Abstract
MicroRNAs (miRNAs) packaged into exosomes mediate cell communication and contribute to the pathogenesis of acute type A aortic dissection (ATAAD) with acute lung injury (ALI). The expression profile of plasma exosomal miRNAs in ATAAD patients with ALI hasn’t been identified. We performed a miRNA-sequencing to analyze the differentially expressed miRNAs (DE-miRNAs) of circulating exosomes in ATAAD patients with ALI compared to patients without ALI, founding 283 specific miRNAs in two groups. We respectively selected the top 10 downregulated and upregulated DE-miRNAs for further studies. The predicted transcription factors (TFs) of these DE-miRNAs were SMAD2, SRSF1, USF1, etc. The Gene Ontology (GO) and Kyoto Encyclopedia Genes and Genomes (KEGG) analysis predicted their target genes mainly involved acute inflammatory response, cell junction, cytoskeleton, NF-κB signaling pathway, etc. Construction and analysis of the PPI network revealed that RHOA and INSR were considered hub genes with the highest connectivity degrees. Moreover, we confirmed two exosomal miRNAs (hsa-miR-485-5p and hsa-miR-206) by real-time quantitative polymerase chain reaction (RT-qPCR) in a validation cohort. Our study identified a plasma exosomal miRNAs signature related to ATAAD with ALI. Certain DE-miRNAs may contribute to the progression of this disease, which help us better understand the pathogenesis of ATAAD with ALI.
Similar content being viewed by others
Introduction
Acute Stanford type A aortic dissection (ATAAD) is a life-threatening cardiovascular disease with urgent onset, rapid progression and high mortality, which often requires emergency surgical treatment1. Acute lung injury (ALI) is one of its preoperative complications that seriously affect the surgical outcome2. In recent years, studies have shown that about 50% of ATAAD patients are complicated with ALI before surgery2. This not only lengthens the duration of postoperative intubation, mechanical ventilation and intensive care unit (ICU) stay but also leads to a death rate of about 25–67%, greatly increasing the perioperative mortality and seriously affecting the prognosis of ATAAD patients3.
Inflammation is involved in the occurrence and development of ATAAD with ALI. The progression of local vascular inflammation caused by intima tear and false lumen formation in aortic dissection can develop into systemic inflammation and result in multiple organ damage including lung4,5. Some studies indicated that inflammatory reactants such as interleukin-6 (IL-6), C-reactive protein (CRP) and monocyte chemoattractant protein-1 (MCP-1) were significantly increased in blood or lung tissues from ATAAD patients with ALI, and these reactants could directly or indirectly promote the apoptosis and barrier dysfunction of pulmonary microvascular endothelial cells (PMVECs), which contributed to the elevation of endothelial permeability and the formation of ALI6,7,8. However, the underlying mechanism has not been well elucidated.
Exosomes are microvesicles (30–100 nm in diameter) that originated in the endosomal membrane compartment and formed in the cytoplasm9. They are secreted by almost all living cells and exist in various body fluids such as plasma, urine and synovial fluid10. Exosomes mediate cell-to-cell communication by transporting their cargo including proteins, lipids, mRNAs and RNAs to target cells10,11. microRNAs (miRNAs) account for 41.72% of all RNAs in exosomes12 and they function as posttranscriptional regulators of gene expression by interacting with target mRNAs, playing a critical role in maintenance tissues and organ homeostasis13,14,15. For instance, miR-208a was shown to directly increase β-myosin heavy chain (MHC), which was associated with arrhythmia, fibrosis and hypertrophic growth14. miR-21and miR-16 were reported to impair barrier integrity and regulating inflammatory responses through downregulating Ras homolog gene family member B (RhoB) and interleukin-6 (IL-6), which were involved in intestinal and pulmonary mucosa, such as ALI or inflammatory bowel disease (IBD)15. Besides, increasing evidence has demonstrated that the potential role of exosomal miRNAs can positively affect the progression of cardiovascular diseases such as acute coronary syndrome, heart failure, pulmonary arterial hypertension, rheumatic valvular disease and aortic disease16. However, little research has been conducted on their influence on ATAAD with ALI. Research into plasma exosomal miRNAs may bring us a new perspective as we consider the underlying mechanism of the multiple organ disorder (e.g. lung) in ATAAD. Thus, we investigated plasma exosomal miRNAs profiles in ATAAD patients with ALI in order to partly explain the disease’s pathophysiology.
Results
Description of study patients
This is a case–control study that included ATAAD patients with and without ALI as a discovery cohort and validation cohort (Fig. 1). We recruited 19 ATAAD patients treated as the discovery cohort for plasma exosomal miRNAs sequencing, and they were categorized ATAAD with ALI (ALI group, n = 11) and ATAAD without ALI (Non-ALI group, n = 8). Additional 20 ATAAD patients were enrolled for each group as the validation cohort. The clinical and demographic characteristics of study patients in the discovery and validation cohort were presented in Supplementary Table S1 and S2 online. There is no significant difference in most clinical parameters between the non-ALI group and the ALI group. In the discovery cohort, patients in the ALI group had a higher respiratory and heart rate and lower arterial oxygen tension (PaO2)/inspiratory oxygen fraction (FiO2) and PaO2. Additionally, left ventricular ejection fraction (LVEF) was statistically different between the two groups, but not clinically significant. In the validation cohort, the value of PaO2 and PaO2/FiO2 were lower in patients in the ALI group, and these patients had more hypertension.
Schematic overview of the strategy for identifying an plasma exosomal miRNAs signature associated with ATAAD with ALI. Our study design involved a discovery phase including ATAAD (n = 19) with ALI or without ALI by small RNA sequencing. In a second validation phase with a large ATAAD cohort (n = 40), including with ALI or without ALI, 2 miRNAs were finally validated by RT-qPCR. ATAAD acute type A aortic dissection; ALI acute lung injury, RT-qPCR real-time quantitative polymerase chain reaction, miRNAs microRNAs.
