Abstract
Cryptosporidium spp. and Giardia duodenalis are commonly detected intestinal protozoa species in humans and animals, contributing to global gastroenteritis spread. The present study examined the prevalence and zoonotic potential of Cryptosporidium spp. and G. duodenalis in Himalayan marmots and Alashan ground squirrels in China's Qinghai-Tibetan Plateau area (QTPA) for the first time. Four hundred ninety-eight intestinal content samples were collected from five counties of QTPA of Gansu province, China. All samples were examined for Cryptosporidium spp. and G. duodenalis by PCR amplification. The resultant data were statistically analyzed by chi-square, Fisher's test and Bonferroni correction using SPSS software 25. 0. Cryptosporidium positive samples were further subtyped through analysis of the 60-kDa glycoprotein (gp60) gene sequence. A total of 11 and 8 samples were positive for Cryptosporidium spp. and G. duodenalis, respectively. Prevalence of Cryptosporidium spp. and G. duodenalis were 2.5% (10/399) and 1.5% (6/399) in Himalayan marmots, 1.0% (1/99) and 2.0% (2/99) in Alashan ground squirrels, respectively. Sequence analysis confirmed the presence of C. rubeyi (n = 2), ground squirrel genotype II (n = 7), chipmunk genotype V (n = 1) and horse genotype (n = 1). The horse genotype was further subtyped as novel subtype VIbA10. G. duodenalis zoonotic assemblages A (n = 1), B (n = 6), E (n = 1) were identified in the present study. This is the first study to identify Cryptosporidium spp. and G. duodenalis in Himalayan marmots and Alashan ground squirrels, suggesting the potential zoonotic transmission of the two pathogens in QTPA.
Similar content being viewed by others
Introduction
Cryptosporidium spp. and Giardia duodenalis are critical protozoan parasites responsible for diarrhea and infect a wide range of hosts including humans worldwide. Typically, contaminated food or water has been identified as the primary vehicle for Cryptosporidium spp. and G. duodenalis transmission1,2. Infection of these pathogens can also be acquired following contact with infected persons or animals directly2,3.
Currently, at least 45 valid Cryptosporidium spp. species and over 120 genotypes have been identified. Over 23 Cryptosporidium species/genotypes have been identified in humans, and C. hominis and C. parvum are the most common species (more than 90%) responsible for human cryptosporidiosis4,5,6,7,8,9,10,11,12. G. duodenalis is a complex protozoan species, and it has been divided into at least eight genetically different assemblages (A–H) based on genetic characterization. Among them, assemblages A and B are considered as critical zoonotic pathogens. Assemblages (C–H) are host-specific: assemblages C and D in canines, assemblage E in cloven-hoofed mammals, assemblage F in cats, assemblage G in rodents, and assemblage H in seals13. However, assemblages C, D, E and F have also been found in humans14.
Rodents can act as reservoirs or carriers for numerous zoonotic pathogens, including bacteria, parasites and viruses. Himalayan marmots (Marmota himalayana) and Alashan ground squirrels (Spermophilus alashanicus) are two common wild rodent species distributed widely in Qinghai-Tibetan Plateau area (QTPA) of China. They typically reside near livestock, water sources and human environments. Among them, infected hosts can play essential roles in environmental contamination by excreting oocysts/cysts via feces15. Some epidemiological studies also revealed the identity of Cryptosporidium spp. and G. duodenalis in numerous investigated hosts in QTPA, such as wild Qinghai voles, plateau pikas, wild birds, cattle, yaks and sheep16,17,18,19,20. Furthermore, the zoonotic species and genotypes of Cryptosporidium spp. and G. duodenalis were also reported in environmental samples in QTPA, including sewage and river water, slaughterhouse water and vegetables from street markets15,21. However, no previously study about the prevalence and transmission of Cryptosporidium spp. and G. duodenalis in Himalayan marmots and Alashan ground squirrels in China was reported. In the present study, a cross-sectional investigation was carried out in Himalayan marmots and Alashan ground squirrels to understand the prevalence of Cryptosporidium spp. and G. duodenalis and assess the zoonotic potential at the genotype and subtype levels.
