Abstract
High-temperature ethanol fermentation by thermotolerant yeast is considered a promising technology for ethanol production, especially in tropical and subtropical regions. In this study, optimization conditions for high-temperature ethanol fermentation of pineapple waste hydrolysate (PWH) using a newly isolated thermotolerant yeast, Saccharomyces cerevisiae HG1.1, and the expression of genes during ethanol fermentation at 40 °C were carried out. Three independent variables, including cell concentration, pH, and yeast extract, positively affected ethanol production from PWH at 40 °C. The optimum levels of these significant factors evaluated using response surface methodology (RSM) based on central composite design (CCD) were a cell concentration of 8.0 × 107 cells/mL, a pH of 5.5, and a yeast extract concentration of 4.95 g/L, yielding a maximum ethanol concentration of 36.85 g/L and productivity of 3.07 g/L. Gene expression analysis during high-temperature ethanol fermentation using RT–qPCR revealed that the acquisition of thermotolerance ability and ethanol fermentation efficiency of S. cerevisiae HG1.1 are associated with genes responsible for growth and ethanol stress, oxidative stress, acetic acid stress, DNA repair, the pyruvate-to-tricarboxylic acid (TCA) pathway, and the pyruvate-to-ethanol pathway.
Similar content being viewed by others
Introduction
Increasing energy demands encourage scientists to find low-cost, clean, renewable, and sustainable alternative energy sources1,2,3. A comparative study of literature on various alternative fuels, such as ethanol, vegetable oils, microbial oils, biomass, glycerol, biodiesel, and hydrogen, has been reported4,5. Commercial ethanol for biofuel is produced from feedstocks such as sugarcane, corn, and cassava. These raw materials, which are also food for human needs and animal feed, are competitively priced6. Agricultural wastes, particularly lignocellulosic materials, have been considered promising for second-generation bioethanol production. Pineapple peel, core, stem, and leaves are byproducts of pineapple processing (approximately 50% (w/w) of the pineapple weight)7. These byproducts are highly biodegradable and rich in proteins and carbohydrates, which are promising raw and abundant materials for ethanol production8,9. Thailand and Vietnam are the top countries in pineapple production, producing 2.21 and 0.59 million metric tons, accounting for 8.91% and 2.38% of the world's production, respectively10.
In summer, the temperature in Thailand and Vietnam dramatically increases, which will increase with global warming. Furthermore, the temperature inside a bioreactor may rise from 30 °C to approximately 40 °C during ethanol fermentation11. High temperatures inhibit cell growth and the metabolic activity of yeast cells, resulting in a reduction in ethanol yield and volumetric ethanol productivity12,13. Therefore, the use of thermotolerant microorganisms is a promising approach to solving the problem of ethanol production at high temperatures. There are several advantages of using high-temperature ethanol fermentation, such as decreased costs associated with a cooling system, higher yields obtained in saccharification, and reduced risk of contamination by bacteria14,15. Even though many thermotolerant yeasts can tolerate and ferment at high temperatures, several stresses, e.g., thermal, ethanol, osmotic, ionic, lignocellulosic inhibitors, and reactive oxygen species (ROS), are unfavorable conditions for yeast growth and fermentation activity. Denaturation of DNA, proteins, lipids, and essential cellular structures of yeast cells under stressful situations has been previously reported16,17,18,19. However, the molecular mechanism conferring thermotolerance acquisition during high-temperature ethanol fermentation using PWH as feedstock has not yet been evaluated.
S. cerevisiae can reproduce in anaerobic and aerobic conditions and accumulate ethanol at high concentrations, making it the preferred choice for starter cultures for beverage and food fermentations20. Recently, S. cerevisiae has become one of the most engineered yeasts for ethanol production from the agricultural, kitchen, industrial, and lignocellulosic wastes21,22. S. cerevisiae HG1.1 is one of several thermotolerant yeasts isolated from soil samples in Vietnam23. This newly isolated yeast can grow and produce ethanol at a temperature up to 45 °C, using a YM medium containing 160 g/L glucose. Furthermore, it can tolerate ethanol and acetic acid up to 14% (v/v) and 4 g/L, respectively, when growing on YM agar at 35 °C23. Since its ethanol production potential using agricultural waste as feedstock has never been elucidated. Therefore, this newly isolated yeast was chosen for ethanol production under high-temperature conditions using PWH as feedstock in this study.
The disadvantages of a single variable optimization technique, for instance, missing the interactions between the experimental factors and requiring a large number of experiments, can be eliminated by a statistical experimental model such as response surface methodology (RSM) based on a central composite design (CCD)24. Statistical tools such as RSM are used for experimental design, determining the positive and negative variables and their interactions, predicting the optimal equation for optimization with the cost-effective process, and reducing experimental runs25,26. Several recent reports have used this statistical method to optimize different medium compositions for bioethanol fermentation27,28,29. This study used a statistical optimization methodology to investigate ethanol production from PWH at high temperatures by the newly isolated thermotolerant yeast S. cerevisiae HG1.1. In addition, reverse transcription quantitative real-time polymerase chain reaction (RT–qPCR) was applied to analyze the expression levels of selected genes responsible for growth and ethanol stress (ATP6, OLE1, ERG8), oxidative stress (GLR1, SOD1), DNA repair (RAD14, MRE11, POL4), the pyruvate-to-tricarboxylic acid (TCA) pathway (PDA1, CIT1, LYS21), the pyruvate-to-ethanol pathway (PDC1, ADH1, ADH2), and acetic acid stress (ACS1, ALD2) in S. cerevisiae HG1.1. This study could provide the optimum conditions for ethanol production from PWH and help better understand the molecular mechanism by which yeast cells acquire thermotolerance and fermentation efficiency during high-temperature ethanol fermentation.
Materials and methods
Strain and culture media
The newly isolated thermotolerant yeast S. cerevisiae HG1.1, isolated from soil samples from Vietnam, was used in this study. Isolation, screening, and selection of this thermotolerant yeast strain were described by Phong et al.23. The yeast culture was stored at the Department of Biotechnology, Faculty of Technology, Khon Kaen University, Thailand.
The medium used was YM medium (0.3% yeast extract, 0.3% malt extract, 0.5% peptone, and 1.0% D-glucose). Yeast inoculum was prepared by transferring one colony of a 24-h culture grown on a slant of YM agar to a test tube containing 10 mL of YM broth and incubating on a rotary shaker at 35 °C and 100 rpm for 18 h. Then, 10 mL of preculture was inoculated into a 500-mL Erlenmeyer flask containing 200 mL of YM broth (pH 5.0) and incubated on a rotary shaker under the same conditions for 18 h. The final yeast cell concentrations were approximately 1.0 × 108–2.5 × 108 cells/mL. The active yeast cells were collected by centrifugation and used as a starter culture.
Plant material
Pineapple (Ananas comosus L. cv. Pattavia) wastes (pineapple peels and core) were collected in May 2018 from the Food Services Center, Khon Kaen University, Khon Kaen province, Thailand, with the permission of the Khon Kaen University Office. The plant used in this study is not wild but cultivated in Nong Khai province, Thailand. A voucher specimen (dried material) was deposited at the Department of Biotechnology, Faculty of Technology, Khon Kaen University, with the code number: KKUDB-PPC-2018-01. All methods were performed following relevant guidelines in the method section.
