Abstract
miR-30c functions as a tumor suppressor gene in the majority of tumors, including gliomas. In our study, we discovered that the expression levels of miR-30c in glioma tissues and plasma prior to surgery were lower than those in normal brain tissue following brain injury decompression and in plasma in healthy volunteers. The low expression of miR-30c was closely aligned with the WHO grade, tumor size, PFS, and OS. Additionally, the miR-30c expression level in tumor tissue was positively correlated with the levels in preoperative plasma. In cell biology experiments, miR-30c inhibited EMT and proliferation, migration, and invasion of glioma cells. Analysis of databases of miRNA target genes, real-time quantitative PCR, western blotting, and dual luciferase reporter assays demonstrated that Notch1 is the direct target gene of miR-30c. An inhibitor and shRNA-Notch1 were cotransfected into glioma cells, and it was found that shRNA-Notch1 reduced the enhancement of inhibitors of EMT and proliferation, migration, and invasion of glioma cells. Therefore, we believe that when utilized as a tumor suppressor gene, miR-30c can inhibit EMT and the proliferation, migration, and invasion of glioma cells by directly acting on Notch1 at the posttranscriptional level and that it is a potential diagnostic and prognostic marker.
Similar content being viewed by others
Gliomas are among the most common malignant tumors of the central nervous system that arise from glial cells or precursor cells. According to World Health Organization (WHO) classifications, gliomas are categorized into four grades. Grade 1 and grade 2 gliomas are considered low grade, and grade 3 and grade 4 gliomas are high grade. Grade 4 gliomas are labeled glioblastomas (GBMs). Despite active and effective treatment, the median survival time of GBM patients is less than 20 months1. Therefore, the treatment of gliomas poses a great challenge. To establish effective treatment methods, clarifying the molecular mechanisms associated with the occurrence and development of gliomas is paramount.
Epithelial mesenchymal transition (EMT) is defined as the biological process through which epithelial cells acquire a mesenchymal phenotype2. In the EMT process, epithelial cells lose cell polarity, decrease contact with surrounding cells and stromal cells, decrease interaction between cells, enhance cell migration and mobility, alter the cell phenotype, and lose the epithelial phenotype. Regarding the latter, one such example is a decrease in E-cadherin levels, which leads to decreased cell adhesion3 and causes cells to obtain the characteristics of invasion and metastasis4. Simultaneously, the number of cells that obtain mesenchymal phenotypes, including the presence of Vimentin and N-cadherin, is increased5,6.
The Notch signaling pathway is a common signaling pathway involved in EMT process control7,8. The Notch signaling pathway comprises the Notch receptor, Notch ligand, CSL-DNA binding protein, and Notch regulatory molecules. A signal transmitted between adjacent cells through the Notch receptor can regulate cell differentiation, proliferation, and apoptosis. The Notch receptor is a single transmembrane protein encoded by the Notch gene. Four kinds of Notch receptors (Notch 1, 2, 3, and 4) are located in mammals, including humans. Notch signaling is activated by the binding of Notch ligands to the corresponding receptors of adjacent cells. The receptors are subsequently hydrolyzed by tumor necrosis factor alpha converting enzyme (TACE) of the metalloproteinase family and release the N-terminal and C-terminal fragments. Then, the N-terminal fragment (extracellular region) is internalized into the ligand-expressing cells, and the C-terminal fragment (intracellular region) is secondarily hydrolyzed by γ-secretase, releasing the Notch intracellular domain (NICD). NICD finally enters the nucleus and binds with recombinant signal binding protein J κ (RBP-J κ) to activate the transcription of downstream target genes9. Therefore, the formation of NICD is a marker of Notch signaling pathway activation10. The target genes downstream of the Notch signaling pathway belong to the HES and HEY families, in which HES1 and HEY1 are mainly expressed on endothelial cells, and their expression level is often used as an important index for the activation of the Notch signaling pathway11.
In the process of malignant tumorigenesis, the Notch signaling pathway plays carcinogenic roles, leading to cell proliferation12, differentiation13, apoptosis14, and process imbalance, all of which contribute to the occurrence and development of malignant tumors. In gliomas, Notch1 is highly expressed15. In glioma cells, Notch1 expression is upregulated and plays a role in tumor progression. Notch1 may also function as an independent prognostic marker that is both separate from and supplementary to the WHO classifications16. Furthermore, Notch1 promotes EMT in a variety of tumors, including gliomas17,18,19.
miRNAs are 19–22-nucleotide-long noncoding RNAs that act as oncogenes or tumor suppressor genes by connecting to the 3′-UTR of a target gene and regulating the expression of related proteins at the posttranscriptional level. miRNAs function in regulating cell proliferation20, apoptosis21, invasion22, angiogenesis23, and the cell cycle24 and in turn participate in the occurrence and progression of an assortment of tumors. miR-30c plays different roles in different tumors25,26,27, but there is no comparable research into the role miR-30c in gliomas. In this study, the mechanism of miR-30c in gliomas is explored, and a theoretical basis for the diagnosis and treatment of gliomas is presented.