Materials and methods
Sample collection
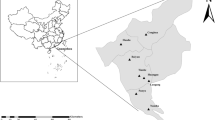
During a period of three months from June to September 2017, 399 Himalayan marmots and 99 Alashan ground squirrels were captured live by mousetraps from QTPA of western China’s Gansu Province (Fig. 1), with the former from Luqu (n = 98), Sunan (n = 100), Xiahe (n = 102) and Zhangye (n = 99) and latter from Huining County (n = 99) (Table 1). These animals were euthanized with a high dose of CO2 following security measures. Intestinal content materials were directly collected from each animal in the local Center for Disease Control and Prevention (CDC) laboratory and placed in 2 ml sterile tubes. They were kept in a freezer and then transported in ice packs to our laboratory in Shanghai for further molecular analysis.
Distribution of five sampling sites from Gansu Province. The map was created with software ArcGIS version 10.0 (URL: https://www.esri.com).
DNA extraction
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Cat. #69506; Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Extracted DNA was stored at − 20 °C in a freezer until further use.
PCR amplification
Cryptosporidium spp. was detected by nested PCR amplification of the fragment (approximately 830 bp) of the small subunit (SSU) rRNA gene22. Subtyping of Cryptosporidium spp. was performed by sequence analysis of the 60 kDa glycoprotein (gp60) gene23. All the isolates of Cryptosporidium-positive samples were selected for further sequence characterization via the actin gene and 70-kDa heat shock protein (HSP70) gene61,62. The assemblages of G. duodenalis were identified and subtyped by amplifying the β-giardin (bg), glutamate dehydrogenase (gdh) and triosephosphate isomerase (tpi)24,25,26. DNA of human-derived C. parvum and C. viatorum were used as positive controls in PCR tests to amplify the SSU rRNA, gp60, actin and HSP70 genes, respectively. Premiers and reaction conditions were shown in Supplementary Table S1. DNA of human-derived G. duodenalis was used as a positive control in PCR tests to amplify the bg, gdh and tpi genes. DNase-free water was used as a negative control in each PCR test. The secondary PCR products were visualized under UV light after electrophoresis on a 1.5% agarose gel containing GelRed (Biotium Inc., Hayward, CA, USA).
Nucleotide analysis
All secondary PCR amplicons of the expected size were sequenced on ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, USA) and Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequence accuracy was confirmed by bi-directional sequencing of all the PCR-positive products. Obtained DNA sequences were aligned with reference sequences deposited in GenBank databases (http://www.ncbi.nlm.nih.gov) using Clustal X (http://www.clustal.org/) to determine the species/subtypes of Cryptosporidium spp. and assemblages of G. duodenalis. Phylogenetic analyses at the SSU rRNA, actin, HSP70 and gp60 gene loci were performed using the neighbor-joining model in MEGA 11 (http://www.megasoftware.net). Bootstrap analysis was used to assess the robustness of the clusters using 1 000 replicates.
Statistical analysis
Differences in prevalence of Cryptosporidium spp. and G. duodenalis in Himalayan marmots and Alashan ground squirrels were compared among species and investigated area using the were processed with chi-square test, Fisher's exact test and pairwise comparisons used a Bonferroni correction to control for multiple testing. All the statistical analyses were performed using SPSS 25. 0 (SPSS Inc., New York, USA). Differences were considered significant at P < 0.05.
Ethics statements
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention. The protocol was approved by the Laboratory Animal Welfare & Ethics Committee (LAWEC), National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention (Permit Number: NIPD-2016-15).
Results
Prevalence of Cryptosporidium spp. and G. duodenalis
Using PCR amplification and sequence analysis, Cryptosporidium spp. and G. duodenalis were found in Himalayan marmots and Alashan ground squirrels. The agarose gel electrophoresis results of PCR amplification products were shown in Supplementary Fig. S1 (partial samples) and Fig. S2 (partial samples). A total of 11 and 8 samples were positive for Cryptosporidium spp. and G. duodenalis, respectively. Prevalence of Cryptosporidium spp. and G. duodenalis were 2.5% (10/399) and 1.5% (6/399) in Himalayan marmots, and 1.0% (1/99) and 2.0% (2/99) in Alashan ground squirrels, respectively (Table 1). The statistical analysis showed no significant difference in the prevalence of Cryptosporidium spp. (P = 0.365) and G. duodenalis (P = 0. 714) between Himalayan marmots and Alashan ground squirrels. Different prevalence of Cryptosporidium spp. and G. duodenalis were observed in five different investigated areas (Table 1): Luqu (0.0% and 0.0%), Sunan (7.0% and 0.0%), Xiahe (2.0% and 2.9%), Zhangye (1.0% and 3.0%) and Huining (1.0% and 2.0%). Moreover, there was no significant difference observed in the prevalence of Cryptosporidium spp. and G. duodenalis in each paired comparison between investigated areas (P > 0.05). No mixed infection of Cryptosporidium spp. and G. duodenalis identified in this study.