PWH preparation and chemical composition analysis
Pineapple wastes were collected and chopped into small pieces. They were dried under natural conditions (sun drying for 3 days) and in a hot air oven for 24 h. The dried pineapple wastes were milled by a laboratory blender, mixed in a single lot, and stored prior to use. The fiber compositions of dried pineapple wastes were analyzed using an Ankom Fiber Analyzer30 at the Animal Science Laboratory, Faculty of Agriculture, Khon Kaen University.
PWH was prepared by transferring dried pineapple wastes to 0.5% (v/v) sulfuric acid (H2SO4) and heating at 121 °C for 15 min31. After hydrolysis, the pellet was removed by centrifugation, and the resulting supernatant was collected and kept at − 20 °C. The sugar compositions, acetic acid, formic acid, and furfural were analyzed using high-performance liquid chromatography (HPLC) at Central Laboratory, Faculty of Technology, Khon Kaen University and Central Laboratory (Thailand) Co., Ltd., Khon Kaen. Minerals, such as nitrogen, phosphorus, and magnesium, were analyzed at the Chemical Analysis Laboratory, Agricultural Development Research Center in Northeast Thailand, Khon Kaen, Thailand.
Effect of inorganic nitrogen sources on ethanol fermentation
Inorganic nitrogen sources have been shown to affect ethanol production under high-temperature fermentation conditions. In this study, based on the literature reviews, various inorganic nitrogen sources, including urea [CO(NH2)2], ammonium sulfate [(NH4)2SO4], ammonium nitrate [NH4NO3], and diammonium phosphate [DAP, (NH4)2HPO4] at different concentrations27,32 were determined for their effect on ethanol production by S. cerevisiae HG1.1. The ethanol fermentation was conducted in triplicate using a 250-mL Erlenmeyer flask containing 100 mL of PWH (pH 5.0) supplemented with various nitrogen sources at different concentrations and an initial yeast cell concentration of 5.0 × 106 cells/mL. All flasks were incubated at 40 °C on a rotary shaker at 100 rpm. Samples were withdrawn every 12 h and subjected to ethanol and total sugar analyses.
Optimization of ethanol production at high temperature
Based on the literature reviews, several environmental factors or variables affect ethanol production under high-temperature conditions. In this study, some influence factors include initial yeast cell concentration, pH of the fermentation medium, manganese (II) sulfate (MnSO4·H2O), zinc sulfate (ZnSO4·7H2O), magnesium sulfate (MgSO4·7H2O), potassium dihydrogen phosphate (KH2PO4), and yeast extract were chosen12,23,27,28,32,33. The significant independent factors positively affecting ethanol production from PWH by S. cerevisiae HG1.1 were screened and selected using PBD. The codes and actual values of the independent factors are presented in Table 1. The batch ethanol fermentation experiments were performed in triplicate using a 250-mL Erlenmeyer flask containing 100 mL of PWH (pH 5.0). The ethanol concentration was set as the response variable in this study.
The significant independent variables selected based on PBD were subjected to an optimization experiment using the RSM based on the CCD. The confirmatory experiment was carried out using the optimized conditions from the response surface analysis.
RT–qPCR analysis of gene expression in S. cerevisiae HG1.1 under high-temperature ethanol fermentation
The yeast inoculum was transferred into a 250-mL Erlenmeyer flask containing 100 mL PWH (pH 5.5) supplemented with 4.95 g/L yeast extract with an initial cell concentration of 8.0 × 107 cells/mL. All flasks were incubated on a rotary shaker at 100 rpm under four different fermentation conditions: (1) unstressed condition (flasks were incubated at 30 °C for 9 h); (2) heat shock condition (flasks were incubated at 30 °C for 9 h, then shifted to 40 °C for 30 min); (3) short-term heat stress (flasks were incubated at 30 °C for 9 h, then shifted to 40 °C for 3 h); and (4) long-term heat stress (flasks were incubated at 40 °C for 9 h). Yeast cells were harvested at specific time points (i.e., 9 h for unstressed, 9 h and 30 min for heat shock, 12 h for short-term heat stress, and 9 h for long-term heat stress) by centrifugation at 5,000 rpm and 4 °C for 5 min, and then subjected to total RNA isolation using an RNA extraction kit (GF-1 Total RNA extraction kit, Vivantis, USA) with some modifications as described by Techaparin et al.34. The RNA concentration in each sample was measured and adjusted using a BioDrop μLITE (BioDrop Ltd, UK). RT–qPCR was performed in triplicate on a 7500 Fast Real-Time PCR System using the qPCRBIO SyGreen One-Step Detect Lo-ROX (PCR Biosystems, London, UK). The reactions were conducted with a total volume of 20 μL containing 1 μL RNA sample (100 ng RNA), 0.8 μL of each specific forward and reverse primer, 1 μL 20 × RTase, 10 μL 2 × qPCRBIO Sygreen One-Step mix, and 6.4 μL RNase-free water. The thermal cycling conditions were as follows: 45 °C for 30 min; 95 °C for 2 min; 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. A list of primer pairs used for RT–qPCR is shown in Table 2. The RNase-free water was used instead of the RNA template for the negative control. The actin gene (ACT1) was used as an internal control. The relative gene expression was calculated using the 2−ΔΔCT method in which the target gene amount was adjusted to the reference gene (ACT1 gene).
Analytical methods and data analysis
Viable cell concentration was determined by a haemacytometer using the methylene blue staining technique35. The total sugars were analyzed by the phenol sulfuric acid method36 using a spectrophotometer (UV-1601, Shimadzu). The ethanol concentration was determined by gas chromatography (GC-14B, Shimadzu) using a packed column of polyethylene glycol (PEG-20 M) with a flame ionization detector37. The following equations were used to calculate the fermentation parameters: ethanol yield (Yp/s, g/g) = PE/[S0 − St]; volumetric ethanol productivity (Qp, g/L.h) = PE/t; yield efficiency (Ey, %) = [Yp/s/0.511] × 100; sugar consumption (Sc, %) = [St/S0] × 100, where PE is ethanol concentration (g/L), S0 is initial sugar concentration (g), St is sugar concentration (g) at time t, and t is fermentation time (h). The data are expressed as the mean ± standard deviation (SD). Analysis of variance was used to evaluate the differences among the treatments using Duncan's multiple range tests (DMRT). The statistical analysis was carried out using Statgraphics Centurion XV (Statpoint Technologies Inc., USA).
Results and discussion
Composition of dried pineapple waste and PWH
The dried pineapple waste had high contents of hemicelluloses (28.81%) and cellulose (16.57%), and the total crude fiber was 48.72%, while lignin comprised only 3.04% of the total dry matter. Previous studies by Niwaswong et al.38 reported that raw pineapple peel comprised 9.43% hemicellulose, 20.44% cellulose, and 41.21% lignin. Choonut et al.8 showed that 51.13% hemicellulose, 37.68% cellulose, and 10.24% lignin were detected in pineapple peel after hot water pretreatment at 100 °C for 240 min. The cellulose content of pineapple waste used in this study was lower than that of other agricultural wastes, such as rice straw (32–47% cellulose)39 and corn stover (38–40% cellulose)40. However, the cellulose and hemicellulose contents were greater than those of yam peel (5.7% cellulose, 5.1% hemicellulose) and cassava peel (12.7% cellulose, 5.5% hemicellulose)41.
