–181 (2004).
Bayon, G. et al. Multi-disciplinary investigation of fluid seepage on an unstable margin: The case of the Central Nile deep sea fan. Mar. Geol. 261, 92–104 (2009).
Yao, H., Panieri, G., Lehmann, M. F., Himmler, T. & Niemann, H. Biomarker and isotopic composition of seep carbonates record environmental conditions in two arctic methane seeps. Front. Earth Sci. 8, 1–12 (2021).
Boetius, A. et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626 (2000).
Hinrichs, K. U., Hayes, J. M., Sylva, S. P., Brewer, P. G. & DeLong, E. F. Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805 (1999).
Beal, E. J., House, C. H. & Orphan, V. J. Manganese- and iron-dependent marine methane oxidation. Science(80-) 325, 184–187 (2009).
Glass, J. B. et al. Geochemical, metagenomic and metaproteomic insights into trace metal utilization by methane-oxidizing microbial consortia in sulphidic marine sediments. Environ. Microbiol. 16, 1592–1611 (2014).
Krüger, M. et al. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426, 878–881 (2003).
Scheller, S., Goenrich, M., Boecher, R., Thauer, R. K. & Jaun, B. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465, 606–608 (2010).
Wang, X. et al. Trace element systematics in cold seep carbonates and associated lipid compounds. Chem. Geol. 528, 119277 (2019).
Kim, Y. G. et al. Occurrence of active gas hydrate mounds in the southwestern slope of the Chukchi plateau. Arctic Ocean. Epis. 43, 811–823 (2020).
Kim, J. H. et al. Upwarding gas source and postgenetic processes in the shallow sediments from the ARAON mounds, Chukchi sea. J. Nat. Gas Sci. Eng. 76, 103223 (2020).
Feng, D. & Roberts, H. H. Geochemical characteristics of the barite deposits at cold seeps from the northern Gulf of Mexico continental slope. Earth Planet. Sci. Lett. 309, 89–99 (2011).
Gontharet, S. et al. Nature and origin of diagenetic carbonate crusts and concretions from mud volcanoes and pockmarks of the nile deep-sea fan (eastern Mediterranean Sea). Deep. Res. Part II Top. Stud. Oceanogr. 54, 1292–1311 (2007).
Pierre, C. & Fouquet, Y. Authigenic carbonates from methane seeps of the Congo deep-sea fan. Geo-Marine Lett. 27, 249–257 (2007).
Roberts, H. H., Feng, D. & Joye, S. B. Cold-seep carbonates of the middle and lower continental slope, northern Gulf of Mexico. Deep. Res. Part II Top. Stud. Oceanogr. 57, 2040–2054 (2010).
Rongemaille, E. et al. Rare earth elements in cold seep carbonates from the Niger delta. Chem. Geol. 286, 196–206 (2011).
Taylor, S. R. & Mclennan, S. M. The Continental Crust: Its Composition and Evolution. An Examination of the Geochemical Record Preserved in Sedimentary Rocks. (Blackwell Scientific Publications, 1985).
Peckmann, J., Goedert, J. L., Thiel, V., Michaelis, W. & Reitner, J. A comprehensive approach to the study of methane-seep deposits from the lincoln creek formation, western Washington State, USA. Sedimentology 49, 855–873 (2002).
Aloisi, G. et al. CH4-consuming microorganisms and the formation of carbonate crusts at cold seeps. Earth Planet. Sci. Lett. 203, 195–203 (2002).
Campbell, K. A., Farmer, J. D. & Des Marais, D. Ancient hydrocarbon seeps from the Mesozoic convergent margin of California: Carbonate geochemistry, fluids and paleoenvironments. Geofluids 2, 63–94 (2002).
Baker, P. A. & Kastner, M. Constraints on the formation of sedimentary dolomite. Science(80-) 213, 214–216 (1981).
Smrzka, D. et al. Trace element distribution in methane-seep carbonates: The role of mineralogy and dissolved sulfide. Chem. Geol. 580, 120357 (2021).
Burton, E. A. & Walter, L. M. Relative precipitation rates of aragonite and Mg calcite from seawater: Temperature or carbonate ion control?. Geology 15, 111–114 (1987).
Aharon, P., Schwarcz, H. P. & Roberts, H. H. Radiometric dating ofsubmarine hydrocarbon seeps in the Gulf of Mexico. Geol. Soc. Am. Bull. 109, 568–579 (1997).
Pierre, C. & Rouchy, J. M. Carbonate replacements after sulfate evaporites in the middle Miocene of Egypt. J. Sediment. Res. 58, 446–456 (1988).
Killops, S. D. & Killops, V. J. Introduction to Organic Geochemistry (Blackwell publishing, 2005).
Whiticar, M. J. Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem. Geol. 161, 291–314 (1999).