Cryptosporidium genotypes and subtypes
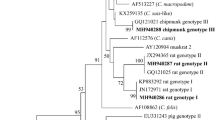
Based on sequence analysis of the SSU rRNA gene, a total four species/genotypes of Cryptosporidium spp. were identified out of 11 isolates, including C. rubeyi (n = 2), ground squirrel genotype II (n = 7), and chipmunk genotype V (n = 1) in Himalayan marmots, and horse genotype (n = 1) in Alashan ground squirrels. Cryptosporidium ground squirrel genotype II was dominant in Himalayan marmots, accounting for 70.0% (7/10) of Cryptosporidium isolates. At the SSU rRNA gene locus, the two identical sequences of C. rubeyi shared the most significant identity (98.43%) with that of C. rubeyi (DQ295012) from California ground squirrels in the USA, with 13 base differences. Seven sequences of ground squirrel genotype II were identical and shared the most prominent similarity (98.28%) to that of the ground squirrel genotype II (KT027480) from black-tailed prairie dogs, with 14 base differences. The sequence of the chipmunk genotype V had 98.90% homology with that (MW521250) of the chipmunk genotype V from chinchillas in China, with nine base differences. The sequence of the horse genotype obtained in the present study had 100% homology with a sequence (MK775040) from a horse in China. The horse genotype isolate was further subtyped by sequence analysis of the gp60 gene. This subtype belonged to the VIb subtype family and was identified as VIbA10 (GenBank: MW531716).
None of the two sequences of C. rubeyi were successfully amplified at the HSP70 gene locus but successfully amplified at the actin gene locus, and the two sequences were identical to each other, had 100% similarity with that of C. rubeyi (GenBank: KT027530) from black-tailed prairie dog. Meanwhile, two of seven isolates of ground squirrel genotype II were successfully amplified at the actin gene locus, and the two isolates shared the same sequence which had 97.68% similarity with that of ground squirrel genotype II (GenBank: KT027545) from black-tailed prairie dog in the USA. The HSP70 sequences have not been reported for ground squirrel genotype II. Three of seven isolates of ground squirrel genotype II were successfully amplified at the HSP70 gene locus and had 93.50% similarity with that of C. viatorum (GenBank: JX978274) from human in Guatemala. The sequence of chipmunk genotype V was only successfully amplified at the actin gene locus and shared 99.69% identity with that of chipmunk genotype V (MW521262) from chinchillas in China. Horse genotype was successfully amplified at the actin gene locus and shared 100% similarity with horse genotype (KU892571) isolated from humans of Kenya.
Phylogenetic analyses of the SSU rDNA, actin, HSP70 and gp60 gene sequences were shown in Figs. 2, 3, 4 and 5.
G. duodenalis assemblages
A total of eight G. duodenalis isolates were amplified and sequenced successfully in Himalayan marmots and Alashan ground squirrels in this study. Assemblages A, B and E were identified in one, four and one Himalayan marmot samples, respectively. Assemblage B was found in two Alashan ground squirrel samples. Meanwhile, assemblage B was observed to show a predominance (75.0%, 6/8) in the detected animals. The gdh and bg genes were successfully amplified in five samples—assemblages B (n = 4) and E (n = 1) and seven samples—assemblages A (n = 1), B (n = 5) and E (n = 1), respectively (Table 1). In this study, PCR amplification failed at the tpi locus.