Glucose and fructose were the principal sugars found in PWH, accounting for 41.11 and 40.87 g/L, respectively, while sucrose and maltose were not detected. Xylose and arabinose were also detected at 5.34 and 4.42 g/L, respectively. Rattanapoltee and Kaewkannetra31 reported that only 18.41 g/L glucose and 24.55 g/L fructose were present in pineapple peel hydrolysate. The total sugar concentration was only 55.91 g/L. Niwaswong et al.38 reported 82.10 g/L reducing sugars by dilute acid hydrolysis of pineapple peel waste. Formic acid, acetic acid, and furfural are considered inhibitors that are derived from acid hydrolysis. The acetic acid concentration of PWH was 8.39 g/L, whereas the concentrations of formic acid and furfural were 0.96 g/L and 0.36 mg/L, respectively, which were lower than those reported by Rattanapoltee and Kaewkannetra31.
The total sugars of PWH were 103.03 g/L, which was as high as the total sugars found in orange peel hydrolysate (101 g/L)33 and banana peel hydrolysate (155 g/L)42. Furthermore, the PWH also contains some minerals, such as nitrogen (686 mg/L), phosphorus (274 mg/L), magnesium (126 mg/L), manganese (34 mg/L), and zinc (5 mg/L), which are essential for yeast growth and metabolic activity. Based on the sugar and mineral contents presented in PWH, it was considered a promising potential feedstock for ethanol and other biochemical production.
Effect of inorganic nitrogen sources on ethanol production
The supplementation of inorganic nitrogen sources into the PWH did not significantly increase the final ethanol concentrations compared to the control treatment without inorganic nitrogen supplementation. The highest ethanol concentrations of 34.60, 34.56, and 33.55 g/L were achieved from the medium supplemented with NH4NO3, (NH4)2SO4, and (NH4)2HPO4, respectively, which were not significantly different from the control treatment (33.54 g/L). Furthermore, supplementation with CO(NH2)2 resulted in a lower ethanol concentration than the control (Table 3). Due to the low sugar content of PWH, S. cerevisiae HG1.1 quickly converted all sugars to reach the maximum ethanol concentration with a low consumption level of nitrogen sources. On the other hand, PWH may contain sufficient nitrogen sources for yeast growth and metabolic activity. Generally, nitrogen is essential when the fermentation process is carried out at a high initial sugar concentration (for example, greater than 200 g/L)43. The results in the present study coincide with those reported by Charoensopharat et al.32 and Arora et al.44.
The C/N ratio of the fermentation medium also played a crucial role in ethanol production. One study reported the optimum C/N ratio of 7.9 for ethanol production from sago starch using recombinant S. cerevisiae YKU 13145, while the other reported the value of 35.2 for ethanol production from tapioca starch using co-culture of Aspergillus niger and S. cerevisiae46. In this study, the C/N ratio of the fermentation medium was not determined; thus, further study is needed to clarify this hypothesis. Based on the results in this study, inorganic nitrogen was not selected as the independent variable in the experiment on screening factors that affected ethanol production using a Plackett–Burman design (PBD).
Screening of significant factors for ethanol production by S. cerevisiae HG1.1 using a Plackett and Burman design (PBD)
The PBD used 7 independent factors and 12 experimental runs. The maximal ethanol concentration of 32.73 g/L and volumetric ethanol productivity of 2.18 g/L.h were achieved after 15 h of fermentation at 40 °C (Table 4). Three factors, including initial cell concentration (A), pH (B), and yeast extract (G), were the most significant variables in ethanol production from PWH by the thermotolerant yeast S. cerevisiae HG1.1, with p values < 0.05 (Table 5). Analysis of the adequate levels of these three crucial factors showed that the most influential factor was pH (p value was 0.0002), and yeast extract was the most negligible influential factor (p value was 0.0485). The selected model was significant (p value < 0.005), with a high confidence level based on the values of R-squared (0.9837) and adjusted R-squared (0.9551). Based on the t value limit on the Pareto chart (Fig. 1), three variables, including initial cell concentration (A), pH (B), and yeast extract (G), were considered significant variables. All three variables positively affected ethanol production from PWH using S. cerevisiae HG1.1.
In PWH with low total sugars (ca. 103 g/L), both inorganic nitrogen sources and other salts were unnecessary. However, the initial cell concentration strongly affects the ethanol production rate. Higher initial cell concentrations can promote the fermentation rate and ethanol production efficiency. Techaparin et al.28 reported that when the initial cell concentration increased from 1.0 × 107 to 3.0 × 108 cells/mL, the ethanol concentrations from sweet sorghum juice were raised from 64.79 to 84.32 g/L using S. cerevisiae KKU-VN8. Greater than ten times higher ethanol productivity from hydrolyzed sugarcane bagasse was achieved when the inoculum size of S. cerevisiae ITV-01 was increased from 0.2 to 10 g/L47.
Yeast growth and fermentation activity are affected directly by the pH of the fermentation medium. The enzymes involved in the yeast growth and ethanol production pathway may be inactivated at a low pH level48. Although S. cerevisiae can grow well at pH values between 4.0 and 6.0, the optimum pH for ethanol production is approximately 5.0–5.5. Singh and Bishnoi49 demonstrated that pH 5.5 was the optimum pH value from statistical optimization of ethanol production from pretreated wheat straw hydrolysate using S. cerevisiae MTCC 174. Izmirlioglu and Demirci50 also found that pH 5.5 was the optimum value for ethanol production from potato mash waste using S. cerevisiae ATCC 24,859, which yielded a 30.99 g/L ethanol concentration. This pH value was also the optimum condition for ethanol production using K. marxianus NIRE-K3 at 45 °C, providing a 93.2% yield efficiency and 0.48 g/g ethanol yield44.
Yeast extract is widely used as the primary organic nitrogen source in several ethanol fermentation processes. It has been recognized as having a highly positive effect on ethanol production44,51. In the present study, 4.95 g/L yeast extract was the optimum concentration for ethanol production from PWH by S. cerevisiae HG1.1. Different optimum concentrations of yeast extract for ethanol production have also been reported. For instance, Schnierda et al.51 demonstrated that 9.43 and 9.24 g/L ethanol were attained from molasses-based medium (20 g/L sugar) supplemented with 0.5 g/L total yeast assimilable nitrogen by S. cerevisiae EC1118 and I. orientalis Y1161, respectively. Yeast extract at 3.0 g/L was determined to be the optimum concentration for ethanol production by S. cerevisiae NP01 using a fermentation medium containing 280 g/L sucrose52.
In comparison, yeast extract at 9.0 g/L promoted ethanol production from sweet sorghum juice containing 270 g/L total sugars using S. cerevisiae NP0153. Yeast extract is essential for efficient ethanol fermentation, especially under very high gravity fermentation conditions and high-temperature fermentation processes, but the most challenging factor is the high cost. Therefore, many scientists have tried to replace other low-cost organic nitrogen sources, such as dried spent yeast, corn-steep liquor, poultry meal, and feather meal. They have also demonstrated their potential application in the production of ethanol and other biochemicals32,52.