Isolated plasma exosomes characterization
Exosomes from plasma were identified by their morphology, diameter distribution and enriched exosome markers such as CD63 and TSG101. The transmission electron microscopy (TEM) image (Supplementary Fig. S1a online) showed that the isolated exosomes were spherical structures with a diameter of about 30–100 nm. Nanoparticle tracking analysis (NTA) (Supplementary Fig. S1b online) revealed that the extracted exosomes had a median diameter of 58.9 nm and their concentration was 4.1 × 109/mL. Besides, these exosomes were positive for enriched exosome markers CD63 and TSG101 and negative for Calnexin (an intracellular compartment marker that was absent in exosomes) on Western blot (Supplementary Fig. S1c online). The results above indicated that the plasma exosomes had been adequately purified.
Exosomal miRNAs signature in ATAAD with ALI
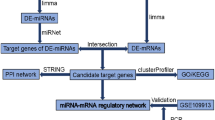
To identify the plasma exosomal miRNAs profile of ATAAD patients with ALI, we used the small RNA sequencing method. As shown in Supplementary Fig. S2 online, we found that 283 exosome-specific miRNAs (163 downregulated and 120 upregulated) were significantly different (Non-ALI group vs ALI group; Cut-off: adjusted p < 0.05, │fold change│ > 1.8). Then, we eliminated those novel miRNAs or miRNAs with low fragments perkiobase million (FPKM) in each sample, and the top 10 downregulated (hsa-miR-181d-5p, hsa-miR-31-3p, hsa-miR-326, hsa-miR-423-5p, hsa-miR-145-5p, hsa-miR-744-5p, hsa-miR-99b-3p, hsa-miR-214-3p, hsa-miR-206, hsa-let-7e-5p) and upregulated (hsa-miR-103a-3p, hsa-miR-26b-3p, hsa-miR-1306-5p, hsa-miR-7-1-3p, hsa-miR-3120-3p, hsa-miR-335-3p, hsa-miR-485-5p, hsa-miR-664a-3p, hsa-miR-29a-3p, hsa-miR-483-3p) miRNAs were shown in Table 1 and Fig. 2.
Heat map of the top 10 downregulated and upregulated DE-miRNAs in Non-ALI group vs ALI group. The color key indicates the expression level of the miRNAs. DE-miRNAs differentially expressed miRNAs; Non-ALI group patients with acute type A aortic dissection without acute lung injury, ALI group patients with acute type A aortic dissection with acute lung injury.
Prediction of transcription factors (TFs) of differentially expressed miRNAs (DE-miRNAs)
Based on the sequencing results above, we concentrated on the following functional study of the top 10 candidates down- and upregulated miRNAs. Their upstream TFs were predicted by FunRich software. We listed the top 10 TFs for these DE-miRNAs, which were shown in Fig. 3a,b. For downregulated DE-miRNAs, the top 10 TFs were SMAD2, MYOCD, MKL1, SMAD4, SMAD3, NR3C1, GTF2I, NFIC, SRC and RREB1. For upregulated DE-miRNAs, the top 10 TFs were CEBPB, IL4, HOXD10, GLI1, SRSF1, ZMYND8, CCNT2, USF1, MKL1 and HMCA1.
Gene Ontology (GO) functional and Kyoto Encyclopedia Genes and Genomes (KEGG) pathway analysis
Initially, we used the TargetScan, miRbase and RNAhybrid websites to predict the target genes of the top 10 DE-miRNAs. Additionally, to better understand the biological function and pathways of the predictive target genes, we performed GO and KEGG pathway analysis. The biological process (BP), cellular component (CC) and molecular functions (MF) of GO analysis were summarized in Fig. 4a–c. The changes of BP were enriched in the response to hypoxia, natural killer (NK) cell proliferation, NK T cell proliferation, acute inflammatory response, etc. (as shown in Fig. 4a), and the changes of CC were enriched in the cell junction, cytoskeleton, etc. (as shown in Fig. 4b). Figure 4c suggested that the changes of MF were manifested in the protein binding, complement component C4b and C3b receptor activity, sulfite oxidase activity, etc. The KEGG pathway analysis was presented in Fig. 4d, and these predicted target genes were primarily involved in adhesion junction, necroptosis, NF-κB signaling pathway, etc.
GO and KEGG pathway analysis for the target genes of the top 10 DE-miRNAs. (A–C) The top 20 associated biological processes (a), cellular components (b) and molecular functions (c) in which the target genes were significantly enriched; (d) the top 20 related KEGG pathways of target genes. GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes.
Protein–protein interaction (PPI) network construction
We submitted those predictive target genes of the candidate DE-miRNAs to the STRING database and constructed PPI networks for them, which was shown in Fig. 5a,b. Using a comprehensive score greater than 0.4, we obtained 55 and 125 node pairs for target genes of downregulated and upregulated DE-miRNAs, respectively. Then, to identify the hub genes of the two PPI networks, we input these node pairs into Cytoscape, and the top 10 hub genes calculated by degree scoring were presented in Table 2. In the target genes of downregulated DE-miRNAs, the hub genes were RHOA, SLC1A2, SCN2A, PLEC, JAK3, GSR, CALM1, RPL13, DNAJC10 and EFR3B; in the target genes of upregulated DE-miRNAs, the hub genes were INSR, PPARA, HNRNPDL, TYR, ATRX, SYNCRIP, PLRG1, HNRNPH2, DCTN2 and PCBD1.
The PPI network of the predictive target genes of the top 10 DE-miRNAs. (a) The red rectangle represents the predicted upregulated gene of downregulated DE-miRNAs; (b) the bule rectangle represents the predicted downregulated gene of upregulated DE-miRNAs. The more node pairs, the darked the color is, which indicates the higher the rank of hub gene is. PPI protein–protein interaction, DE-miRNAs differentially expressed miRNAs.