At the gdh locus, two assemblage B sequences had 100% homology with beaver-derived assemblage B isolated (KM977648) from China. Another two different assemblage B sequences were 100% identical to golden monkey-derived assemblage B isolate (MK952602) from China, and one assemblage E sequence was 100% identical to a pig-derived assemblage E isolate (MK426742) from South Korea. At the bg locus, five assemblage B sequences shared 100% homology with squirrel monkey-derived assemblage B isolate (KJ888974) from China, one assemblage A sequence had 100% homology with human-derived assemblage A isolates (GQ329671) from Sweden and chipmunk-derived isolate (MF671918) from China, one assemblage E sequence (GenBank: MZ494459) shared the most considerable similarity (99.79%) to that (KY633473) from a Tibetan sheep in China, with only one base difference.
Discussion
In this study, the overall prevalence of Cryptosporidium spp. were 2.2% (11/498), with 2.5% in Himalayan marmots, and 1.0% in Alashan ground squirrels. There was no significant difference in the prevalence of Cryptosporidium spp. and G. duodenalis, and we will enlarge the research sample size for further verification. Other studies reported much higher prevalence of Cryptosporidium spp. in wild rodent species in China than this study, including in house mice (3.2%, 1/31), long-tailed rats (3.6%, 4/111 and 55.3%, 21/38), brown rats (6.3%, 4/64; 9.1%, 22/242 and 28.6%, 16/56), wild plateau pikas (6.3%, 4/64), Qinghai voles (8.9%, 8/90), Asian house rats (18.0%, 21/117; 18.2%, 6/33 and 73.9%, 4/46), Brandt’s voles (18.7%, 127/678), Muridae (40.0%, 4/10)20,27,28,29,30,31,32. The prevalence in this study was also lower than that in some pet rodent species, including in bamboo rats (3.3%, 3/92), Siberian hamsters (7.8%, 4/51), red squirrels (8.6%, 27/314 and 26.3%, 5/19), chinchillas (9.3%, 26/280 and 10.0%, 14/140), campbell hamsters (10.0%, 3/30 and 22.2%, 6/27), Siberian chipmunks (30.0%, 6/20), gold hamsters (32.0%, 16/50), chipmunks (50.0%, 1/2 and 75.0%, 3/4), guinea pigs (52.3%, 162/310 and 85.0%, 34/40), Roborovski dwarf hamsters (100.0%,1/1), and higher than that in pet red-bellied tree squirrels (1.4%, 4/287)29,33,34,35,36,37,38. In addition, there was difference between prevalence in different farmed and laboratory rodent species, including farmed bamboo rats (2.1%, 9/435 and 29.5%, 209/709), farmed brown rats (7.1%, 12/168), experimental brown rats (0.6%, 2/355), laboratory mice (1.7%, 4/229), laboratory rats (4.0%, 1/25)27,29,39,40,41. These variations in the prevalence of Cryptosporidium spp. in different studies may be explained by many factors, including the population densities, the health status of hosts, management systems, experimental methods and source region42.
To date, including Cryptosporidium species/genotypes obtained in this study, a total of 14 Cryptosporidium species and 17 genotypes have been detected in 16 studies of various rodents in China (Table 2)20,27,28,29,30,31,32,33,34,35,36,37,39,40,41. Among them, 11 species/genotypes have been detected in humans: C. parvum, C. muris, C. ubiquitum, C. andersoni, C. occultus, C. viatorum, C. canis, C. suis, C. erinaceid, C. tyzzeri and horse genotype4, indicating rodents may play essential roles in the transmission of zoonotic cryptosporidiosis.
Altogether, four Cryptosporidium species/genotypes were identified in this study: C. rubeyi, ground squirrel genotype II, chipmunk genotype V in Himalayan marmots, and horse genotype in Alashan ground squirrels. C. rubeyi was characterized by numerous wild rodent hosts such as golden-mantled ground squirrels, California ground squirrels, Belding's ground squirrels, and black-tailed prairie dogs43,44. Previously ground squirrel genotype II and chipmunk genotype V were only identified in black-tailed prairie dogs in the USA43 and chinchillas in China38, respectively. Our identification of ground squirrel genotype II and chipmunk genotype V expanded the host range of the two genotypes. Horse genotype was initially isolated from a Przewalski wild horse at the Prague Zoo in the Czech Republic, and commonly detected in horses and donkeys, occasionally found in neonatal calves and hedgehogs45,46. Horse genotype has also been found in human patients with diarrhea in the UK and the USA, suggesting its zoonotic potential47,48,49. In the present study, the horse genotype was identified in rodents for the first time, indicating it has a broader range of host than initially anticipated. Horse genotype isolated from Alashan ground squirrels was further identified as novel subtype VIbA10. Currently, two subtype families are recognized within the Cryptosporidium horse genotype by sequence analysis targeting the gp60 gene: the VIa subtype family in animals (horses, donkeys and calves, etc.) and the VIb subtype family in humans and hedgehogs.