Optimization conditions for ethanol production by S. cerevisiae HG1.1 using CCD
The experimental design codes and actual values of the significant independent factors, including initial cell concentration (5.0 × 106 to 1.0 × 108 cells/mL), pH (4.0 to 6.5), and yeast extract (3.0 to 12.0 g/L), are shown in Table 6. The observed ethanol concentrations from the CCD with 20 experimental runs were 19.10–33.54 g/L, and the predicted ethanol concentrations were 19.49–33.78 g/L (Table 7). The ethanol productivities were 1.59–2.80 g/L.h. The quadratic polynomial regression model and a second-order polynomial equation to predict the final ethanol concentration (PE) as a function of the fermentation variables were established, and the prediction equation was as follows:
The results revealed that the model was statistically significant (p value < 0.0001) (Table 8). The model was reliable because the p value of lack of fit was not statistically significant (p value > 0.005), the R-squared was 0.9808, and the adjusted R-squared was 0.9635, which was close to the R-squared. The standard deviation and coefficient values were only 0.71 and 2.40%, respectively. ANOVA also demonstrated that all these factors strongly affected ethanol production from PWH by S. cerevisiae HG1.1 at 40 °C. The p values of these significant factors were less than 0.0001.
The 3-D response surfaces and contour plots for ethanol are presented in Fig. 2. The most fixed model was achieved when the yeast extract value was fixed at 7.50 g/L, and the cell concentration and pH levels were varied. The ethanol concentration was strongly affected by both cell concentration and pH. The maximum ethanol concentration of 33.54 g/L was achieved after 15 h of fermentation at the center pH value (pH 5.5) and cell concentration of 1.0 × 108 cells/mL. The maximum ethanol productivity (2.80 g/L.h) was also attained. Based on the three-factor quadratic polynomial equation, the maximum predicted ethanol concentration was 33.67 g/L under the optimum conditions: cell concentration of 8.0 × 107 cells/mL, pH of 5.4, and yeast extract concentration of 4.9 g/L.
Based on the result of the CCD experiment and the solution of the three-factor quadratic polynomial equation, three runs of experiments that gave high levels of ethanol were selected for a confirmatory experiment. A cell concentration of 8.0 × 107 cells/mL, pH values in the range of 5.39–5.50, and yeast extract concentrations of 4.90–4.97 g/L were chosen for the confirmation test. The maximum ethanol concentration of 36.85 g/L, the productivity of 3.07 g/L.h, the ethanol yield of 0.48 g/g, corresponding to a yield efficiency of 93.61%, and sugar consumption of 74.81% were achieved under the optimum conditions, i.e., cell concentration of 8.0 × 107 cells/mL, pH of 5.5, and yeast extract concentration of 4.95 g/L. The ethanol concentrations of these three confirmatory runs were not significantly different (36.07–36.85 g/L) (Table 9).
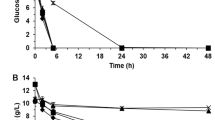
Figure 3 shows the time profile of ethanol production from PWH at 40 °C using S. cerevisiae HG1.1. The ethanol concentration quickly reached the maximal value (36.85 g/L) after 12 h of fermentation, corresponding to the dramatic decrease in total sugars (from 102.98 to 25.93 g/L). The ethanol content was slightly decreased after it reached the maximum concentration due to the oxidation of ethanol by yeast when the sugar in the fermentation medium was depleted. The remaining sugars, mostly C-5 sugars, such as xylose and arabinose, were almost unchanged since S. cerevisiae could not consume this type of sugar. The remaining total sugars in the fermented medium were 21.79 g/L. Although PWH contained some fermentation inhibitors, such as acetic acid (8.23 g/L), formic acid (0.96 g/L), and furfural (0.68 mg/L), the growth and fermentation activity of S. cerevisiae HG1.1 were not affected. The ethanol concentration, productivity, and yield efficiency achieved from PWH by S. cerevisiae HG1.1 at 40 °C were relatively high compared to several previous studies summarized in Table 10. This finding suggests that pineapple waste is a promising agricultural waste for second-generation bioethanol production.
Time profile of ethanol production from pineapple waste hydrolysate using S. cerevisiae HG1.1 under optimum conditions. Symbols: filled black rhombus, ethanol; unfilled rhombus, total sugars; filled black square, glucose; unfilled square, fructose and xylose; unfilled circle, arabinose; unfilled triangle, formic acid; filled black triangle, acetic acid; filled black circle, yeast cells.
RT–qPCR analysis of gene expression in thermotolerant S. cerevisiae HG1.1
Hundreds of genes possess different expressions in response to heat stress in yeast cells60,61,62. However, most previous gene expression studies were carried out using the synthetic medium. Only a few studies have shown the gene expression pattern using lignocellulosic materials as a feedstock. Thus, this study evaluated the expression of some groups of genes related to growth and ethanol production pathway, ethanol, oxidative and acetic acid stress, and DNA repair.
The expression levels of sixteen genes responsible for growth and ethanol stress (ATP6, OLE1, ERG8), oxidative stress (GLR1, SOD1), DNA repair (RAD14, MRE11, POL4), the pyruvate-to-TCA pathway (PDA1, CIT1, LYS21), the pyruvate-to-ethanol pathway (PDC1, ADH1, ADH2), and acetic acid stress (ACS1, ALD2) in S. cerevisiae HG 1.1 were successfully evaluated using RT–qPCR. As shown in Table 11, ERG8, RAD14, ADH2, and ALD2 were up-regulated under a control growth condition (30 °C) and down-regulated under heat stress. ATP6, GLR1, SOD1, CIT1, LYS21, ADH1, and ACS1 genes were highly expressed under heat shock at 40 °C for 30 min and markedly decreased under short- and long-term stress conditions. In contrast, the expression levels of OLE1, MRE11, POL4, PDA1, and PDC1 were highly increased when yeast cells were shifted from 30 °C to 40 °C for 3 h (short-term stress) and then dramatically decreased when cells were shifted from 30 °C to 40 °C for 9 h (long-term stress). Under heat stress, the expression levels of the PDC1 gene were more remarkable than that under a control condition. Relatively low expression levels of the RAD14 gene responsible for DNA repair and the ALD2 gene responsible for acetic acid stress were detected in yeast cells under long-term stress. It should be noted from the present study that genes involved in the same stress condition exhibited different expression patterns, suggesting their unique expression profile during high-temperature ethanol fermentation.
ATP6, OLE1, and ERG8 are essential genes responsible for yeast growth and ethanol stress63. In response to heat and ethanol stresses, more ATP is needed for several biosynthesis processes that produce critical components to protect microbial cells, such as trehalose, glycogen, unsaturated fatty acids and heat shock proteins63. As shown in the present study, the expression of ATP6 was triggered by a heat shock condition, and its expression was slightly decreased under short- and long-term heat stresses. ATP6 mitochondrially encoded subunit a of the F0 sector of mitochondrial F1F0 ATP synthase. It integrates into the F0F1-ATPase complex and completes the process to yield a functional ATPase64. Thus, a high level of ATP6 expression in S. cerevisiae HG1.1 might be correlated with ATP production under heat shock conditions. In S. cerevisiae sun049T and K. marxianus DMKU 3–1042, the ATP6 gene is also highly up-regulated during high-temperature ethanol production at 38°C65 and 45°C60, respectively.