Validation of DE-miRNAs by real-time quantitative polymerase chain reaction (RT-qPCR)
To confirm the disorder of specific plasma exosomes miRNAs in response to ATAAD with ALI, we selected 10 miRNAs from the top 10 DE-miRNAs signature and previous bibliography retrieval for RT-qPCR in a validation cohort (20 ATAAD patients with or without ALI, respectively): hsa-miR-181d-5p, hsa-miR-423-5p, hsa-miR-145-5p, hsa-miR-206, hsa-let-7e-5p, hsa-miR-103a-3p, hsa-miR-26b-3p, hsa-miR-485-5p, hsa-miR-29a-3p and hsa-miR-483-3p (Supplementary Table S3 online).
As shown in Fig. 6a–h, of the ten candidate miRNAs, the relative expression levels of 3 miRNAs (hsa-miR-485-5p, hsa-miR-206 and hsa-miR-483-3p) were significantly different between the non-ALI group and the ALI group. The expression of hsa-miR-485-5p (upregulated in ALI group, P < 0.05) and hsa-miR-206 (downregulated in ALI group, P < 0.01) were consistent with sequencing results. Hsa-miR-181d-5p and hsa-miR-26b-3p could not be detected due to their extremely low expression levels in all exosome samples.
Validation of candidate DE-miRNAs by RT-qPCR in the Non-ALI and ALI groups. (a–h) Relative expression levels of selected miRNAs in the Non-ALI and ALI groups were detected by RT-qPCR (n = 20 in each group). The results were normalized to cel-miR-39. Data were analyzed by Mann–Whitney U test. Hsa-miR-181d-5p and hsa-miR-26b-3p could not be detected owing to their low expression levels. DE-miRNAs differentially expressed miRNAs; RT-qPCR real-time quantitative polymerase chain reaction; Non-ALI group patients with acute type A aortic dissection without acute lung injury, ALI group patients with acute type A aortic dissection with acute lung injury. *P < 0.05; **P < 0.01.
Discussion
In this study, we analyzed the profile of plasma exosomal miRNAs in ATAAD patients with ALI and selected the top 10 downregulated and upregulated DE-miRNAs in 283 specific miRNAs for further functional analysis. Initially, SMAD2, MYOCD, MKL1, SMAD4, SMAD3, NR3C1, GTF2I, NFIC, SRC, and RREB1 were predicted as the TFs of the downregulated DE-miRNAs and CEBPB, IL4, HOXD10, GLI1, SRSF1, ZMYND8, CCNT2, USF1, MKL1 and HMCA1 were predicted as the TFs of the upregulated DE-miRNAs. Additionally, the biological functions and pathways of predictive target genes of these DE-miRNAs were involved in the regulation of response to hypoxia, NK cell proliferation, NK T cell proliferation, acute inflammatory response, cell junction, cytoskeleton, HIF-1 signaling pathway, NF-κB signaling pathway, necroptosis and adherent junction. Finally, RHOA and INSR were considered hub genes in predictive target genes of DE-miRNAs. For further validation, we found that the expression of hsa-miR-485-5p and hsa-miR-206 showed the same trend with regard to sequencing results, which indicated their regulatory role in the pathogenesis of ATAAD with ALI.
Preoperative ALI is a frequent and severe complication in ATAAD, and hypoxemia is the most typical symptom in these patients. It has been reported that ATAAD patients complicated with ALI have longer the duration of ventilation and ICU and hospital stays, which leads to a poor clinical prognosis17. As a result, a better understanding of the pathophysiology of the ATAAD with ALI can improve clinical prophylaxis and treatments in these patients. Exosomes are small membrane vesicles that are rich in miRNAs and can protect miRNAs from the degradation of RNase in circulation, which act as the carriers of intercellular communication18. Recent studies revealed that plasma exosomal miRNAs are closely related to the onset of ALI19,20. Jiang et al.20 explored the influence of circulating exosomes on macrophage activation of sepsis-related ALI and demonstrated that exosomal miR-155 promoted macrophages proliferation and inflammation by targeting SHIP1 and SOCS1. Another study showed that exosome-shuttling miR-1-3p increased PMVECs permeability and membrane injury by targeting SERP1, leading to vascular barrier dysfunction19. However, the association between the plasma exosomal miRNAs and ATAAD with ALI has not been reported.
In our present study, we conducted RNA sequencing and identified a total of 283 specific plasma exosomal miRNAs in ATAAD patients with ALI. Based on its pathophysiological process, these miRNAs might be released from a variety of activated inflammatory cells (e.g. neutrophils, lymphocytes, monocytes, etc.), aortic and pulmonary vascular endothelial cells, and even alveolar epithelial cells via exosomes, which were transferred into the circulation21,22. In these DE-miRNAs, we eliminated those novel miRNAs or low expression level miRNAs in each sample and finally, selected the top 10 downregulated (hsa-miR-181d-5p, hsa-miR-31-3p, hsa-miR-326, hsa-miR-423-5p, hsa-miR-145-5p, hsa-miR-744-5p, hsa-miR-99b-3p, hsa-miR-214-3p, hsa-miR-206, hsa-let-7e-5p) and upregulated (hsa-miR-103a-3p, hsa-miR-26b-3p, hsa-miR-1306-5p, hsa-miR-7-1-3p, hsa-miR-3120-3p, hsa-miR-335-3p, hsa-miR-485-5p, hsa-miR-664a-3p, hsa-miR-29a-3p, hsa-miR-483-3p) DE-miRNAs for further studies.