The present study detected the infection of Cryptosporidium spp. in wild rodent species of the genus Marmota and genus Spermophilus. Further, eight previous studies have reported the occurrence of Cryptosporidium species/genotypes in other three species of the genus Marmota and other four species of genus Spermophilus: including C. ubiquitum in woodchucks (Marmota monax) in the USA50,51; C. parvum in yellow-bellied marmots (Marmota flaviventris) in the USA52; C. andersoni in Bobak marmots (Marmota bobac) in the Czech Republic45; C. rubeyi in California ground squirrels (Spermophilus beecheyi) in the USA, Belding's ground squirrels (Spermophilus beldingi) and golden-mantled ground squirrels (Spermophilus lateralis) in the USA44,53,54; ground squirrel genotype I and ground squirrel genotype III in thirteen-lined ground squirrels (Spermophilus tridecemlineatus) in USA43.
In this study, the overall prevalence of G. duodenalis were 1.6% (8/498), with 1.5% (6/399) for Himalayan marmots and 2.0% (2/99) for Alashan ground squirrels. This study reported much lower prevalence of G. duodenalis than other studies in wild rodent species in China: house mouse (3.2%, 1/31); Asian house rat (6.1%, 2/33); brown rat (6.6%, 11/168 and 9.3%, 33/355); pet chipmunks (8.6%, 24/279); bamboo rat (10.8%, 52/480); coypus (12.3%, 38/308); pet chinchillas (27.1%, 38/140)27,39,55,56,57,58 (Table 3).
In this study, the sequences of amplicons from G. duodenalis-positive samples were determined to be assemblages A, B, and E, with assemblages B showing dominance in the detected animals. Assemblages A, B and E were identified in Himalayan marmots and assemblage B in Alashan ground squirrels. G. duodenalis assemblages in Himalayan marmots were richer than Alashan ground squirrels. As we know, in previous studies, G. duodenalis infections of Chinese rodents were reported to be caused by assemblages A, B and G27,39,55,56,57,58. Among them, assemblages A and B have a broad host range and are commonly found in humans56. Some recent studies in China also reported the occurrence of assemblage A in pet chipmunks, coypus and pet chinchillas, while assemblage B in bamboo rats, coypus and pet chinchillas55,56,57,58. These two assemblages were detected in this study suggest that Himalayan marmots and Alashan ground squirrels can play roles in the zoonotic dissemination of G. duodenalis. Assemblage E is commonly found in a range of hoofed livestocks and occasionally found in rodent species, and it has also been found in human cases, indicating that this assemblage is of zoonotic significance59,60.
In the investigated areas of QTPA, wild rodent species Himalayan marmots and Alashan ground squirrels have strong migration habits and often share pasture with humans, herbivorous animals and other wild animals. Results of this study suggest that these two wild rodent species may play a role in the transmission cycle of Cryptosporidium spp. oocysts and G. duodenalis cysts among humans, animals, water sources and fresh produce in QTPA grassland ecosystem.
Conclusion
This study examined the prevalence and zoonotic potential of Cryptosporidium spp. and G. duodenalis in Himalayan marmots and Alashan ground squirrels in the Qinghai-Tibetan Plateau area (QTPA) of China for the first time. Four Cryptosporidium species/genotypes were identified, including C. rubeyi, ground squirrel genotype II, chipmunk genotype V and horse genotype (novel subtype VIbA10). These two rodent species identified G. duodenalis zoonotic assemblages A, B, and E. The results expanded the host range of Cryptosporidium spp. and G. duodenalis, providing more information on the prevalence, epidemiology and genetic characterizations of the two pathogens in Himalayan marmots and Alashan ground squirrels. Further surveys are also required to understand the prevalence and transmission dynamics of the two pathogens.