OLE1, encoding a fatty acid desaturase, synthesizes monounsaturated fatty acids, such as palmitoleic acid and oleic acid, from saturated fatty acids, such as palmitic acid and stearic acid66. These unsaturated fatty acids and ergosterol maintain membrane fluidity as an adaptive response to the physiochemical interaction of both temperature and ethanol stresses67. It has been reported in S. cerevisiae that the overexpression of the OLE1 gene enhanced acetic acid tolerance and other stresses, such as ethanol, H2O2, NaCl, benzoic acid, diamide, and menadione68. In this study, the OLE1 gene of S. cerevisiae HG1.1 was highly expressed under heat shock and short-term heat stress, which was different from that of Qiu and Jiang69, who demonstrated that the OLE1 gene of S. cerevisiae M1 was approximately 2.2–3.0-fold overexpressed at 30 °C under very high gravity ethanol production. The overexpression of the OLE1 gene at 30 °C was also observed in S. cerevisiae K-9 under shaking and static sake fermentation70. Based on the present study, OLE1 is a heat-shock responsive gene in S. cerevisiae HG1.1. The ERG8 gene encodes phosphomevalonate kinase, which converts phosphomevalonate to diphosphomevalonate using ATP in ergosterol biosynthesis71. In the present study, the ERG8 gene was down-regulated under heat stress conditions, which coincided with Rossignol et al.72, who pointed out that most of the genes encoding proteins involved in ergosterol biosynthesis in S. cerevisiae EC1118, including ERG8, were down-regulated at high-temperature fermentation. In S. cerevisiae YZ1 and YF3, low levels of ERG8 expression were also observed under high-temperature conditions (42 °C), resulting in a reduction in ergosterol accumulation73.
DNA damage, including base disruption, base loss, and strand breaks, is not only induced by exposure to environmental agents, such as heat, UV rays, ROS, and oxidizing agents but also spontaneously generated during cellular metabolism18. RAD14, MRE11 and POL4 are common genes that encode proteins or enzymes involved in the DNA repair of yeast74,75. The expression of the RAD14 gene in S. cerevisiae HG1.1 was decreased under heat stress, similar to that reported by Boiteux and Jinks-Robertson76. The RAD14 gene is recognized as a DNA damage binding factor for nucleotide excision repair in the UV-damaged DNA of S. cerevisiae. This gene is not induced by heat stress. The other two genes, i.e., MRE11 and POL4, were up-regulated under heat shock and short-term heat stress conditions, and their expression was slightly reduced under long-term heat stress. The present results were similar to those of the MRE11 and POL4 genes in K. marxianus DMKU 3–1042, in which both genes were up-regulated under high-temperature stress60. It was proposed from the present study that MRE11 and POL4 are involved in the DNA repair of S. cerevisiae HG1.1 under heat stress conditions.
Oxidative stress, by increasing the accumulation of reactive oxygen species (ROS), such as superoxide anions, hydrogen peroxide, and hydroxyl radicals, has been shown to cause denaturation of macromolecules, such as DNA, RNA, proteins, and lipids, in yeast cells. Several genes, including SOD1 and GLR1, are responsible for oxidative stress in yeasts. The SOD1 gene encodes superoxide dismutase, while the GLR1 gene encodes glutathione reductase. Both proteins have been shown to scavenge the superoxide anion radical and hydrogen peroxide, which can then be converted to water by the action of catalases or peroxidases19. The up-regulation of SOD1 and GLR1 genes under heat stress might correlate with high ROS accumulation. The overproduction of superoxide dismutase and glutathione reductase in S. cerevisiae HG1.1 might be needed to convert oxidative substrates such as superoxide anion radicals to hydrogen peroxide and finally to H2O. Overexpression of SOD1 and GLR1 has also been reported in K. marxianus DMKU 3–1042 and S. cerevisiae YZ1 and YF3 under heat stress60,77. In S. cerevisiae M1, up-regulation of the SOD1 gene and down-regulation of the GLR1 gene under high ethanol and high osmotic pressure have been reported69. Based on this information, the SOD1 gene, but not GLR1, can be activated by heat, ethanol, and osmotic stresses, depending on the yeast species.
PDA1 and CIT1 are involved in the pyruvate-to-TCA pathway. These genes in S. cerevisiae HG1.1 were up-regulated under heat stress conditions, particularly under heat shock and short-term heat stress. Under long-term heat stress, the expression levels of both genes were slightly reduced. The expression of the PDA1 gene of S. cerevisiae HG1.1 was similar to that of S. cerevisiae Y-5031663. However, they differed somewhat from those reported in S. cerevisiae M1, where the PDA1 gene was down-regulated while CIT1 was up-regulated under heat stress69. In S. cerevisiae Y-50316, the expression of the PDA1 gene is activated not only by heat but also by ethanol stress63. A high expression level of the CIT1 gene has also been reported in S. cerevisiae when cells are exposed to a high temperature of 35 °C for 10 min. The increasing expression level of CIT1 increased the conversion of acetyl-CoA into the TCA pathway, leading to the accumulation of metabolic intermediates involved in the stress response16.
LYS21 encodes homocitrate synthase, which functions to synthesize homocitrate from acetyl-CoA and oxoglutarate. Homocitrate is a precursor for the biosynthesis of L-lysine, which plays an essential protective role in response to oxidative stress induced by hydrogen peroxide in S. cerevisiae78. Furthermore, the homocitrate synthase enzyme is also associated with the mechanism of DNA repair in the nucleus79. In the present study, the expression of LYS21 was enhanced under heat shock and short-term heat stress, and its expression slightly decreased after exposure to long-term heat stress. In K. marxianus DMKU 3–1042, the LYS21 gene is also up-regulated under heat stress at 45°C60. Therefore, it was proposed from this finding that the S. cerevisiae HG1.1 The LYS21 gene may be involved in DNA repair under heat stress.
In yeast cells, PDC1, ADH1, and ADH2 are involved in a pyruvate-to-ethanol pathway. These genes are highly expressed in the stationary growth phase of K. marxianus DBKKU Y-102 under heat stress at 45°C32. Down-regulation of the ADH2 gene has been reported in S. cerevisiae KKU-VN8 under heat stress at 40°C34. In S. cerevisiae Y-50316, the expression of ADH1 and ADH2 is also induced by ethanol stress63. In this study, PDC1 and ADH1, but not ADH2, were up-regulated under heat stress at 40 °C, suggesting that heat stress could trigger the expression of PDC1 and ADH1 genes while suppressing the expression of ADH2 in S. cerevisiae HG1.1 during high-temperature ethanol fermentation. A high ethanol concentration produced by S. cerevisiae HG1.1 at 40 °C might also be correlated with the overexpression of PDC1 and ADH1 genes.