By predicting TFs of these DE-miRNAs, we found that SMAD2, MYOCD, MKL1, SMAD4, SMAD3, NR3C1, GTF2I, NFIC, SRC, RREB1 were predicted to be the downregulated DE-miRNAs TF, while CEBPB, IL4, HOXD10, GLI1, SRSF1, ZMYND8, CCNT2, USF1, MKL1 and HMCA1 proteins were predicted to be the upregulated DE-miRNAs TF. Among these TFs, the SMAD proteins family might potentially modulate the expression of the main downregulated DE-miRNAs. Previous studies showed that SMAD proteins could control DROSHA-mediated miRNA maturation in human smooth muscle cells23. Recent research expanded the results and indicated that they were involved in miRNA transcription. For instance, Du et al.24 demonstrated that SMAD4 regulated miR-425-TGF-β signaling pathway, resulting in granulosa cell apoptosis. Their influence on miRNAs in osteoclast differentiation was also well documented25. In the predictive TFs of upregulated DE-miRNAs, SRSF1, an RNA binding protein, had been shown to modulate miRNAs processing such as miR-7, miR-10b, miR-29b26. Studies indicated that SRSF1 regulated the expression of miR-7 through binding to the pri-miR-7 sequence and promoting DROSHA cleavage27. It can also modulate miR-10b and miR-29b in the same way, which played a critical role in immune response and tumorigenesis28,29. Besides, USF1 can activate the expression of miR-132 by binding to the E-box in the miR-132 promoter and influence its synthesis in oxygen and glucose deprivation-induced cell apoptosis30. More researches about the role of these predictive TFs in ATAAD with ALI need to be further conducted.
According to GO and KEGG functional analysis, the top 20 enrichments in BP, CC and MF of GO were listed and showed that certain miRNAs participate in biological processes or functions associated with response to hypoxia, natural killer (NK) cell proliferation, NK T cell proliferation, acute inflammatory response, cell junction and cytoskeleton. Numerous evidence has supported that ATAAD with ALI is characterized by widespread uncontrolled inflammation in the lungs, a functional or structural breakdown in pulmonary vascular endothelial junction and barrier damage, resulting in severe oxygenation impairment in body31. Hypoxia can be further pro-inflammatory and lead to a breakdown of vascular barriers. But the adaptive response to hypoxia, especially hypoxia-inducible transcription factors (HIF) stabilization and induction of its downstream genes, bears anti-inflammatory and lung tissue-protective aspects, which highlight the functional role of hypoxia signaling in ALI32. It has been reported that the expression of many miRNAs changes under hypoxia, and these miRNAs may modulate HIF expression or switch, but the results are controversial33. Besides, most of these studies have focused on cancer cell lines only and often ignore endothelial or epithelial cells, so that the exact role of hypoxia-induced miRNAs in them remains unclear33. Our results above further, suggested that specific plasma exosomal miRNAs were involved in the pathogenesis of ATAAD with ALI by recruiting immune cells, expanding inflammatory response, destroying cell junction and skeleton (a model figure was shown in Supplementary Fig. S3). In addition, HIF-1 signaling pathway, NF-κB signaling pathway, necroptosis and adherens junction were listed in the top 20 results of KEGG analysis, and these biological pathways were also closely related to ATAAD with ALI34. Recent evidence revealed that the activation of the HIF-1 signaling pathway could amplify the inflammatory response by stimulating the release of inflammatory factors, and promote the apoptosis of alveolar type II epithelial (AT-II) cells, which exacerbated the ALI35. The NF-κB family controls various processes such as immunity and inflammation36. Chen et al.37 confirmed that NF-κB signaling pathway was abnormally activated in the AT-II of patients with ALI and promoted the production of inflammatory factors including IL-1β, IL-6 and TNF-α, which initiated the inflammatory cascade. Besides, necroptosis might be involved in endothelial injury38 and adherens junction were critical for basal pulmonary microvascular integrity39. These results were similar to those of GO analysis, indicating the potential role of selected DE-miRNAs in ATTAD with ALI.
Next, we constructed the PPI network of those predicted target genes and identified the top 10 hub genes. Inspiringly, the changes in the 20 genes expression were generally consistent with previous studies’ results. For the upregulated genes, we found that RAS homolog family member A (RHOA) ranked first. Recent studies showed that RHOA and its downstream effector were closely related to the pathogenesis of ALI40. Upregulation of the RHOA/Rho kinase (ROCK) signaling pathway promotes immune cell migration and adhesion, accelerates pulmonary endothelial cells’ apoptosis, and these effects result in endothelium barrier dysregulation and edema—the hallmarks of ALI26. Besides, other hub genes such as janus kinase 3 (JAK3)41, calmodulin 1 (CALM1) 42 and ribosomal protein L13 (RPL13) 43 have also been reported to act as risk factors involved in the onset of ALI. For the downregulated genes, insulin receptors (INSR) had the highest connectivity degrees. It binds insulin to produce biological effects and is positively correlated with insulin level in the body44. Emerging evidence revealed that hyperinsulinemia had a protective effect on obese rats with lipopolysaccharide (LPS)-induced ALI by increasing their alveolar fluid clearance45. Furthermore, peroxisome proliferator-activated receptor-alpha (PPARA), as the predictive downregulated hub gene, has also been reported that it reduces inflammation and vascular leakage in a murine model of ALI46. These hub genes mentioned above may play a potential role in exploring novel mechanisms and therapeutic targets in ATAAD with ALI.