Chen, Z. et al. Discovery of seep carbonate nodules as new evidence for gas venting on the northern continental slope of South China Sea. Chinese Sci. Bull. 51, 1228–1237 (2006).
Ritger, S., Carson, B. & Suess, E. Methane-derived authigenic carbonates formed by subduction-induced pore-water expulsion along the Oregon/Washington margin: Reply. Bull. Geol. Soc. Am. 98, 147–156 (1987).
Andreassen, K. et al. Massive blow-out craters formed by hydrate-controlled methane expulsion from the Arctic seafloor. Science(80-) 356, 948–953 (2017).
Feng, D. et al. Time integrated variation of sources of fluids and seepage dynamics archived in authigenic carbonates from Gulf of Mexico Gas Hydrate Sea floor observatory. Chem. Geol. 385, 129–139 (2014).
Naehr, T. H. et al. Authigenic carbonate formation at hydrocarbon seeps in continental margin sediments: A comparative study. Deep. Res. Part II Top. Stud. Oceanogr. 54, 1268–1291 (2007).
Haley, B. A., Klinkhammer, G. P. & McManus, J. Rare earth elements in pore waters of marine sediments. Geochim. Cosmochim. Acta 68, 1265–1279 (2004).
Smrzka, D. et al. The behavior of trace elements in seawater, sedimentary pore water, and their incorporation into carbonate minerals: a review. Facies vol. 65 (Springer Berlin Heidelberg, 2019).
Peketi, A. et al. Tracing the Paleo sulfate-methane transition zones and H2S seepage events in marine sediments: An application of C-S-Mo systematics. Geochem. Geophys. Geosystems 13, 1–11 (2012).
Algeo, T. J. & Maynard, J. B. Trace-element behavior and redox facies in core shales of upper pennsylvanian kansas-type cyclothems. Chem. Geol. 206, 289–318 (2004).
Smrzka, D. et al. Trace elements in methane-seep carbonates: Potentials, limitations, and perspectives. Earth-Sci. Rev. 208, 103263 (2020).
Barton, L. L. et al. The bacterial metallome: Composition and stability with specific reference to the anaerobic bacterium desulfovibrio desulfuricans. Biometals 20, 291–302 (2007).
Glass, J. B. & Orphan, V. J. Trace metal requirements for microbial enzymes involved in the production and consumption of methane and nitrous oxide. Front. Microbiol. 3, 1–20 (2012).
Zerkle, A. L., House, C. H. & Brantley, S. L. Biogeochemical signatures through time as inferred from whole microbial genomes. Am. J. Sci. 305, 467–502 (2005).
Ferry, J. G. Methanogenesis: Ecology, physiology, biochemistry & genetics (Springer Science & Business Media, 2012).
Glass, J. B. et al. Trace metal imaging of sulfate-reducing bacteria and methanogenic archaea at single-cell resolution by synchrotron X-Ray fluorescence imaging. Geomicrobiol. J. 35, 81–89 (2018).
Sushkevich, V. L., Palagin, D., Ranocchiari, M. & Van Bokhoven, J. A. Selective anaerobic oxidation of methane enables direct synthesis of methanol. Science(80-) 356, 523–527 (2017).
Bayon, G. et al. Microbial utilization of rare earth elements at cold seeps related to aerobic methane oxidation. Chem. Geol. 555, 119832 (2020).
Niemann, H. & Elvert, M. Diagnostic lipid biomarker and stable carbon isotope signatures of microbial communities mediating the anaerobic oxidation of methane with sulphate. Org. Geochem. 39, 1668–1677 (2008).
Lee, D.-H. et al. Chemosynthetic bacterial signatures in frenulata tubeworm (Oligobrachia sp.) in an active mud volcano of the Canadian Beaufort Sea. Mar. Ecol. Prog. Ser. 628, 95–104 (2019).
Niemann, H. et al. Novel microbial communities of the Håkon Mosby mud volcano and their role as a methane sink. Nature 443, 854–858 (2006).
Peckmann, J. & Thiel, V. Carbon cycling at ancient methane-seeps. Chem. Geol. 205, 443–467 (2004).
Lee, D.-H. et al. Biogeochemical evidence of anaerobic methane oxidation on active submarine mud volcanoes on the continental slope of the Canadian Beaufort Sea. Biogeosciences 15, 7419–7433 (2018).
Langworthy, T. A., Holzer, G., Zeikus, J. G. & Tornabene, T. G. Iso-and anteiso-branched glycerol diethers of the thermophilic anaerobe Thermodesulfotobacterium commune. Syst. Appl. Microbiol. 4, 1–17 (1983).
Sturt, H. F., Summons, R. E., Smith, K., Elvert, M. & Hinrichs, K.-U. Intact polar membrane lipids in prokaryotes and sediments deciphered by high-performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Commun. Mass Spectrom. 18, 617–628 (2004).