Results
The expression of miR-30c was decreased in tissues and plasma from patients with gliomas
The expression of miR-30c was determined using qRT‒PCR. As detailed in Fig. 1, the relative expression level of miR-30c in 66 glioma tissues was 3.077 ± 0.282, while the relative expression level of miR-30c in 9 patients with traumatic brain injury was 9.352 ± 0.190. The expression levels of miR-30c in glioma tissues were significantly lower than those in normal brain injury tissues (P < 0.01) (Fig. 1A). As detailed in Table 1, the expression level of miR-30c in glioma tissues was closely correlated with the WHO grade and tumor size (P < 0.01) but not with the sex, age, extent of resection, or KPS scores of the patients (P > 0.05).
The expression levels of miR-30c in human glioma tissue, preoperative plasma, and paraffin specimens. (A) The expression levels of miR-30c in glioma tissues were significantly lower than the levels in normal brain injury tissue (**P < 0.01). (B) The expression levels of miR-30c in the plasma of patients with glioma prior to operation were significantly lower than the levels in healthy volunteers (**P < 0.01). (C) Correlation analysis of miR-30c in glioma tissue and plasma. (D, E) The PFS and OS of patients with high miR-30c expression were significantly longer than those of patients with low miR-30c expression (P < 0.01).
We also identified the expression levels of miR-30c in the presurgery plasma of patients with gliomas and healthy volunteers. The expression level of miR-30c in the glioma patients was 0.813 ± 0.066, and that in the healthy volunteers was 2.403 ± 0.136. The difference was statistically significant (P < 0.01) (Fig. 1B). As illustrated in Table 1, the expression level of miR-30c in the plasma of the glioma patients was closely related to the WHO grade and tumor size (P < 0.01) but not to the sex, age, extent of resection, or KPS scores of the patients (P > 0.05). The expression levels of miR-30c in tumor tissue were positively correlated with the levels found in plasma according to the Pearson correlation test (r = 0.868, P = 0.002) (Fig. 1C). The expression levels of miR-30c in tissues and plasma were consistent.
To discern whether miR-30c can be used to determine the prognosis of gliomas in patients, we determined the expression levels of miR-30c in paraffin-embedded glioma tissues, and we analyzed the correlation between the expression level of miR-30c, progression-free survival (PFS), and overall survival (OS) using Kaplan‒Meier curves. Figure 1D,E shows that the PFS and OS of patients with low expression levels of miR-30c were significantly shorter than those of patients with higher expression levels of miR-30c. Thus, it can be inferred that miR-30c could be used to predict the prognosis of patients with gliomas.
miR-30c inhibits EMT and the proliferation, migration, and invasion of glioma cells
We then transfected mimics or a negative control (NC) or an inhibitor or an inhibitor NC into A172 and U251 glioma cells. The expression level of miR-30c in the mimics group was significantly higher than that in the NC group (P < 0.01). The expression level of miR-30c in the inhibitor group was significantly lower than that in the inhibitor NC group (P < 0.01) (Fig. 2A,B).
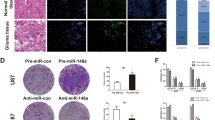
miR-30c inhibits the EMT process of glioma cells. (A, B) qRT‒PCR was used to verify the change in miR-30c expression levels in A172 and U251 cells after transfection with the mimics and the inhibitor (**P < 0.01). (C, D) The EMT process of glioma cells was changed after the transfection of the mimics and the inhibitor (*P < 0.05, **P < 0.01).
In the Western blot experiment, the protein expression level of N-cadherin and Vimentin in the mimics group was lower than that in the NC group (P < 0.05), and the protein expression level of E-cadherin in the mimics group was higher than that in the NC group (P < 0.01). Regarding the inhibitor group, the protein quantity of N-cadherin and Vimentin was higher than that of the inhibitor NC group (P < 0.05), and the protein expression of E-cadherin was lower than that in the inhibitor NC group (P < 0.01) (Fig. 2C,D). E-cadherin, N-cadherin, and Vimentin are marker genes for the process of EMT. Therefore, we are confident that miR-30c can inhibit the EMT process in glioma cells.
In the cell proliferation experiment, as shown in Fig. 3A,B, the proliferation of glioma cells in the mimics group was less than that in the NC group (P < 0.05, T = 48 h; P < 0.01, T = 72 h). The cell proliferation of the inhibitor group was greater than that of the inhibitor NC group (P < 0.05, T = 48 h; P < 0.01, T = 72 h). Therefore, we have determined that miR-30c can potentially inhibit the proliferation of glioma cells.
miR-30c reduces the proliferation, migration, and invasion of glioma cells. (A, B) CCK8 assay revealed that miR-30c could impact the proliferation of A172 and U251 cells (*P < 0.05, **P < 0.01). (C, E) The migration and invasion of the mimics and inhibitor groups were significantly changed in A172 cells (**P < 0.01). (D, F) The migration and invasion of U251 cells were significantly changed after transfection with the mimics and the inhibitor (**P < 0.01).
As shown in Fig. 3C–F, the migration and invasion of A172 and U251 cells transfected with mimics were significantly less than those in the NC group (P < 0.01), whereas the migration and invasion of A172 and U251 cells transfected with the inhibitor were significantly greater than those in the inhibitor NC group (P < 0.01). Our results suggested that miR-30c has the potential to inhibit the migration and invasion of glioma cells.