Moreover, based on sequencing results and previous bibliography retrieval, we selected 10 candidate miRNAs for validation. The RT-qPCR confirmed that hsa-miR-485-5p was significantly upregulated and hsa-miR-206 was significantly downregulated in ALI group. Of the two specific miRNAs, miR-206 has been previously reported to be associated with ALI. Zhou et al. 47 demonstrated that miR-206 expression was significantly decreased in lung tissue of sepsis-induced ALI mouse model, showing the same trend as our findings in plasma exosomes in ALI group, and the phenotype of ALI including the lung tissue inflammatory response, wet to dry weight ratio (W/D) and bronchoalveolar lavage fluid (BALF) could be attenuated by injection of miR-206 agomir. Further in vitro experiments revealed that downregulated miR-206 increased the permeability of alveolar air-blood barrier and promoted the development of ALI by targeting CX43 mRNA and upregulating CX43 expression in AT-II cells. These pathological changes above are also common in ATAAD-induced ALI4, suggesting a potential role of miR-206 in its pathogenesis. What’s more, miR-206 was found to be related to inflammation, apoptosis and autophagy8,48,49. For miR-485-5p, a relative study with reference to ATAAD or ALI is lacking. Existing evidence showed that it mediated inflammatory factors (TNF-α, IL-6, IL-8, etc.) involved in inflammation50. Other studies have reported its association with suppression of cell proliferation and migration51,52. Thus, we speculate that miR-485-5p may be involved in ATAAD with ALI by regulating inflammation and promoting pulmonary capillary endothelial damage. However, its precise function in this process remains to be elucidated.
As far as we know, this study is the first to explore the DE-miRNAs in plasma exosomes from ATAAD patients with ALI. We measured the plasma exosomal miRNA profile of these patients, which provided us with a novel source of biomarkers for this disease. In addition, plasma exosomal miRNAs bind and enter their target cells through the circulation to regulate the post-transcriptional process. Such mediation can help us better clarify the underlying mechanisms of ATAAD with ALI. However, there are still several limitations. Firstly, the validation cohort in this study was relatively small. The results need to be verified in a larger population. Secondly, the present study was lack of in vitro and in vivo experimental validation of the functions of the two confirmed exosomal miRNAs in ATAAD with ALI. Corresponding experiments will be performed in the future to gain insight into its pathogenesis.
Conclusion
In summary, this study has identified differential expression profile of plasma exosomal miRNAs in ATAAD with ALI, which were related to inflammation and dysfunction of cells junction. Further research is required to explore their potential role in pathogenic mechanisms.
Methods
Patients and ethics
59 patients diagnosed with ATAAD confirmed by computed tomographic angiography (CTA) of aorta according to 2014 ESC Guidelines53 were recruited at Xiangya Hospital of Central South University, from July to December in 2020. 19 Patients were enrolled as the discovery cohort (Non-ALI group, n = 8; ALI group, n = 11) and 40 patients were enrolled as the validation cohort (Non-ALI group, n = 20; ALI group, n = 20). Plasma exosome miRNAs were evaluated by next-generation sequencing (NGS) in the discovery cohort, and then, selected miRNA candidates from the differential expression profile of plasma exosomal miRNAs were validated by RT-qPCR in the validation cohort.
The diagnosis of ALI was based on the American-European Consensus Conference (AECC) definition54. The following four criteria were required to be present: (1) acute onset; (2) PaO2/FiO2 ≤ 300 mmHg [regardless of positive end-expiratory pressure (PEEP) level]; (3) bilateral infiltrates seen on frontal chest radiograph; (4) pulmonary artery wedge pressure (PAWP) ≤ 18 mmHg when measured or no clinical evidence of left atrial hypertension. Patients with the following conditions were excluded: those with chronic aortic dissection, chronic lung diseases, chronic renal or liver failure, immune system diseases, connective tissue diseases or malignant diseases.
The present study was approved by the Medical Ethical Committee of the Xiangya Hospital of Central South University (Hunan, China) and conducted following their guidelines and regulations. All patients provided written informed consent.
Exosome isolation
Whole blood (5–10 ml) was sampled from each patient on admission, centrifuged at 3000 rpm for 10 min at 4 °C. The collected plasma samples were stored − 80 °C until required. Exosomes were extracted by differential ultra-centrifugation as follows. Briefly, plasma (4 ml) was centrifuged at 12,000 rpm for 10 min at 4 °C to remove cell debris. The supernatant fluid was diluted with 20 ml phosphate-buffered saline (PBS) and filtered through a 0.22 µm disposable filter. Then, it was transferred to a ultracentrifuge (Beckman Coulter, Inc., Brea, CA) and ultracentrifuged using a Type 70Ti rotor (Beckman Coulter, Inc., Brea, CA) at 120,000×g for 90 min at 4 °C twice. The pellets containing exosomes were suspended in 200 µl PBS for further application.
TEM
For electron microscopy analysis, isolated exosomes suspended in PBS were dropped on copper-coated grids and stained with 2% phosphotungstic acid. After the grids were completely dried at room temperature, they were visualized using a Hitachi 7800 TEM (H-7800, Hitachi Ltd., Tokyo, Japan).
NTA
NTA were performed using a NanoFCM N30E instrument (N30E, Fujian, China) according to the manufacturer’s instruction. The exosome samples were diluted in PBS. The NanoFCM instrument captured the Brownian motion of the extracted nanoparticles and measured their size and concentration.
Western blot
The isolated exosomes were lysed in RIPA lysis buffer containing a protease inhibitor to extract protein and the samples were quantified by a BCA protein assay kit. Equal amounts of protein were separated on a 10% or 15% sodium dodecyl sulfate (SDS)—polyacrylamide gel electrophoresis (PAGE), and the bands were transferred to polyvinylidene fluoride (PVGF) membranes. After blocking with 5% skimmed milk for 1 h, the membranes were incubated primary antibodies at 4 °C overnight and then incubated with peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies for 1 h at room temperature. Finally, the membranes were visualized and analyzed using the ChemiDoc MP Imaging system (Bio-Rad, Hercules, CA, USA).
The primary antibodies included anti-tumor susceptibility gene 101 protein (TSG 101) antibody (ab125011, 1:1000, Abcam), anti-CD63 antibody (A5271, 1:1000, Abclonal) and anti-Calnexin antibody (ab22595, 1:1000, Abcam).