Data availability
Nucleotide sequences of this article were deposited in the GenBank database under following accession numbers: MZ478131-MZ478133, ON384432 (SSU rRNA), ON419488-ON419491 (actin), ON456466 (HSP70), MW531716 (gp60) for Cryptosporidium spp.; MZ494459 (bg) for G. duodenalis.
References
Fayer, R. Cryptosporidium: A water-borne zoonotic parasite. Vet. Parasitol. 126, 37–56 (2004).
Feng, Y. & Xiao, L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 24, 110–140 (2011).
Xiao, L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 124, 80–89 (2010).
Feng, Y., Ryan, U. M. & Xiao, L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 34, 997–1011 (2018).
Holubová, N. et al. Cryptosporidium proventriculi sp. N. (Apicomplexa: Cryptosporidiidae) in Psittaciformes birds. Eur. J. Protistol. 69, 70–87 (2019).
Holubová, N. et al. Description of Cryptosporidium ornithophilus n. sp. (Apicomplexa: Cryptosporidiidae) in farmed ostriches. Parasit. Vectors. 13, 340 (2020).
Bolland, S. J., Zahedi, A., Oskam, C., Murphy, B. & Ryan, U. Cryptosporidium bollandi n. sp. (Apicomplexa: Cryptosporidiiae) from angelfish (Pterophyllum scalare) and Oscar fish (Astronotus ocellatus). Exp. Parasitol. 217, 107956 (2020).
Ježková, J. et al. Cryptosporidium ratti n. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitology 148, 84–97 (2021).
Zahedi, A., Bolland, S. J., Oskam, C. L. & Ryan, U. Cryptosporidium abrahamseni n. sp. (Apicomplexa: Cryptosporidiiae) from red-eye tetra (Moenkhausia sanctaefilomenae). Exp. Parasitol. 223, 108089 (2021).
Ježková, J. et al. Cryptosporidium myocastoris n. sp. (Apicomplexa: Cryptosporidiidae), the Species Adapted to the Nutria (Myocastor coypus). Microorganisms 9, 813 (2021).
Fan, Y. et al. Subtyping Cryptosporidium xiaoi, a common pathogen in sheep and goats. Pathogens 10, 800 (2021).
Naguib, D., Roellig, D. M., Arafat, N. & Xiao, L. Genetic characterization of Cryptosporidium cuniculus from rabbits in Egypt. Pathogens 10(6), 775 (2021).
Xiao, L. & Feng, Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food. Waterborne. Parasitol. 8–9, 14–32 (2017).
Dixon, B. R. Giardia duodenalis in humans and animals: Transmission and disease. Res. Vet. Sci. 135, 283–289 (2021).
Ma, L. et al. Detection of Cryptosporidium and Giardia in the slaughterhouse, sewage and river waters of the Qinghai Tibetan plateau area (QTPA), China. Parasitol. Res. 118, 2041–2051 (2019).
Jian, Y. et al. Occurrence of Cryptosporidium and Giardia in wild birds from Qinghai Lake on the Qinghai-Tibetan Plateau, China. Parasitol. Res. 120, 615–628 (2021).
Wu, Y. et al. Genotyping and identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from free–range Tibetan yellow cattle and cattle–yak in Tibet, China. Acta Trop. 212, 05671 (2020).
Jian, Y. et al. Prevalence and molecular characterization of Giardia duodenalis in cattle and sheep from the Qinghai-Tibetan Plateau Area (QTPA), northwestern China. Vet. Parasitol. 250, 40–44 (2018).
Zhang, X. et al. Molecular detection and prevalence of Cryptosporidium spp. infections in two types of domestic farm animals in the Qinghai-Tibetan Plateau Area (QTPA) in China. Parasitol. Res. 117, 233–239 (2018).
Zhang, X. et al. The first report of Cryptosporidium spp. in Microtus fuscus (Qinghai vole) and Ochotona curzoniae (wild plateau pika) in the Qinghai-Tibetan Plateau area, China. Parasitol. Res. 117, 1401–1407 (2018).
Li, X. et al. Detection of Cryptosporidium oocysts and Giardia cysts in vegetables from street markets from the Qinghai Tibetan Plateau Area in China. Parasitol. Res. 119, 847–1855 (2020).