Notch1 is a direct target gene of miR-30c in gliomas
According to the TargetScan, miRWalk, and miRanda databases, we predicted that Notch1 might be the direct target gene of miR-30c, and its action site is shown in Fig. 4A. When mimics were transfected into A172 and U251 cells, the expression levels of the Notch1 protein in the mimics group were significantly lower than those in the NC group (P < 0.01). The expression level of NICD protein, which indicates the activation of Notch1 in the mimics group, was also significantly lower than that in the NC group, and the expression levels of downstream target genes of Notch1, such as HES1 and HEY1 proteins, in the mimics group were also significantly lower than those in the NC group (P < 0.01) (Fig. 4B,C). However, there was no significant change in Notch1, HES1 and HEY1 mRNA levels (Fig. 4D). When the inhibitor was transfected into glioma cells, the expression levels of the Notch1, NICD, HES1 and HEY1 proteins in the inhibitor group were significantly higher than those in the inhibitor NC group (P < 0.05) (Fig. 4B,C). There was no significant change in Notch1, HES1 and HEY1 mRNA levels (Fig. 4D). In the dual luciferase experiment, as shown in Fig. 4E,F, miR-30c reduced the luciferase activity of Notch1-WT in A172 and U251 cells, but it had no effect on Notch1-Mut. Our findings suggest that Notch1 is a direct target gene of miR-30c in gliomas.
Notch1 is the direct target gene of miR-30c in gliomas. (A) The 3′-UTR of Notch1 binds to miR-30c. (B, C) Changes in the Notch1, NICD, HES1 and HEY1 proteins after transfection with the mimics and the inhibitor (*P < 0.05, **P < 0.01). (D) There was no alteration in Notch1, HES1 and HEY1 mRNAs after the transfection of the mimics and the inhibitor. (E, F) Dual luciferase assay of two kinds of glioma cells (**P < 0.01).
Notch1 can promote EMT and the proliferation, migration, and invasion of gliomas
shRNA can hinder the expression of Notch1 in glioma cells. In Fig. 5A, qRT-PCR detection revealed that the expression level of Notch1 mRNA following shRNA-Notch1 transfection was notably less than that in the NC group (P < 0.01), which provided important information for subsequent experiments. As shown in Fig. 5B,C, the protein levels of N-cadherin and Vimentin in the shRNA-Notch1 group were lower than those in the NC group (P < 0.01), and the protein levels of E-cadherin in the shRNA-Notch1 group were higher than those in the NC group (P < 0.01). Figure 6A,B shows that inhibition of Notch1 expression can reduce the proliferation of A172 and U251 cells (P < 0.01, T = 48h; P < 0.01, T = 72h) and reduce the expression of Notch1 in glioma cells, which can significantly curtail cell migration and invasion (P < 0.01) (Fig. 6C,D). Therefore, Notch1 can promote EMT and the proliferation, migration, and invasion of gliomas.
miR-30c inhibited EMT and the proliferation, migration, and invasion of gliomas by reducing Notch1 expression
To more fully discern the mechanism of miR-30c in gliomas and to clarify that miR-30c can inhibit the EMT process of gliomas by reducing the expression levels of Notch1, we implemented a cotransfection experiment. By cotransfecting the miR-30c inhibitor and shRNA-Notch1 into A172 and U251 cells, we revealed that the miR-30c inhibitor could enhance the proliferation of glioma cells, while shRNA-Notch1 could attenuate this change (P < 0.05, T = 48h; P < 0.01, T = 72h) (Fig. 7A,B). Moreover, we discovered that shRNA-Notch1 can attenuate the effect of the miR-30c inhibitor on the migration and invasion of A172 and U251 glioma cells (Fig. 7C–F). The miR-30c inhibitor enhanced the Notch1, NICD, HES1 and HEY1 proteins in A172 and U251 glioma cells, and shRNA-Notch1 weakened this enhancement (Fig. 8A,B). The expression of the key factors N-cadherin increased, and the expression of E-cadherin decreased in the EMT process induced by the inhibitor. However, cotransfection of shRNA-Notch1 attenuated the changes in key gene proteins in EMT. But the expression of Vimentin was not affected in the cotransfection experiments (Fig. 8A,B). Our results suggest that miR-30c could inhibit EMT and the proliferation, migration, and invasion of glioma cells by directly acting on Notch1.
miR-30c hindered proliferation, migration, and invasion by downregulating the expression levels of Notch1. (A, B) The miR-30c inhibitor facilitated an increase in the proliferation of glioma cells, while shRNA-Notch1 attenuated this change (*P < 0.05, **P < 0.01). (C, D, E, F) shRNA-Notch1 attenuated the enhancement of migration and invasion of glioma cells by the miR-30c inhibitor (**P < 0.01).
The miR-30c-mediated EMT depended on Notch1. (A, B) shRNA-Notch1 attenuated the effect of the miR-30c inhibitor on the Notch1, NICD, HES1 and HEY1 proteins of A172 and U251 glioma cells. The miR-30c inhibitor triggered an increase in N-cadherin and a decrease in E-cadherin, while cotransfection with shRNA-Notch1 partially eliminated these effects. However, the expression of Vimentin was not affected (*P < 0.05, **P < 0.01).