Several genes, including ACS1 (encoded acetate-CoA ligase) and ALD2 (encoded aldehyde dehydrogenase), are responsible for acetic acid stress. The expression of these genes in S. cerevisiae HG1.1 under heat stress was investigated in this study. The results revealed that ACS1 was up-regulated under heat shock and short-term heat stress, whereas ALD2 was down-regulated under all stress conditions. The up-regulation of ACS1 in S. cerevisiae HG1.1 under heat stress may lead to a high formation of acetic acid from acetyl-CoA but not from acetaldehyde because aldehyde dehydrogenase also utilizes NAD(P) + . The conversion of acetyl-CoA to acetic acid might generate more ATP, which can be used as an energy source for the biosynthesis of essential components or enzymes critical for yeast adaptation under heat stress. In K. marxianus DMKU 3–1042, ACS1 and ALD2 are up-regulated under heat stress at 45°C60, while they are highly expressed in S. cerevisiae M1 under normal growth condition (30 °C)69, suggesting that their expression profiles depend on the yeast species.
Conclusion
The maximum ethanol concentration of 36.85 g/L, the productivity of 3.07 g/L.h, and yield efficiency of 93.61% were achieved after fermentation of PWH using S. cerevisiae HG1.1 at 40 °C under the optimum yeast inoculum concentration of 8.0 × 107 cells/mL, pH of 5.5, and yeast extract concentration of 4.95 g/L. The expression of genes during high-temperature ethanol fermentation using RT–qPCR revealed that most of the genes, except ERG8, RAD14, ADH2, and ALD2, were up-regulated under heat stress conditions, particularly under heat shock and short-term heat stress. Interestingly, up-regulation of the SOD1, PDA1, CIT1, PDC1, and ADH1 genes was observed under all stresses compared to a control treatment (unstress). Although the gene expression profiles were distinctive depending on the nature and characteristics of the yeasts, the thermotolerance acquisition and fermentation efficiency of S. cerevisiae HG1.1 during high-temperature ethanol fermentation correlated with genes responsible for growth and ethanol stress, oxidative stress, acetic acid stress, DNA repair, pyruvate-to-TCA, and pyruvate-to-ethanol pathway. These results provide useful information for further advanced research to explore the regulation of the genes to benefit this potential thermotolerant yeast for producing ethanol or other valuable bio-products under high-temperature fermentation conditions.
Submission declaration and verification
Submission of an article implies that the work described has not been published previously in any form.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Ho, D. P., Ngo, H. H. & Guo, W. A mini review on renewable sources for biofuel. Bioresour. Technol. 169, 742–749 (2014).
Zabed, H., Sahu, J. N., Suely, A., Boyce, A. N. & Faruq, G. Bioethanol production from renewable sources: Current perspectives and technological progress. Renew. Sustain. Energy Rev. 71, 475–501 (2017).
Liu, C. M. & Wu, S. Y. From biomass waste to biofuels and biomaterial building blocks. Renew. Energy. 96, 1056–1062 (2016).
Moka, S. et al. Alternative fuels: An overview of current trends and scope for future. Renew. Sustain. Energy Rev. 32, 697–712 (2014).
Naseeruddin, S., Desai, S. & Venkateswar, R. L. Ethanol production from lignocellulosic substrate Prosopis juliflora. Renew. Energy. 103, 701–707 (2017).
Farrell, A. E. et al. Ethanol can contribute to energy and environmental goals. Science 311, 506–508 (2006).
Ketnawa, S., Chaiwut, P. & Rawdkuen, S. Pineapple wastes: A potential source for bromelain extraction. Food Bioprod. Process. 90, 385–391 (2012).
Choonut, A., Saejong, M. & Sangkharak, K. The production of ethanol and hydrogen from pineapple peel by Saccharomyces cerevisiae and Enterobacter aerogenes. Energy Procedia. 52, 242–249 (2014).
Aditiya, H. B. et al. Second generation bioethanol potential from selected Malaysia’s biodiversity biomasses: A review. Waste Manag. 47, 46–61 (2016).
FAO regional office for Asia and the Pacific. FAO Statistical Yearbook 2014: Asia and the Pacific Food and Agriculture. (Bangkok, 2014).
Sree, K. N., Sridhar, M., Suresh, K., Banat, I. M. & Venkateswar, R. L. Isolation of thermotolerant, osmotolerant, flocculating Saccharomyces cerevisiae for ethanol production. Bioresour. Technol. 72, 43–46 (2000).
Yuangsaard, N., Yongmanitchai, W., Yamada, M. & Limtong, S. Selection and characterization of a newly isolated thermotolerant Pichia kudriavzevii strain for ethanol production at high temperature from cassava starch hydrolysate. Antonie Van Leeuwenhoek 103, 577–588 (2013).
Kwon, Y. J., Wang, F. & Liu, C. Z. Deep-bed solid state fermentation of sweet sorghum stalk to ethanol by thermotolerant Issatchenkia orientalis IPE 100. Bioresour. Technol. 102, 11262–11265 (2011).
Abdel-Banat, B. M., Hoshida, H., Ano, A., Nonklang, S. & Akada, R. High-temperature fermentation: How can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl. Microbiol. Biotechnol. 85, 861–867 (2010).
Limtong, S., Sringiew, C. & Yongmanitchai, W. Production of fuel ethanol at high temperature from sugar cane juice by a newly isolated Kluyveromyces marxianus. Bioresour. Technol. 98, 3367–3374 (2007).
Richter, K., Haslbeck, M. & Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell. 40, 253–266 (2010).
Cadet, J. & Wagner, J. R. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect Biol. 5, a012559 (2013).
Tiwari, S., Thakur, R. & Shankar, J. Role of heat-shock proteins in cellular function and in the biology of fungi. Biotechnol. Res. Int. 2015, 132635 (2015).
Palma, M., Guerreiro, J. F. & Sá-Correia, I. Adaptive response and tolerance to acetic acid in Saccharomyces cerevisiae and Zygosaccharomyces bailii: A physiological genomics perspective. Front. Microbiol. 9, 274 (2018).
Azhar, S. H. M. et al. Yeasts in sustainable bioethanol production: A review. Biochem. Biophys. Rep. 10, 52–61 (2017).
Khatun, M. M., Yu, X., Kondo, A., Bai, F. & Zhao, X. Improved ethanol production at high temperature by consolidated bioprocessing using Saccharomyces cerevisiae strain engineered with artificial zinc finger protein. Bioresour. Technol. 245, 1447–1454 (2017).
Thammasittirong, S. N. R., Thirasaktana, T., Thammasittirong, A. & Srisodsuk, M. Improvement of ethanol production by ethanol-tolerant Saccharomyces cerevisiae UVNR56. Springerplus 2, 583 (2013).
Phong, H. X., Klanrit, P., Dung, N. T. P., Yamada, M. & Thanonkeo, P. Isolation and characterization of thermotolerant yeasts for the production of second-generation bioethanol. Ann. Microbiol. 69, 765–776 (2019).
Singh, V. et al. Strategies for fermentation medium optimization: An in-depth review. Front. Microbiol. 7, 2087 (2016).
Antony, J. Design of Experiments for Engineers and Scientists 2nd edn. (Elsevier Ltd., 2014).