Exosomal RNAs extraction and NGS
Total RNA was extracted from plasma exosomes samples using miRNeasy® Mini kit (Qiagen, Cat. No. 217004). The RNA quality and quantity were evaluated by the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 System (Agilent Technologies, CA, USA). Then, the samples were used to establish a sequencing library and the quality of the library was tested with an Agilent 2100 bioanalyzer before the NGS.
Small RNA sequencing (16-30nt) was performed using the BGISEQ-500 sequencer (BGI Park, Shenzhen, China). Low-quality data were removed, and a substantial amount of small RNAs, which mapped perfectly to the human genome (ftp://ncbi.nlm.nih.gov/genomes/Homo_sapiens) were obtained for further analysis. The miRNA expression levels were estimated by the value of fragments perkiobase million (FPKM) and their relative expression levels were assessed by the DEGseq method. Quantitative data for each miRNA from the ALI and non-ALI group was compared to obtain absolute fold change (FC) values, and then log2 FC transformation were performed. Differentially expressed miRNAs (DE-miRNAs) was defined by an absolute value of log2 FC > 0.8, the Q-value (adjusted P-value) < 0.05 and the value of FPKM in each sample > 0.
Prediction of miRNAs potential TFs
The upstream TFs of DE-miRNAs were predicted using FunRich software55. The selected up-regulated or down-regulated miRNAs were inputted into the software, and the results showed the top 10 predictive TFs.
Prediction of miRNAs target genes and functional and pathway analysis
The downstream target genes of DE-miRNAs were predicted using three online database: TargetScan (http://www.targetscan.org/vert_72/) 56, miRbase (https://www.mirbase.org/) 57 and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) 58. The classification of main function annotation of predictive target genes was performed by GO analysis (www.geneontology.org) 59 and the identification of molecular pathways was applied by KEGG analysis (www.genome.jp/kegg) 60.
PPI network construction and analysis
The predicted target genes of the top 10 down- and upregulated DE-miRNAs were submitted to the STRING database which was used for a PPI network construction (http://string-db.org) 61. A composite score > 0.4 was defined as a significant interaction. Subsequently, the results were loaded into Cytoscape for visualization, and the top 10 hub genes were calculated by degree scoring62.
RT-qPCR of DE-miRNAs
Total RNA was reverse transcribed by using PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara, RR037A). The miRNA expression was examined by RT-qPCR using TaqMan® probe, and the sequence of primers and probes were shown in Supplementary Table S4 online. Each sample was examined in triplicate. The relative expression levels of exosomal miRNAs was analyzed by the ΔΔCt method relative to cel-miR-39.
Statistical analysis
Shapiro–Wilk tests were applied to test if the continuous variables conform to the normal distribution. Normally distributed continuous variables were presented as the mean ± SD while non-normally distributed continuous variables were presented as the median and interquartile range (IQR). Categorical variables were presented by number and percentage. Continuous data were compared using the Student t test or Mann–Whitney U test depending on the data’s distribution, and categorical data were compared using the chi-squared test. A two-tailed P value < 0.05 was considered statistically significant. Statistical analyses were performed using SPSS 25.0 software (SPSS, Inc., Chicago, IL, USA).
Data availability
The data analyzed in this study are available from the corresponding author on reasonable request.
Abbreviations
- ATAAD:
-
Acute type A aortic dissection
- ALI:
-
Acute lung injury
- ICU:
-
Intensive care unit
- miRNAs:
-
MicroRNAs
- MHC:
-
Myosin heavy chain
- RhoB:
-
Ras homolog gene family member B
- IBD:
-
Inflammatory bowel disease
- IL-6:
-
Interleukin-6
- CTA:
-
Computed tomographic angiography
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- AECC:
-
American-European Consensus Conference
- PaO2 :
-
Arterial oxygen tension
- FiO2 :
-
Inspiratory oxygen fraction
- PEEP:
-
Positive end-expiratory pressure
- PAWP:
-
Pulmonary artery wedge pressure
- PBS:
-
Phosphate buffered saline
- TEM:
-
Transmission electron microscopy
- NTA:
-
Nanoparticle tracking analysis
- SDS:
-
Sodium dodecyl sulfate
- PAGE:
-
Polyacrylamide gel electrophoresis
- PVGF:
-
Polyvinylidene fluoride
- TSG 101:
-
Anti-tumor susceptibility gene 101 protein
- NGS:
-
Next-generation sequencing
- FPKM:
-
Fragments perkiobase million
- FC:
-
Fold change
- DE-miRNAs:
-
Differentially expressed miRNAs
- TFs:
-
Transcription factors
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia Genes and Genomes
- PPI:
-
Protein–protein interaction
- 3′-UTR:
-
3′-Untranslated region
- LVEF:
-
Left ventricular ejection fraction
- BP:
-
Biological process
- CC:
-
Cellular component
- MF:
-
Molecular functions
- RHOA:
-
Ras homolog family member A
- ROCK:
-
Rho kinase
- JAK3:
-
Janus kinase 3
- CALM1:
-
Calmodulin 1
- RPL13:
-
Ribosomal protein L13
- INSR:
-
Insulin receptor
- PPARA:
-
Peroxisome proliferator-activated receptor-alpha
- W/D:
-
Wet to dry weight ratio
- BALF:
-
Bronchoalveolar lavage fluid
- AT-II:
-
Alveolar type II epithelial cells
References
Zhu, Y. et al. Type A aortic dissection-experience over 5 decades: JACC historical breakthroughs in perspective. J. Am. Coll. Cardiol. 76, 1703–1713 (2020).
Pan, X. et al. Independent factors related to preoperative acute lung injury in 130 adults undergoing Stanford type-A acute aortic dissection surgery: a single-center cross-sectional clinical study. J. Thorac. Dis. 10, 4413–4423 (2018).