Discussion
miRNA target genes are large in number and wide in variety and are involved in multiple cellular signal transduction pathways. They not only create a complex signal regulatory network but also serve an assortment of biological purposes28,29,30. In recent years, research has indicated that miR-30c is actively involved in the regulation of malignant tumor signaling pathways and that it acts as a tumor suppressor gene in the majority of malignant tumors, such as renal cell carcinoma, colorectal cancer, gastric cancer, and breast cancer31,32,33,34. Sox9 is a high-mobility group box transcription factor that plays critical roles during embryogenesis, differentiation, tumor initiation, invasion and stem cell self-renewal35,36. Sox9 is critical for tumorigenesis and is overexpressed in gliomas37,38. In glioblastoma, miR-30c can impede the proliferation, migration and invasion of tumor cells by affecting Sox939. The latest research showed that Notch1 activation induced strong Sox9 expression40. Therefore, Notch1 is more important in the pathogenesis of gliomas. Apoptosis is triggered by Bcl-2 superfamily members. miR-30c targets Bcl-2 and activates the caspase pathway in glioma cells41. Glioma cell proliferation is inhibited, and cell apoptosis is promoted through the suppression of Notch142. Therefore, if the apoptosis of tumor cells is decreased in gliomas, the expression of Notch1 needs to be inhibited first. Based on the important role of Notch1, we were eager to determine which miRNA mainly regulates it in gliomas. Our study explored all levels of gliomas and discovered that even in low-grade gliomas, the expression levels of miR-30c decreased to a significant extent and that those with a higher grade had lower expression levels of miR-30c. The expression level of miR-30c in glioma tissues and preoperative plasma was closely related to the WHO grade and tumor size, and the expression level of miR-30c in tissues and plasma remained consistent. Moreover, in paraffin sections, with lower miR-30c levels, we noted reduced PFS and OS, and such information can be used to predict the prognosis of glioma patients.
miR-30c is closely related to a variety of cell biological functions. Unlimited cell proliferation is an important biological feature of malignant tumor cells. In this experiment, miR-30c was discovered to hinder the proliferation of glioma cells. EMT is defined as a process by which epithelial tumor cells lose their adhesive ability and obtain mesenchymal cell migration ability to promote metastasis and drug resistance. Our research indicated that increasing the expression level of miR-30c can reduce the protein expression of N-cadherin and Vimentin, just as it can increase the protein expression of E-cadherin, thereby inhibiting the EMT processes of glioma cells. Additionally, tumor metastasis is a multistage and multistep orderly process, and tumor migration and invasion are the premise of tumor metastasis. We determined that miR-30c can inhibit the migration and invasion of glioma cells.
Based on a database analysis, we predicted that Notch1, which functions as an important element in glioma, may be the direct target gene of miR-30c. By increasing the expression level of miR-30c, the expression level of Notch1 protein decreased; conversely, when the expression level of miR-30c was inhibited, the expression of Notch-1 protein increased. However, there was no effect on Notch1 mRNA. In the dual luciferase assay, miR-30c reduced the levels of luciferase activity of Notch1-WT in glioma cells but had no effect on Notch1-Mut. Therefore, we believe that Notch1 is the direct target gene of miR-30c and functions posttranscriptionally.
Notch1 plays an instrumental role in glioma cells, with the functions of enhancing stemness and tumorigenicity43,44, promoting tumor growth45 and EMT46, and increasing chemotherapy drug resistance47. In this study, we used shRNA to inhibit the expression levels of Notch1 in glioma cells, and we revealed that EMT (N-cadherin decreased, E-cadherin increased) and the proliferation, migration, and invasion of cancer cells were significantly decreased. This further confirmed the carcinogenic effect of Notch1. Afterward, we cotransfected the miR-30c inhibitor and shRNA-Notch1 into two glioma cell lines. shRNA-Notch1 attenuated the enhancement effect of the inhibitor on EMT and the proliferation, migration, and invasion of glioma cells, thus confirming that miR-30c regulates these processes in glioma cells posttranscriptionally by targeting Notch1. The expression of Vimentin was not affected, we considered that there may be the following two reasons: First, transcription factors such as SNAI1, SNAI2, TWIST1, and ZEB-1 play the important roles in mesenchymal differentiation48,49. The depletion of TWIST1/SNAI1 resulted in reduced Bcl-w-induced Vimentin expression and invasion50. Second, we speculate that the occurrence of this situation may also be related to Notch1 expression. This may be related to the heterogeneity of tumor cells.
In conclusion, the expression level of miR-30c in glioma tissue and preoperative plasma was decreased, and the low expression level was closely tied to the WHO grade, tumor size, and prognosis. miR-30c can inhibit EMT and the proliferation, migration, and invasion of glioma cells. miR-30c acts as a tumor suppressor gene by targeting Notch1 at the posttranscriptional level. Therefore, miR-30c may act as a marker for tumor diagnosis, treatment, and prognosis.