Huang, D. et al. Establishment and application of PCR method for detection of Cryptosporidium, China [in Chinese]. Trop. Med. 16, 8 (2016).
Alves, M. et al. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41, 2744–2747 (2003).
Cacciò, S. M., Beck, R., Lalle, M., Marinculic, A. & Pozio, E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int. J. Parasitol. 38, 1523–1531 (2008).
Abe, N., Kimata, I. & Iseki, M. Identification of genotypes of Giardia intestinalis isolates from dogs in Japan by direct sequencing of the PCR amplified glutamate dehydrogenase gene. J. Vet. Med. Sci. 65, 29–33 (2003).
Sulaiman, I. M. et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg. Infect. Dis. 9, 1444–1452 (2003).
Zhao, Z. et al. Genotyping and subtyping of Giardia and Cryptosporidium isolates from commensal rodents in China. Parasitology 142, 800–806 (2015).
Chen, Y. W. et al. Identification of Cryptosporidium viatorum XVa subtype family in two wild rat species in China. Parasit. Vectors. 12, 502 (2019).
Lv, C. et al. Cryptosporidium spp. in wild, laboratory, and pet rodents in china: Prevalence and molecular characterization. Appl. Environ. Microbiol. 75, 7692–7699 (2009).
Zhao, W. et al. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasit. Vectors 11, 313–313 (2018).
Feng, S. et al. Molecular characterization of Cryptosporidium spp. in Brandt’s Vole in China. Front. Vet. Sci. 7, 300 (2020).
Zhao, W. et al. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: Molecular detection, species/genotype identification and implications for public health. Int. J. Parasitol. Parasites Wildl. 9, 317–321 (2019).
Chai, Y. et al. First detection of Cryptosporidium spp. in red-bellied tree squirrels (Callosciurus erythraeus) in China. Parasite 26, 28 (2019).
Liu, X. et al. Occurrence of novel and rare subtype families of Cryptosporidium in bamboo rats (Rhizomys sinensis) in China. Vet. Parasitol. 207, 144–148 (2015).
Deng, L. et al. Occurrence and genetic characteristics of Cryptosporidium spp. and Enterocytozoon bieneusi in pet red squirrels (Sciurus vulgaris) in China. Sci. Rep. 10, 1026 (2020).
Qi, M. et al. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitol. Int. 64, 339–341 (2015).
Li, Q. et al. Molecular investigation of Cryptosporidium in small caged pets in northeast China: Host specificity and zoonotic implications. Parasitol. Res. 115, 2905–2911 (2016).
Chen, J. et al. Genetic characterizations of Cryptosporidium spp. from pet rodents indicate high zoonotic potential of pathogens from chinchillas. One Health 13, 100269 (2021).
Li, J. et al. Molecular characterization of Cryptosporidium spp. and Giardia duodenalis in experimental rats in China. Parasitol. Int. 77, 102127 (2020).
Zhao, W. et al. Prevalence and diversity of Cryptosporidium spp. in bamboo rats (Rhizomys sinensis) in South Central China. Int. J. Parasitol. Parasites Wildl. 9, 312–316 (2019).
Li, F. et al. Common occurrence of divergent Cryptosporidium species and Cryptosporidium parvum subtypes in farmed bamboo rats (Rhizomys sinensis). Parasit. Vectors 13, 149 (2020).
Feng, Y. & Xiao, L. Molecular epidemiology of Cryptosporidiosis in China. Front. Microbiol. 8, 1701 (2017).
Stenger, B. L. et al. North American tree squirrels and ground squirrels with overlapping ranges host different Cryptosporidium species and genotypes. Infect. Genet. Evol. 36, 287–293 (2015).
Li, X. et al. Cryptosporidium rubeyi n. sp. (Apicomplexa: Cryptosporidiidae) in multiple Spermophilus ground squirrel species. Int. J. Parasitol. Parasites Wildl. 4, 343–350 (2015).
Ryan, U. et al. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl. Environ. Microbiol. 69, 4302–4307 (2003).
Li, F. et al. Different distribution of Cryptosporidium species between horses and donkeys. Infect. Genet. Evol. 75, 103954 (2019).