Guo, Z. et al. Preoperative hypoxemia in patients with type A acute aortic dissection: A retrospective study on incidence, related factors and clinical significance. J. Thorac. Dis. 11, 5390–5397 (2019).
Wu, Z., Ruan, Y., Chang, J., Li, B. & Ren, W. Angiotensin II is related to the acute aortic dissection complicated with lung injury through mediating the release of MMP9 from macrophages. Am. J. Translat. Res. 8, 1426–1436 (2016).
Wu, Z. et al. A novel finding: Macrophages involved in inflammation participate in acute aortic dissection complicated with acute lung injury. Curr. Mol. Med. 17, 568–579 (2017).
Duan, X. Z. et al. Inflammation is related to preoperative hypoxemia in patients with acute Stanford type A aortic dissection. J. Thorac. Dis. 10, 1628–1634 (2018).
Wu, Z. et al. Dephosphorylation of Y685-VE-cadherin involved in pulmonary microvascular endothelial barrier injury induced by angiotensin II. Mediat. Inflamm. 2016, 8696481 (2016).
Wu, Z. et al. Angiotensin II induces apoptosis of human pulmonary microvascular endothelial cells in acute aortic dissection complicated with lung injury patients through modulating the expression of monocyte chemoattractant protein-1. Am. J. Translat. Res. 8, 28–36 (2016).
Van Deun, J. et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesic. 3, 248258 (2014).
Gao, Y. & Raj, J. U. Extracellular vesicles as unique signaling messengers: Role in lung diseases. Comprehens. Physiol. 11, 1351–1369 (2020).
Hessvik, N. P. & Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. (CMLS) 75, 193–208 (2018).
Yu, X., Odenthal, M. & Fries, J. W. Exosomes as miRNA carriers: Formation-function-future. Int. J. Mol. Sci. 17, 20160274 (2016).
Ferrari, D., Bianchi, N., Eltzschig, H. K. & Gambari, R. MicroRNAs modulate the purinergic signaling network. Trends Mol. Med. 22, 905–918 (2016).
Neudecker, V., Brodsky, K. S., Kreth, S., Ginde, A. A. & Eltzschig, H. K. Emerging roles for microRNAs in perioperative medicine. Anesthesiology 124, 489–506 (2016).
Neudecker, V., Yuan, X., Bowser, J. L. & Eltzschig, H. K. MicroRNAs in mucosal inflammation. J. Mol. Med. (Berlin, Germany) 95, 935–949 (2017).
Zheng, D. et al. The role of exosomes and exosomal microRNA in cardiovascular disease. Front. Cell Dev. Biol. 8, 616161 (2020).
Tamura, Y. et al. Predictors of hypoxemia in type-B acute aortic syndrome: A retrospective study. Sci. Rep. 11, 23413 (2021).
Zhao, W., Zheng, X. L. & Zhao, S. P. Exosome and its roles in cardiovascular diseases. Heart Fail. Rev. 20, 337–348 (2015).
Gao, M. et al. Sepsis plasma-derived exosomal miR-1-3p induces endothelial cell dysfunction by targeting SERP1. Clin. Sci. (London, England: 1979) 135, 347–365 (2021).
Jiang, K. et al. Peripheral circulating exosome-mediated delivery of miR-155 as a novel mechanism for acute lung inflammation. Mol. Ther. J. Am. Soc. Gene Ther. 27, 1758–1771 (2019).
Lee, L. K., Medzikovic, L., Eghbali, M., Eltzschig, H. K. & Yuan, X. The role of microRNAs in acute respiratory distress syndrome and sepsis, from targets to therapies: A narrative review. Anesth. Analgesia 131, 1471–1484 (2020).
Lee, T. J. et al. Strategies to modulate microRNA functions for the treatment of cancer or organ injury. Pharmacol. Rev. 72, 639–667 (2020).
Davis, B. N., Hilyard, A. C., Nguyen, P. H., Lagna, G. & Hata, A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell 39, 373–384 (2010).
Du, X., Pan, Z., Li, Q., Liu, H. & Li, Q. SMAD4 feedback regulates the canonical TGF-β signaling pathway to control granulosa cell apoptosis. Cell Death Dis. 9, 151 (2018).
Zhao, H. et al. Transforming growth factor β1/Smad4 signaling affects osteoclast differentiation via regulation of miR-155 expression. Mol. Cells 40, 211–221 (2017).
Abedi, F., Hayes, A. W., Reiter, R. & Karimi, G. Acute lung injury: The therapeutic role of Rho kinase inhibitors. Pharmacol. Res. 155, 104736 (2020).
Akalay, I. et al. Targeting WNT1-inducible signaling pathway protein 2 alters human breast cancer cell susceptibility to specific lysis through regulation of KLF-4 and miR-7 expression. Oncogene 34, 2261–2271 (2015).
Sokół, E. et al. microRNA-mediated regulation of splicing factors SRSF1, SRSF2 and hnRNP A1 in context of their alternatively spliced 3’UTRs. Exp. Cell Res. 363, 208–217 (2018).
Steiner, D. F. et al. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 35, 169–181 (2011).
Wang, R. et al. USF-1 inhibition protects against oxygen-and-glucose-deprivation-induced apoptosis via the downregulation of miR-132 in HepG2 cells. Biochem. Biophys. Res. Commun. 446, 1053–1059 (2014).
Wu, Z. et al. Obesity is a risk factor for preoperative hypoxemia in Stanford A acute aortic dissection. Medicine 99, e19186 (2020).
Yuan, X. et al. Targeting hypoxia signaling for perioperative organ injury. Anesth. Analgesia 126, 308–321 (2018).
Serocki, M. et al. miRNAs regulate the HIF switch during hypoxia: A novel therapeutic target. Angiogenesis 21, 183–202 (2018).
Ren, W. et al. JAK2/STAT3 pathway was associated with the protective effects of IL-22 on aortic dissection with acute lung injury. Dis. Mark. 2017, 1917804 (2017).