Materials and methods
Clinical specimens
Sixty-six patients with brain glioma who were first diagnosed at Taian Central Hospital (Taian, Shandong, China) from June 2018 to May 2020 were selected, and 2 ml of peripheral venous plasma was collected from each patient prior to surgery. The other 9 normal, nonglioma brain tissue samples were obtained from patients after decompression surgery to treat traumatic brain injury, which was confirmed as normal brain tissue by postoperative pathology. Control plasma samples were collected from 9 healthy volunteers.
Brain glioma paraffin specimens from 72 patients were collected from Taian Central Hospital from June 2013 to May 2018. Eight of the 72 patients were lost during the follow-up stage, and 64 patients were ultimately assessed. The included patients were diagnosed with glioma by postoperative pathology and were graded according to the WHO classification and the grading standard of nervous system tumors (2016). None of the patients received any form of radiotherapy, chemotherapy, targeted therapy, immunotherapy, or any other anticancer treatments prior to operation, and each study participant signed the informed consent form before sampling. The ethics committee of the Taian Central Hospital approved this study. All methods were performed in accordance with the relevant guidelines and regulations.
Cell culture
U251 cells were purchased from Shanghai Cell Bank at the Chinese Academy of Medical Sciences (Shanghai, China), and A172 cells were from the Central Laboratory of Taian Central Hospital. Both types of cells were cultured in DMEM (GIBCO, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS, GIBCO) and 0.2% penicillin streptomycin (Invitrogen, Carlsbad, California, USA). The cells were cultured in an incubator at 37 °C with 95% humidity and 5% CO2.
Real-time quantitative PCR (qRT‒PCR)
Following the instructions of the miRNeasy Mini Kit (Qiagen, Duesseldorf, Nordrhein-Westfalen, Germany), total RNA was extracted from glioma tissue, normal brain tissue, and glioma cells. Total RNA was extracted from plasma by using the miRNeasy Serum/Plasma Kit (Qiagen), and FFPE tissue samples were extracted using the miRNeasy FFPE Kit (Qiagen). The reverse transcription experiment was carried out by using the miScript II RT Kit (Qiagen) to synthesize cDNA. The miScript SYBR Green PCR Kit (Qiagen) was implemented to measure the qRT-PCR of miRNA. Primer sequences were used as follows: miR-30c forward, 5′-TGTGTTTTTATTGTTTTTGTTGTCCCA-3′; miR-30c reverse, 5′-GGGACAGAACAGGTTAATGGGAA-3′; U6-forward, 5′-GCTTCGGCAGCACATATACT- AAAAT-3′; and U6-reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
The reaction was performed in a LightCycler 480 qRT‒PCR system (Roche molecular systems, Indianapolis, USA) according to the instructions. After amplification, the cycle threshold (CT) of each PCR was calculated by measuring the number of cycles required when the cDNA template reached the set fluorescence intensity by PCR. Three multiple wells were set in each sample, and U6 was used as an internal reference to normalize the target gene. The relative expression of miR-1 was determined according to 2−∆∆Ct. TB Green™ Premix Ex Taq™ II (Tli RNaseH plus) (Takara, Japan) was used to analyze the PCR of mRNA. β-actin was used as an internal reference. The primer sequences used were as follows:
Notch1 forward, 5′-GCTACAACTGCGT GTGTGTC-3′; Notch1 reverse, 5′-GTTGGTGTC- GCAGTTGGAGC-3′; HES1 forward, 5′-CTGAGCACAGACCCAAGTGT-3′; HES1 reverse, 5′-GAGTGCGCACCTCGGTATTA-3′; HEY1 forward, 5′-GTGCGGACGAGAATGGAAAC-3′; HEY1 reverse, 5′-TTGCTCCATTACCTGCTTCTCA-3′; β-actin forward, 5′-CAAAGGCCAACAGAGAGAAGAT-3′; and β-actin reverse, 5′-TGAGACACACCATCACCAGAAT-3′. The relative expression of Notch1 was determined according to 2−∆∆Ct.
Cell transfection
In six-well plates, as glioma cells reached 70–80% confluence, transfection was executed. Mimics that increased the expression levels of miR-30c, negative control (NC), inhibitor suppressed the expression levels of miR-30c or inhibitor negative control (inhibitor NC), and shRNA inhibiting Notch1 expression or shRNA negative control (shRNA NC) (Gemma, Shanghai, China) were diluted by using Opti-MEM™ to a suitable concentration. Next, Lipofectamine™ 3000 transfection reagent (Invitrogen) was added. After being incubated for 10–15 min at room temperature, the mixture was added to the well plates. After transfection for 24–72 h, the cells were collected and saved for later experiments.
Cell proliferation, migration, and invasion experiments
The cell proliferation of miR-30c was detected by using the CCK-8 (Cell Counting Kit-8, Beyotime, Shanghai, China) assay. Logarithmic growth phase glioma cells were transferred into 96-well plates, with approximately 2 × 103 cells per well. At 0, 24, 48, and 72 h posttransfection, 10 μl of CCK-8 was added to each well. After adding CCK-8, the cells were placed in an incubator at 37 °C with 5% CO2. After 2 h, the absorbance was detected at 450 nm by using a microplate reader.