Khuri, A. I. & Mukhopadhyay, S. Response surface methodology. Wiley Interdiscip. Rev. Comput. Stat. 2, 128–149 (2010).
Nuanpeng, S., Thanonkeo, S., Yamada, M. & Thanonkeo, P. Ethanol production from sweet sorghum juice at high temperatures using a newly isolated thermotolerant yeast Saccharomyces cerevisiae DBKKU Y-53. Energies 9, 253 (2016).
Techaparin, A., Thanonkeo, P. & Klanrit, P. High-temperature ethanol production using thermotolerant yeast newly isolated from Greater Mekong Subregion. Braz. J. Microbiol. 48, 465–475 (2017).
Aruwajoye, G. S., Faloye, F. D. & Kana, E. G. Process optimisation of enzymatic saccharification of soaking assisted and thermal pretreated cassava peels waste for bioethanol. Waste Biomass Valor. 11, 2409–2420 (2020).
Vogel, K. P. et al. Evaluation of a filter bag system forage analysis. Crop Sci. 39, 276–279 (1999).
Rattanapoltee, P. & Kaewkannetra, P. Utilization of agricultural residues of pineapple peels and sugarcane bagasse as cost-saving raw materials in Scenedesmus acutus for lipid accumulation and biodiesel production. Appl. Biochem. Biotechnol. 173, 1495–1510 (2014).
Charoensopharat, K., Thanonkeo, P., Thanonkeo, S. & Yamada, M. Ethanol production from Jerusalem artichoke tubers at high temperature by newly isolated thermotolerant inulin-utilizing yeast Kluyveromyces marxianus using consolidated bioprocessing. A. Van Leeuw. J. Microb. 108, 173–190 (2015).
Koutinas, M., Patsalou, M., Stavrinou, S. & Vyrides, I. High temperature alcoholic fermentation of orange peel by the newly isolated thermotolerant Pichia kudriavzevii KVMP10. Lett. Appl. Microbiol. 62, 75–83 (2015).
Techaparin, A., Thanonkeo, P. & Klanrit, P. Gene expression profiles of the thermotolerant yeast Saccharomyces cerevisiae strain KKU-VN8 during high-temperature ethanol fermentation using sweet sorghum juice. Biotechnol. Lett. 39, 1521–1527 (2017).
Zoecklein, B. W., Fugelsang, K. C., Gump, B. H. & Nury, F. S. Laboratory Procedures. In Wine Analysis and Production (eds Zoecklein, B. W. et al.) 310–516 (Springer, 1995).
Mecozzi, M. Estimation of total carbohydrate amount in environmental samples by the phenol-sulphuric acid method assisted by multivariate calibration. Chemom. Intell. Lab. Syst. 79, 84–90 (2005).
Laopaiboon, L., Nuanpeng, S., Srinophakun, P., Klanrit, P. & Laopaiboon, P. Ethanol production from sweet sorghum juice using very high gravity technology: Effects of carbon and nitrogen supplementations. Bioresour. Technol. 100, 4176–4182 (2009).
Niwaswong, C., Chaiyamate, P., Chotikosaikanon, P. & Ruangviriyachai, C. Simple and enhanced production of lignocellulosic ethanol by diluted acid hydrolysis process of pineapple peel (Ananas comosus) waste. African. J. Biotechnol. 13, 3928–3934 (2014).
Sarkar, N., Ghosh, S. K., Bannerjee, S. & Aikat, K. Bioethanol production from agricultural wastes: An overview. Renew. Energy. 37, 19–27 (2012).
Saini, J. K., Saini, R. & Tewari, L. Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: Concepts and recent developments. Biotech. 5, 337–353 (2014).
Thomsen, S. T., Kádár, Z. & Schmidt, J. E. Compositional analysis and projected biofuel potentials from common West African agricultural residues. Biomass Bioenergy. 63, 210–217 (2014).
Deepshika, K. & Chauhan, K. Chemo-enzymatic conversion of biomass into bio-ethanol. J. Integr. Sci. Technol. 2, 34–36 (2014).
Khongsay, N., Lin, Y. H., Laopaiboon, P. & Laopaiboon, L. Improvement of very-high-gravity ethanol fermentation from sweet sorghum juice by controlling fermentation redox potential. J. Taiwan Inst. Chem. Eng. 45, 302–307 (2014).
Arora, R., Behera, S., Sharma, N. K. & Kumar, S. Augmentation of ethanol production through statistically designed growth and fermentation medium using novel thermotolerant yeast isolates. Renew. Energy. 109, 406–421 (2017).
Abd-Aziz, S. et al. Effect of C/N ratio and starch concentration on ethanol production from sago starch using recombinant yeast. World J. Microbiol. Biotechnol. 17, 713–719 (2001).
Manikandan, K. & Viruthagiri, T. Optimization of C/N ratio of the medium and fermentation conditions of ethanol production from tapioca starch using co-culture of Aspergillus niger and Saccharomyces cerevisiae. Int. J. ChemTech Res. 2, 947–955 (2010).
Gutiérrez-Rivera, B. et al. Bioethanol production from hydrolyzed sugarcane bagasse supplemented with molasses “B” in a mixed yeast culture. Renew. Energy. 74, 399–405 (2015).
Narendranath, N. V. & Power, R. Relationship between pH and medium dissolved solids in terms of growth and metabolism of Lactobacilli and Saccharomyces cerevisiae during ethanol production. Appl. Environ. Microbiol. 71, 2239–2243 (2005).
Singh, A. & Bishnoi, N. R. Ethanol production from pretreated wheat straw hydrolyzate by Saccharomyces cerevisiae via sequential statistical optimization. Ind. Crops Prod. 41, 221–226 (2013).
Izmirlioglu, G. & Demirci, A. Ethanol production from waste potato mash by using Saccharomyces cerevisiae. Appl. Sci. 2, 738–753 (2012).
Schnierda, T., Bauer, F. F., Divol, B., van Rensburg, E. & Görgens, J. F. Optimization of carbon and nitrogen medium components for biomass production using non-Saccharomyces wine yeasts. Lett. Appl. Microbiol. 58, 478–485 (2014).
Chan-u-tit, P., Laopaiboon, L., Jaisil, P. & Laopaiboon, P. High level ethanol production by nitrogen and osmoprotectant supplementation under very high gravity fermentation conditions. Energies 6, 884–899 (2013).
Deesuth, O., Laopaiboon, P., Klanrit, P. & Laopaiboon, L. Improvement of ethanol production from sweet sorghum juice under high gravity and very high gravity conditions: Effects of nutrient supplementation and aeration. Ind. Crops Prod. 74, 95–102 (2015).
Oberoi, H. S., Vadlani, P. V., Saida, L., Bansal, S. & Hughes, J. D. Ethanol production from banana peels using statistically optimized simultaneous saccharification and fermentation process. Waste Manag. 31, 1576–1584 (2011).
Huang, R. et al. Enhanced ethanol production from pomelo peel waste by integrated hydrothermal treatment, multienzyme formulation, and fed-batch operation. J. Agric. Food Chem. 62, 4643–4651 (2014).