Liu, Y., Xiang, D., Zhang, H., Yao, H. & Wang, Y. Hypoxia-inducible factor-1: A potential target to treat acute lung injury. Oxid. Med. Cell. Longev. 2020, 8871476 (2020).
Albensi, B. C. What is nuclear factor kappa B (NF-κB) doing in and to the mitochondrion?. Front. Cell Dev. Biol. 7, 154 (2019).
Chen, R., Xie, F., Zhao, J. & Yue, B. Suppressed nuclear factor-kappa B alleviates lipopolysaccharide-induced acute lung injury through downregulation of CXCR4 mediated by microRNA-194. Respir. Res. 21, 144 (2020).
Matthay, M. A., Ware, L. B. & Zimmerman, G. A. The acute respiratory distress syndrome. J. Clin. Invest. 122, 2731–2740 (2012).
Matthay, M. A. et al. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers 5, 18 (2019).
Chen, G., Sun, X. & Dong, C. RhoA regulates lipopolysaccharide-induced lung cell injury via the Wnt/β-catenin pathway. Mol. Med. Rep. 16, 8501–8506 (2017).
Cao, F. et al. Network-based integrated analysis of transcriptomic studies in dissecting gene signatures for LPS-induced acute lung injury. Inflammation 21, 1–13 (2021).
Birukova, A. A. et al. Role of Rho GTPases in thrombin-induced lung vascular endothelial cells barrier dysfunction. Microvasc. Res. 67, 64–77 (2004).
Dong, J. et al. Ribosomal protein S3 gene silencing protects against cigarette smoke-induced acute lung injury. Mol. Ther. Nucleic Acids 12, 370–380 (2018).
Vigneri, R., Goldfine, I. D. & Frittitta, L. Insulin, insulin receptors, and cancer. J. Endocrinol. Invest. 39, 1365–1376 (2016).
Deng, J. et al. An increase in alveolar fluid clearance induced by hyperinsulinemia in obese rats with LPS-induced acute lung injury. Respir. Physiol. Neurobiol. 279, 103470 (2020).
Schaefer, M. B. et al. Peroxisome proliferator-activated receptor-alpha reduces inflammation and vascular leakage in a murine model of acute lung injury. Eur. Res. J. 32, 1344–1353 (2008).
Zhou, J., Fu, Y., Liu, K., Hou, L. & Zhang, W. miR-206 regulates alveolar type II epithelial cell Cx43 expression in sepsis-induced acute lung injury. Exp. Ther. Med. 18, 296–304 (2019).
Huang, C. Y., Deng, J. S., Huang, W. C., Jiang, W. P. & Huang, G. J. Attenuation of lipopolysaccharide-induced acute lung injury by hispolon in mice, through regulating the TLR4/PI3K/Akt/mTOR and Keap1/Nrf2/HO-1 pathways, and suppressing oxidative stress-mediated ER stress-induced apoptosis and autophagy. Nutrients 12, 1742 (2020).
Huang, Y. et al. Bone marrow mesenchymal stem cell-derived exosomal miR-206 promotes osteoblast proliferation and differentiation in osteoarthritis by reducing Elf3. J. Cell. Mol. Med. 25, 7734–7745 (2021).
Chen, H. O., Zhang, L., Tang, Z. Y. & Gong, Z. M. MiR-485-5p promotes the development of osteoarthritis by inhibiting cartilage differentiation in BMSCs. Eur. Rev. Med. Pharmacol. Sci. 22, 3294–3302 (2018).
Liu, Q. et al. miR-485-5p/HSP90 axis blocks Akt1 phosphorylation to suppress osteosarcoma cell proliferation and migration via PI3K/AKT pathway. J. Physiol. Biochem. 76, 279–290 (2020).
Zhang, Z., Li, X., Li, A. & Wu, G. miR-485-5p suppresses Schwann cell proliferation and myelination by targeting cdc42 and Rac1. Exp. Cell Res. 388, 111803 (2020).
Erbel, R. et al. 2014 ESC guidelines on the diagnosis and treatment of aortic diseases: Document covering acute and chronic aortic diseases of the thoracic and abdominal aorta of the adult. The Task Force for the Diagnosis and Treatment of Aortic Diseases of the European Society of Cardiology (ESC). Eur. Heart J. 35, 2873–2926 (2014).
Bernard, G. R. et al. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am. J. Respir. Crit. Care Med. 149, 818–824 (1994).
Pathan, M. et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicl. 6, 1321455 (2017).
Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, 5005 (2015).
Griffiths-Jones, S., Grocock, R. J., van Dongen, S., Bateman, A. & Enright, A. J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34, D140–D144 (2006).
Krüger, J. & Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–W454 (2006).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 11, R14 (2010).
Kanehisa, M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484 (2008).
Szklarczyk, D. et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
We would like to thank for Kai Nie statistical assistance. This work was supported by grant from the Hunan Provincial Natural Science Foundation of China (Hunan, China; Grant No. 2021JJ41041) and the ChangSha City Natural Science Foundation of China (ChangSha, Hunan, China; Grant No. kq2202363).
Author information
Authors and Affiliations
Contributions
G.Q.L. and Q.X. conceptualized this research aim, planned the analyses and guided the literature review. C.Y.Z. and H.B. extracted and analyzed the data from RNA-sequencing. Y.F.Z. and L.Z. participated in collecting clinical materials. C.Y.Z. wrote the first draft of the paper. L.Z., X.L.C., R.Z.S. and G.G.Z. revised and commented on the draft and overall responsibility. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, C., Bai, H., Zhang, L. et al. Differential expression profile of plasma exosomal microRNAs in acute type A aortic dissection with acute lung injury. Sci Rep 12, 11667 (2022). https://doi.org/10.1038/s41598-022-15859-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-15859-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.