The cell migration experiment was carried out in a Transwell chamber with 24-pore plates and 8-µm holes. DMEM containing 20% fetal bovine serum was added to the plates. Next, 1 × 105 transfected glioma cells (200 µl, serum-free medium) were added to the upper chamber. After 24 h, the glioma cells reached the opposite side of the membrane through the pores on the filter membrane and were then fixed with methanol for 15 min and stained with 0.1% crystal violet for 20 min. Five visual fields were selected to count at random. Matrigel (Matrigel/DMEM = 8:1, BD, Franklin, NJ, USA) was added to the upper chamber, with each chamber containing 60 µl. The same method was employed for the cell invasion experiment, and fixation and staining were performed at 48 h.
Luciferase reporter assay
A172 and U251 glioma cells were placed into 96-well plates. The Notch1 wild-type vector (pGL3-Notch1-3′-UTR-WT) or mutant vector (pGL3-Notch1-3′-UTR-Mut) and miR-30c mimics or NC were cotransfected with Lipofectamine™ 3000 (Invitrogen). At 48 h after transfection, the luciferase activity was measured using the dual luciferase reporter system (Promega, Madison, WI). The ratio of firefly to Renilla luciferase activity was defined as the normalized luciferase activity.
Western blotting
Cell lysate was used to treat cells and tissues. The protease inhibitor PMSF (Beyotime) was added. The BCA method was used to measure the protein concentration. Proteins were extracted by SDS‒PAGE and then transferred to nitrocellulose membranes. The nitrocellulose membranes were blocked with 5% skimmed milk prepared with TBST and subsequently incubated with the diluted primary antibodies, namely, anti-Notch1 (1:1000), anti-Vimentin (1:1000), anti-N-cadherin (1:1000), anti-E-cadherin (1:1000), anti-NICD (1:1000), and anti-HES1 (1:1000) (Cell Signaling Technology, CST, Boston, USA), anti-HEY1(1:1000) (Abcam, Cambridge, UK) at 4 °C overnight. After TBST was eluted on three occasions, horseradish peroxidase-labeled anti-rabbit or anti-mouse secondary antibodies (CST) were added and incubated on a shaking table for 1 h at room temperature. Then, the target bands were imaged using a chemiluminescence detection system (EMD, Millipore, Billerica, MA, USA) and analyzed quantitatively by using ImageJ software (National Institutes of Health, Bethesda, MD, USA). β-actin (1:5000) was used as an internal reference.
Statistical analysis
The experimental results are expressed as the means ± SEMs. An independent-samples T test was implemented to detect the differences among glioma tissues and normal tissues after brain trauma decompression, preoperative plasma of glioma patients, and plasma of healthy volunteers. ANOVA was used to evaluate the expression of miR-30c according to various clinicopathological features. Next, Pearson analysis was used to evaluate the connection between miR-30c expression and clinicopathological features. Cytological experiments were then analyzed with a paired-samples T test. A result of P < 0.05 was considered statistically significant, and P < 0.01 was considered remarkably statistically significant.
References
Brandao, M., Simon, T., Critchley, G. & Giamas, G. Astrocytes, the rising stars of the glioblastoma microenvironment. Glia 67, 779–790. https://doi.org/10.1002/glia.23520 (2019).
Lu, W. & Kang, Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell 49, 361–374. https://doi.org/10.1016/j.devcel.2019.04.010 (2019).
Sommariva, M. & Gagliano, N. E-cadherin in pancreatic ductal adenocarcinoma: A multifaceted actor during EMT. Cells https://doi.org/10.3390/cells9041040 (2020).
Wong, S. H. M., Fang, C. M., Chuah, L. H., Leong, C. O. & Ngai, S. C. E-cadherin: Its dysregulation in carcinogenesis and clinical implications. Crit. Rev. Oncol. Hematol. 121, 11–22. https://doi.org/10.1016/j.critrevonc.2017.11.010 (2018).
Loh, C. Y. et al. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells https://doi.org/10.3390/cells8101118 (2019).
Tian, H. et al. AKT-induced lncRNA VAL promotes EMT-independent metastasis through diminishing Trim16-dependent Vimentin degradation. Nat. Commun. 11, 5127. https://doi.org/10.1038/s41467-020-18929-0 (2020).
Arefin, B., Parvin, F., Bahrampour, S., Stadler, C. B. & Thor, S. Drosophila neuroblast selection is gated by Notch, Snail, SoxB, and EMT gene interplay. Cell Rep. 29, 3636–3651. https://doi.org/10.1016/j.celrep.2019.11.038 (2019).
Yuan, X. et al. Notch signaling and EMT in non-small cell lung cancer: Biological significance and therapeutic application. J. Hematol. Oncol. 7, 87. https://doi.org/10.1186/s13045-014-0087-z (2014).
Arumugam, T. V. et al. Notch signaling and neuronal death in stroke. Prog. Neurobiol. 165–167, 103–116. https://doi.org/10.1016/j.pneurobio.2018.03.002 (2018).
Schmidl, B. et al. NOTCH1 intracellular domain and the tumor microenvironment as prognostic markers in HNSCC. Cancers (Basel) https://doi.org/10.3390/cancers14041080 (2022).
Katoh, M. & Katoh, M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int. J. Oncol. 31, 461–466 (2007).
Hibdon, E. S. et al. Notch and mTOR Signaling pathways promote human gastric cancer cell proliferation. Neoplasia 21, 702–712. https://doi.org/10.1016/j.neo.2019.05.002 (2019).