Robinson, G., Elwin, K. & Chalmers, R. M. Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg. Infect. Dis. 14, 1800–1802 (2008).
Xiao, L. et al. Subtype analysis of Cryptosporidium specimens from sporadic cases in Colorado, Idaho, New Mexico, and Iowa in 2007: Widespread occurrence of one Cryptosporidium hominis subtype and case history of an infection with the Cryptosporidium horse genotype. J. Clin. Microbiol. 47, 3017–3020 (2009).
Elwin, K. et al. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000–2008. Epidemiol. Infect. 140, 673–683 (2012).
Feng, Y. et al. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 73, 6475–6483 (2007).
Ziegler, P. E. et al. Prevalence of Cryptosporidium species in wildlife populations within a watershed landscape in southeastern New York State. Vet. Parasitol. 147, 176–184 (2007).
Montecino-Latorre, D., Li, X., Xiao, C. & Atwill, E. R. Elevation and vegetation determine Cryptosporidium oocyst shedding by yellow-bellied marmots (Marmota flaviventris) in the Sierra Nevada Mountains. Int. J. Parasitol. Parasites Wildl. 4, 171–177 (2015).
Atwill, E. R., Phillips, R., Pereira, M. D., Li, X. & McCowan, B. Seasonal shedding of multiple Cryptosporidium genotypes in California ground squirrels (Spermophilus beecheyi). Appl. Environ. Microbiol. 70, 6748–6752 (2004).
Pereira, M. D., Li, X., McCowan, B., Phillips, R. L. & Atwill, E. R. Multiple unique Cryptosporidium isolates from three species of ground squirrels (Spermophilus beecheyi, S. beldingi, and S. lateralis) in California. Appl. Environ. Microbiol. 76, 8269–8276 (2010).
Deng, L. et al. First identification and multilocus genotyping of Giardia duodenalis in pet chipmunks (Eutamias asiaticus) in Sichuan Province, southwestern China. Parasit. Vectors 11, 199 (2018).
Ma, X., Wang, Y., Zhang, H. J., Wu, H. X. & Zhao, G. H. First report of Giardia duodenalis infection in bamboo rats. Parasit. Vectors 11, 520 (2018).
Qi, M. et al. Multilocus genotyping of potentially zoonotic Giardia duodenalis in pet chinchillas (Chinchilla lanigera) in China. Vet. Parasitol. 208, 113–117 (2015).
Cui, Z. et al. Occurrence and multi-Locus analysis of Giardia duodenalis in coypus (Myocastor coypus) in China. Pathogens 10, 179 (2021).
Fantinatti, M., Bello, A. R., Fernandes, O. & Da-Cruz, A. M. Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J. Infect. Dis. 214, 1256–1259 (2016).
Gherman, C. M., Kalmár, Z., Györke, A. & Mircean, V. Occurrence of Giardia duodenalis assemblages in farmed long-tailed chinchillas Chinchilla lanigera (Rodentia) from Romania. Parasit. Vectors 11, 86 (2018).
Sulaiman, I. M., Lal, A. A. & Xiao, L. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 88(2), 388–394 (2002).
Sulaiman, I. M., Morgan, U. M., Thompson, R. A., Lal, A. A. & Xiao, L. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microb. 66(6), 2385–2391 (2000).
Acknowledgements
We are grateful to Dr. Daqin Xu at Gansu Provincial Centre for Disease Control and Prevention for help in collecting the samples.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 82072307 and 81772224 to YS), the National Science and Technology Major Program of China (No. 2018ZX10713001-004 to YS), and the Fifth Round of Three-Year Public Health Action Plan of Shanghai, China (No. GWV-10.1-XK13 to JC). The funders had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.S. and X.W. designed the study. J.X., H.L., Y.J., L.T. and Y.S. participated in the sample collection and methodology. J.X., H.L., H.J. and J.Y. contributed to data analysis. Y.S. and J.C. contributed reagents and materials. J.X. wrote the manuscript. Y.S. and X.W. revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, J., Liu, H., Jiang, Y. et al. Genotyping and subtyping of Cryptosporidium spp. and Giardia duodenalis isolates from two wild rodent species in Gansu Province, China. Sci Rep 12, 12178 (2022). https://doi.org/10.1038/s41598-022-16196-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-16196-1
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.