Niemann, H. et al. Methane emission and consumption at a north sea gas seep (Tommeliten area) methane emission and consumption at a North Sea gas seep (Tommeliten area). Biogeosciences 2, 1197–1241 (2005).
Gontharet, S., Stadnitskaia, A., Bouloubassi, I., Pierre, C. & Damsté, J. S. S. Palaeo methane-seepage history traced by biomarker patterns in a carbonate crust, Nile deep-sea fan (Eastern mediterranean Sea). Mar. Geol. 261, 105–113 (2009).
Wegener, G., Niemann, H., Elvert, M., Hinrichs, K. & Boetius, A. Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ. Microbiol. 10, 2287–2298 (2008).
Blumenberg, M., Seifert, R., Reitner, J., Pape, T. & Michaelis, W. Membrane lipid patterns typify distinct anaerobic methanotrophic consortia. Proc. Natl. Acad. Sci. 101, 11111–11116 (2004).
Knittel, K. & Boetius, A. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334 (2009).
Lazar, C. S., Dinasquet, J., L’Haridon, S., Pignet, P. & Toffin, L. Distribution of anaerobic methane-oxidizing and sulfate-reducing communities in the G11 Nyegga pockmark, Norwegian Sea. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 100, 639–653 (2011).
Pancost, R. D., Hopmans, E. C., Sinninghe Damsté, J. S., The M. S. Scientific Party. Archaeal lipids in mediterranean cold seeps: Molecular proxies for anaerobic methane oxidation. Geochim Cosmochim Acta 65, 1611–1627 (2001).
Orphan, V. J., House, C. H., Hinrichs, K.-U., McKeegan, K. D. & DeLong, E. F. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. 99, 7663–7668 (2002).
Nauhaus, K., Albrecht, M., Elvert, M., Boetius, A. & Widdel, F. In vitro cell growth of marine archaeal-bacterial consortia during anaerobic oxidation of methane with sulfate. Environ. Microbiol. 9, 187–196 (2007).
Thauer, R. K., Kaster, A. K., Seedorf, H., Buckel, W. & Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591 (2008).
Mayr, S. et al. Structure of an F430 variant from archaea associated with anaerobic oxidation of methane. J. Am. Chem. Soc. 130, 10758–10767 (2008).
Kaneko, M. et al. Quantitative analysis of coenzyme F430 in environmental samples: A new diagnostic tool for methanogenesis and anaerobic methane oxidation. Anal. Chem. 86, 3633–3638 (2014).
Knittel, K. et al. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71, 467–479 (2005).
Barrat, J. A. et al. Determination of rare earth elements in sixteen silicate reference samples by ICP-MS after TM addition and ion exchange separation. Geostand. Newsl. 20, 133–139 (1996).
Bayon, G. et al. Multi-disciplinary investigation of fl uid seepage on an unstable margin: The case of the central Nile deep sea fan. Mar. Geol. 261, 92–104 (2009).
Comeau, A. M., Li, W. K. W., Tremblay, J. É., Carmack, E. C. & Lovejoy, C. Arctic ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One 6(11), e27492 (2011).
Walters, W. et al. Improved bacterial 16s rrna gene (v4 and v4–5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. MSystems 1(1), e00009-e00015 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17(1), 10–12 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Schloss, P. D. et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Yoon, S. H. et al. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617 (2017).
Felsenstein, J. Evolutionary trees from DNA sequences : A maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Yunker, M. B., Belicka, L. L., Harvey, H. R. & Macdonald, R. W. Tracing the inputs and fate of marine and terrigenous organic matter in Arctic ocean sediments: A multivariate analysis of lipid biomarkers. Deep. Res. Part II Top. Stud. Oceanogr. 52, 3478–3508 (2005).
Acknowledgements
We would like to thank the captain and crew of the R/V ARAON for their help at sea. This study was supported by the Korea Institute of Marine Science and Technology Promotion (KIMST) (KIMST Grant 20210632; KOPRI-PM22050) and a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (NRF-2021M1A5A1075512; KOPRI-PN22013).
Author information
Authors and Affiliations
Contributions
D.H.L. and J.H.K. designed and coordinated the study. D.H.L., J.H.K., Y.M.L., Y.J.J., and Y.K.J. conducted the field work during the ARAON expedition. G.B. and X.W. conducted the trace element analysis. D.H.L. and D.K. carried out the lipid biomarker analysis. Y.M.L. conducted the nucleic acid analysis. D.H.L. and J.H.K. led the writing and interpretation of the data. All authors commented on the manuscript and contributed to the writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lee, DH., Kim, JH., Lee, Y.M. et al. Metalloenzyme signatures in authigenic carbonates from the Chukchi Borderlands in the western Arctic Ocean. Sci Rep 12, 16597 (2022). https://doi.org/10.1038/s41598-022-21184-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-21184-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.