Bigas, A. & Espinosa, L. The multiple usages of Notch signaling in development, cell differentiation and cancer. Curr. Opin. Cell Biol. 55, 1–7. https://doi.org/10.1016/j.ceb.2018.06.010 (2018).
Chen, L., Wang, J., Li, J. W., Zhao, X. W. & Tian, L. F. LncRNA MEG3 inhibits proliferation and promotes apoptosis of osteosarcoma cells through regulating Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 24, 581–590. https://doi.org/10.26355/eurrev_202001_20034 (2020).
Baisiwala, S. et al. LNX1 modulates Notch1 signaling to promote expansion of the glioma stem cell population during Temozolomide therapy in glioblastoma. Cancers (Basel) https://doi.org/10.3390/cancers12123505 (2020).
Li, J. et al. Notch1 is an independent prognostic factor for patients with glioma. J. Surg. Oncol. 103, 813–817. https://doi.org/10.1002/jso.21851 (2011).
Li, J. et al. Targeting the Notch1 oncogene by miR-139-5p inhibits glioma metastasis and epithelial-mesenchymal transition (EMT). BMC Neurol. 18, 133. https://doi.org/10.1186/s12883-018-1139-8 (2018).
Natsuizaka, M. et al. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat. Commun. 8, 1758. https://doi.org/10.1038/s41467-017-01500-9 (2017).
Zeng, et al. Inhibition of Notch1 reverses EMT and chemoresistance to cisplatin via direct downregulation of MCAM in triple-negative breast cancer cells. Int. J. Cancer 147, 490–504. https://doi.org/10.1002/ijc.32911 (2020).
Liu, J. et al. Mir-758-5p suppresses glioblastoma proliferation, migration and invasion by targeting ZBTB20. Cell Physiol. Biochem. 48, 2074–2083. https://doi.org/10.1159/000492545 (2018).
Villanova, L. et al. miR-1285–3p controls colorectal cancer proliferation and escape from apoptosis through DAPK2. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21072423 (2020).
Liu, W. et al. MicroRNA-1 suppresses proliferation, migration and invasion by targeting Notch2 in esophageal squamous cell carcinoma. Sci. Rep. 8, 5183. https://doi.org/10.1038/s41598-018-23421-3 (2018).
Lu, G. D., Cheng, P., Liu, T. & Wang, Z. BMSC-derived exosomal miR-29a promotes angiogenesis and osteogenesis. Front. Cell Dev. Biol. 8, 608521. https://doi.org/10.3389/fcell.2020.608521 (2020).
Zhu, C., Huang, Q. & Zhu, H. miR-383 inhibited the cell cycle progression of gastric cancer cells via targeting cyclin E2. DNA Cell Biol. 38, 849–856. https://doi.org/10.1089/dna.2019.4624 (2019).
Baraniskin, A. et al. MicroRNA-30c as a novel diagnostic biomarker for primary and secondary B-cell lymphoma of the CNS. J. Neurooncol. 137, 463–468. https://doi.org/10.1007/s11060-018-2749-0 (2018).
Pei, B. et al. Downregulation of microRNA-30c-5p was responsible for cell migration and tumor metastasis via COTL1-mediated microfilament arrangement in breast cancer. Gland Surg. 9, 747–758. https://doi.org/10.21037/gs-20-472 (2020).
Song, S. et al. Urinary exosome miR-30c-5p as a biomarker of clear cell renal cell carcinoma that inhibits progression by targeting HSPA5. J. Cell Mol. Med. 23, 6755–6765. https://doi.org/10.1111/jcmm.14553 (2019).
Chen, P., Chen, F., Lei, J., Li, Q. & Zhou, B. Activation of the miR-34a-mediated SIRT1/mTOR signaling pathway by urolithin A attenuates D-galactose-induced brain aging in mice. Neurotherapeutics 16, 1269–1282. https://doi.org/10.1007/s13311-019-00753-0 (2019).
Jafarzadeh, M., Mohammad Soltani, B., Ekhteraei Tousi, S. & Behmanesh, M. Hsa-miR-497 as a new regulator in TGFbeta signaling pathway and cardiac differentiation process. Gene 675, 150–156. https://doi.org/10.1016/j.gene.2018.06.098 (2018).
Qian, Z. et al. Excess administration of miR-340-5p ameliorates spinal cord injury-induced neuroinflammation and apoptosis by modulating the P38-MAPK signaling pathway. Brain Behav. Immun. 87, 531–542. https://doi.org/10.1016/j.bbi.2020.01.025 (2020).
Yuan, R. X., Bao, D. & Zhang, Y. Linc00707 promotes cell proliferation, invasion, and migration via the miR-30c/CTHRC1 regulatory loop in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 24, 4863–4872. https://doi.org/10.26355/eurrev_202005_21175 (2020).
Onyshchenko, K. V. et al. Expression of micro-RNA hsa-miR-30c-5p and hsa-miR-138-1 in renal cell carcinoma. Exp. Oncol. 42, 115–119. https://doi.org/10.32471/exp-oncology.2312-8852.vol-42-no-2.14632 (2020).