Wang, G. et al. Production of bioethanol from rice straw by simultaneous saccharification and fermentation of whole pretreated slurry using Saccharomyces cerevisiae KF-7. Environ. Prog. Sustain. Energy. 34, 582–588 (2015).
Yu, C. Y., Jiang, B. H. & Duan, K. J. Production of bioethanol from carrot pomace using the thermotolerant yeast Kluyveromyces marxianus. Energies 6, 1794–1801 (2013).
Yuan, S. F., Guo, G. L. & Hwang, W. S. Ethanol production from dilute-acid steam exploded lignocellulosic feedstocks using an isolated multistress-tolerant Pichia kudriavzevii strain. Microb. Biotechnol. 10, 1581–1590 (2017).
Kwon, Y. J. et al. Effect of lignocellulosic inhibitory compounds on growth and ethanol fermentation of newly-isolated thermotolerant Issatchenkia orientalis. Bioresour. Technol. 102, 8099–8104 (2011).
Lertwattanasakul, N. et al. Genetic basis of the highly efficient yeast Kluyveromyces marxianus: Complete genome sequence and transcriptome analyses. Biotechnol. Biofuels. 8, 47 (2015).
Varol, D., Purutçuoglu, V. & Yilmaz, R. Transcriptomic analysis of the heat stress response for a commercial baker’s yeast Saccharomyces cerevisiae. Genes Genom. 40, 137–150 (2018).
Mühlhofer, M. et al. The heat shock response in yeast maintains protein homeostasis by chaperoning and replenishing proteins. Cell Rep. 29, 4593–4607 (2019).
Ma, M. & Liu, L. Z. Quantitative transcription dynamic analysis reveals candidate genes and key regulators for ethanol tolerance in Saccharomyces cerevisiae. BMC Microbiol. 10, 169 (2010).
García, J. J., Ogilvie, I., Robinson, B. H. & Capaldi, R. A. Structure, functioning, and assembly of the ATP synthase in cells from patients with the T8993G mitochondrial DNA mutation. J. Biol. Chem. 275, 11075–11081 (2000).
Ismail, K. S. K., Sakamoto, T., Hatanaka, H., Hasunuma, T. & Kondo, A. Gene expression cross-profiling in genetically modified industrial Saccharomyces cerevisiae strains during high-temperature ethanol production from xylose. J. Biotechnol. 163, 50–60 (2013).
Kim, H. S., Kim, N. R. & Choi, W. Total fatty acid content of the plasma membrane of Saccharomyces cerevisiae is more responsible for ethanol tolerance than the degree of unsaturation. Biotechnol. Lett. 33, 509–515 (2011).
Vanegas, J. M., Contreras, M. F., Faller, R. & Longo, M. L. Role of unsaturated lipid and ergosterol in ethanol tolerance of model yeast biomembranes. Biophys. J. 102, 507–516 (2012).
Nasution, O. et al. Overexpression of OLE1 enhances stress tolerance and constitutively activates the MAPK HOG pathway in Saccharomyces cerevisiae. Biotechnol. Bioeng. 114, 620–631 (2017).
Qiu, Z. & Jiang, R. Improving Saccharomyces cerevisiae ethanol production and tolerance via RNA polymerase II subunit Rpb7. Biotechnol. Biofuels. 10, 125 (2017).
Shobayashi, M., Ukena, E., Fujii, T. & Iefuji, H. Genome-wide expression profile of sake brewing yeast under shaking and static conditions. Biosci. Biotechnol. Biochem. 71, 323–335 (2007).
Chemler, J., Yan, Y. & Koffas, M. Biosynthesis of isoprenoids, polyunsaturated fatty acids and flavonoids in Saccharomyces cerevisiae. Microb. Cell Fact. 5, 20 (2006).
Rossignol, T., Dulau, L., Julien, A. & Blondin, B. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20, 1369–1385 (2003).
Zhang, M., Shi, J. & Jiang, L. Modulation of mitochondrial membrane integrity and ROS formation by high temperature in Saccharomyces cerevisiae. Electron. J. Biotechnol. 18, 202–209 (2015).
Liu, Y. et al. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 35, 743–758 (2016).
Galli, A., Chan, C. Y., Parfenova, L., Cervelli, T. & Schiestl, R. H. Requirement of POL3 and POL4 on non-homologous and microhomology-mediated end joining in rad50/xrs2 mutants of Saccharomyces cerevisiae. Mutagenesis 30, 841–849 (2015).
Boiteux, S. & Jinks-Robertson, S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 193, 1025–1064 (2013).
Landolfo, S., Politi, H., Angelozzi, D. & Mannazzu, I. ROS accumulation and oxidative damage to cell structures in Saccharomyces cerevisiae wine strains during fermentation of high-sugar-containing medium. Biochim. Biophys. Acta. Gen. Subj. 1780, 892–898 (2008).
O’Doherty, P. J. et al. Transcriptomic and biochemical evidence for the role of lysine biosynthesis against linoleic acid hydroperoxide-induced stress in Saccharomyces cerevisiae. Free Radic. Res. 48, 1454–1461 (2014).
Scott, E. M. & Pillus, L. Homocitrate synthase connects amino acid metabolism to chromatin functions through Esa1 and DNA damage. Genes. Dev. 24, 1903–1913 (2010).
Acknowledgements
The authors would like to thank the Faculty of Technology, Khon Kaen University, for a scholarship for HXP for Academic Year 2014 and the International Research Funding by the Department of Biotechnology, Faculty of Technology, Khon Kaen University, and the Graduate School, Khon Kaen University, Thailand for partial financial support. The authors also thank the Japan Society for the Promotion of Science (JSPS, Japan) and the International Collaboration Research Project Core-to-Core Program (CCP, 2014-2019) for support. The authors also thank the Center for Alternative Energy Research and Development (AERD), Khon Kaen University, for financial support.
Funding
This research was financially supported by the Center for Alternative Energy Research and Development (AERD), Khon Kaen University (R03-63).
Author information
Authors and Affiliations
Contributions
H.X.P. conducted the experiments, analyzed the data, and prepared the manuscript. P.K., S.T., and P.T. conceived the ideas, designed the experiments, analyzed the data, and revised and submitted the manuscript. N.T.P.D. and M.Y. supervised the experimental work and contributed some chemicals, reagents, and analysis tools. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Phong, H.X., Klanrit, P., Dung, N.T.P. et al. High-temperature ethanol fermentation from pineapple waste hydrolysate and gene expression analysis of thermotolerant yeast Saccharomyces cerevisiae. Sci Rep 12, 13965 (2022). https://doi.org/10.1038/s41598-022-18212-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-18212-w
This article is cited by
-
Systemic metabolic engineering of Enterobacter aerogenes for efficient 2,3-butanediol production
Applied Microbiology and Biotechnology (2024)
-
Adaptive laboratory evolution under acetic acid stress enhances the multistress tolerance and ethanol production efficiency of Pichia kudriavzevii from lignocellulosic biomass
Scientific Reports (2023)
-
Isolation and identification of thermotolerant yeast strains producing bioethanol from agro-food wastes
Biomass Conversion and Biorefinery (2023)
-
The potential of multistress tolerant yeast, Saccharomycodes ludwigii, for second-generation bioethanol production
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.