Herichova, I., Reis, R., Hasakova, K. & Vician, M. Downregulation of miR-30c-5p expression in colorectal cancer tissue is sex-dependent. Physiol. Res. 69, S479–S487. https://doi.org/10.33549/physiolres.934598 (2020).
Cao, J. M., Li, G. Z., Han, M., Xu, H. L. & Huang, K. M. MiR-30c-5p suppresses migration, invasion and epithelial to mesenchymal transition of gastric cancer via targeting MTA1. Biomed. Pharmacother. 93, 554–560. https://doi.org/10.1016/j.biopha.2017.06.084 (2017).
Sekido, R. & Lovell-Badge, R. Sex determination and SRY: Down to a wink and a nudge?. Trends Genet. 25, 19–29. https://doi.org/10.1016/j.tig.2008.10.008 (2009).
Larsimont, J. C. et al. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell 17, 60–73. https://doi.org/10.1016/j.stem.2015.05.008 (2015).
Hiraoka, K. et al. SOX9-mediated upregulation of LGR5 is important for glioblastoma tumorigenicity. Biochem. Biophys. Res. Commun. 460, 216–221. https://doi.org/10.1016/j.bbrc.2015.03.012 (2015).
Liu, N. et al. MicroRNA-101 inhibits proliferation, migration and invasion of human glioblastoma by targeting SOX9. Oncotarget 8, 19244–19254. https://doi.org/10.18632/oncotarget.13706 (2017).
Liu, S., Li, X. & Zhuang, S. miR-30c impedes glioblastoma cell proliferation and migration by targeting SOX9. Oncol. Res. 27, 165–171. https://doi.org/10.3727/096504018X15193506006164 (2019).
Augustus, M. et al. Identification of CRYAB(+) KCNN3(+) SOX9(+) astrocyte-like and EGFR(+) PDGFRA(+) OLIG1(+) oligodendrocyte-like tumoral cells in diffuse IDH1-mutant gliomas and implication of NOTCH1 signalling in their genesis. Cancers (Basel) https://doi.org/10.3390/cancers13092107 (2021).
Yuan, L. Q., Zhang, T., Xu, L., Han, H. & Liu, S. H. miR-30c-5p inhibits glioma proliferation and invasion via targeting Bcl2. Transl. Cancer Res. 10, 337–348. https://doi.org/10.21037/tcr-19-2957 (2021).
Wang, B. Q. et al. MicroRNA-499a decelerates glioma cell proliferation while accelerating apoptosis through the suppression of Notch1 and the MAPK signaling pathway. Brain Res. Bull. 142, 96–106. https://doi.org/10.1016/j.brainresbull.2018.06.005 (2018).
Man, J. et al. Hypoxic induction of vasorin regulates Notch1 turnover to maintain glioma stem-like cells. Cell Stem Cell 22, 104–118. https://doi.org/10.1016/j.stem.2017.10.005 (2018).
Sun, Z. et al. Glioblastoma stem cell-derived exosomes enhance stemness and tumorigenicity of glioma cells by transferring Notch1 protein. Cell Mol. Neurobiol. 40, 767–784. https://doi.org/10.1007/s10571-019-00771-8 (2020).
Gu, G. et al. NKAP alters tumor immune microenvironment and promotes glioma growth via Notch1 signaling. J. Exp. Clin. Cancer Res. 38, 291. https://doi.org/10.1186/s13046-019-1281-1 (2019).
Vinchure, O. S. et al. Polycomb complex mediated epigenetic reprogramming alters TGF-beta signaling via a novel EZH2/miR-490/TGIF2 axis thereby inducing migration and EMT potential in glioblastomas. Int. J. Cancer 145, 1254–1269. https://doi.org/10.1002/ijc.32360 (2019).
Zhao, C. et al. MicroRNA-128-3p enhances the chemosensitivity of temozolomide in glioblastoma by targeting c-Met and EMT. Sci. Rep. 10, 9471. https://doi.org/10.1038/s41598-020-65331-3 (2020).
Thiery, J. P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454. https://doi.org/10.1038/nrc822 (2002).
Yang, J. & Weinberg, R. A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829. https://doi.org/10.1016/j.devcel.2008.05.009 (2008).
Lee, W. S. et al. Bcl-w enhances mesenchymal changes and invasiveness of glioblastoma cells by inducing nuclear accumulation of beta-catenin. PLoS ONE 8, e68030. https://doi.org/10.1371/journal.pone.0068030 (2013).
Acknowledgements
This study was supported by the Shandong Medicine and Health Science Technology Expansion Plan, China [No. 2017WS069] and the Taian Science Technology Expansion Plan, Shandong, China [Nos. 2018NS0117 and 2017NS0147].
Author information
Authors and Affiliations
Contributions
J.X. designed the study, supervised the research and reviewed the manuscript. M.K.L., W.Z.L., J.L. and H.Z. finished the experiments and wrote the manuscript. M.K.L. performed the statistical analysis of the data. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, M., Liu, W., Li, J. et al. miR-30c plays diagnostic and prognostic roles and mediates epithelial–mesenchymal transition (EMT) and proliferation of gliomas by affecting Notch1. Sci Rep 12, 16404 (2022). https://doi.org/10.1038/s41598-022-19326-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-19326-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.