Abstract
Currently, the humanity is in a fierce battle against various health-related challenges especially those associated with human malignancies. This created the urge to develop potent and selective inhibitors for tumor cells through targeting specific oncogenic proteins possessing crucial roles in cancer progression and survive. In this respect, new series of pyrazole-thiazol-4-one hybrids (9a–p) were synthesized as potential anticancer agents. All the synthesized molecules exhibited potent antiproliferative actions against breast cancer (BC) T-47D and MDA-MB-231 cell lines with IC50 ranges 3.14–4.92 and 0.62–58.01, respectively. Moreover, the most potent anti-proliferative counterparts 9g and 9k were assessed against EGFR. They displayed nanomolar inhibitory activity, IC50 267 ± 12 and 395 ± 17 nM, respectively. Worth noting, both compounds 9g and 9k induced apoptosis in MDA-MB-231 cells, and resulted in a cell cycle arrest at G2/M phase. Furthermore, an in silico analysis including docking and molecular dynamic simulations was performed.
Similar content being viewed by others
Introduction
In 2020, 2.3 million women were affected by breast cancer with nearly 685,000 deaths killed by the same disease worldwide. As the year ended, a report of the past 5 years, revealed 7.8 million alive women who diagnosed with breast cancer, giving it rise to be the most prevalent cancer globally1. More than 90% of BCs are not metastatic upon diagnosis. For breast cancer patients that diagnosed without metastatic disease, the main therapeutic objectives are eradication of the tumor and preventing recurrence. In many cases, patients with nonmetastatic breast cancer can be treated with chemotherapy as a systemic therapy2.
RTKs (receptor tyrosine kinases) are found on the cells surface and represent a type of receptor that plays a significant role in the genesis of tumors. RTKs are known to influence cancer stemness, angiogenesis, and metastasis through a variety of downstream signaling pathways3. RTKs are a good target for breast cancer treatment because of their several activities4. However, anti-RTK therapy is complicated by the mutations that are created in these biological targets5. Different types of RTKs are over-expressed in various human malignancies, including breast cancer, they are namely epidermal growth factor receptors (EGFRs), platelet-derived growth factor receptors (PDGFRs), vascular endothelial growth factor receptors (VEGFRs), fibroblast growth factor receptors (FGFRs) and insulin-like growth factor receptors (IGFRs)3,6,7,8. Overexpression of RTKs are associated with increased breast cancer aggressiveness8.
EGFR belongs to the ErbB family of RTK. These proteins are stimulated after binding with EGF-family of proteins. Many reports confirmed that the EGFR is involved in the pathogenesis of different cancer types9. The EGFR and its agonists are over-expressed in human malignancies producing cell transformation10. All EGFR types are expressed in breast cancer. EGFR has been reported to be over-expressed in about 14% of human breast tumors11.
Many EGFRs small molecule inhibitors (EGFRIs) have been developed. In comparison to other ErbB receptors, Gefitinib I (Fig. 1) is a reversible EGFRI with a 200-fold higher affinity for EGFR. It's been approved for the management of individuals with breast tumors and NSCLC who haven't responded to chemotherapies12,13. Erlotinib II (Fig. 1) is a reversible EGFRI that has been approved by the FDA for the management of NSCLC and metastatic pancreatic malignancy14. Gefitinib as well as Erlotinib work as ATP analogues through competing with the EGFR receptors' ATP binding spaces15. Despite the fact that EGFRIs have significant clinical outcomes in treating patients with breast cancer, tumor cells rapidly develop acquired resistance, in which the efficacy of EGFR-targeted therapy is highly limited16 (Fig. 1).
As depicted by Fig. 1, some pharmacophoric features of EGFR inhibitors are required for maximum affinity against the EGFR ATP binding site. These features include: (1) a central flat aromatic heterocyclic moiety that should be accommodated within the adenine binding pocket to interact with Leu768, Met769, Gln767 and Thr76617, (2) a terminal hydrophobic head which should occupy the hydrophobic region I to interact with Glu738, (3) a spacer of one atom (NH, spacer) to allow the terminal hydrophobic motif to access the hydrophobic region I18, (4) a hydrophobic tail that can occupy the hydrophobic region II19,20. Research about the ribose binding pocket has been limited21 (Fig. 1).
Literature investigation revealed that pyrazole moiety is a lead building block in many anticancer molecules through the inhibition of diverse RTKs including EGFR22,23. In addition, thiazol-4(5H)-one was reported as promising scaffold in the discovery of new antineoplastic agents targeting RTKs especially EGFR22,24.
Accordingly, the aim of this study is summarized on the design and synthesis of pyrazole- thiazol-4(5H)-one hybrids (9a-p, Fig. 1) having the essential pharmacophoric features of EGFRIs. In such design, the 2-(4,5-dihydro-1H-pyrazol-1-yl)thiazol-4-one moiety was utilized as an aromatic heterocyclic system to be convenient for the adenine binding pocket. In addition, diverse substituted phenyl rings were used as a hydrophobic head to accommodate within the hydrophobic pocket. Furthermore, the naphthyl moiety was used as a hydrophobic tail to occupy a second hydrophobic pocket. In this design, we gave two modifications on the reported EGFRIs. The first one, the NH linker was replaced by –CH= group as a chemical isostere. The second modification was the grafting of different aromatic substituents to explore the effect of such substituents on the biological activities (Fig. 1).
The synthesized molecules were examined as anti-proliferative agents against 2 BC MDA-MB-231 and T-47D cell lines. Then, the most promising members that showed superior cytotoxic activity were further evaluated for their EGFR inhibitory activity. In addition, the effects of these active members against cell cycle and apoptosis induction were examined. Finally, Molecular modelling studies including docking and molecular dynamic simulations were carried out to examine the binding mode of herein reported pyrazole- thiazol-4(5H)-one hybrids against their proposed biological target (EGFR).
Results and discussion
Chemistry
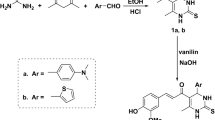
The adopted strategy to prepare the herein proposed pyrazole- thiazol-4(5H)-one hybrids 9a-p was illustrated in Scheme 1. 1-(Naphthalen-1-yl)ethan-1-one 1 was condensed with the appropriate 4-fluorobenzaldehyde 2a or 4-chlorobenzaldehyde 2b in a basic medium through the Claisen-Schmidt condensation reaction to furnish 3-(4-fluorophenyl)-1-(naphthalen-1-yl)propen-1-one 3a and 3-(4-chlorophenyl)-1-(naphthalen-1-yl)propen-1-one 3b, respectively.
Chalcones 3a-b were then subjected to heterocyclization to produce 3-(naphthalen-1-yl)-4,5-dihydro-pyrazole-1-carbothioamide derivatives 5a-b via refluxing in absolute ethanol with thiosemicarbazide and sodium hydroxide. The previously prepared thioamide compounds 5a-b were heated under reflux in absolute ethanol with ethyl bromoacetate 6 to get the two key intermediates 3-(naphthalen-1-yl)-4,5-dihydropyrazol-1-yl)thiazol-4(5H)-one derivatives 7a-b. Finally, the pyrazole- thiazol-4(5H)-one intermediates 7a-b were reacted with different aldehydes 8a-h via the Knoevenagel condensation by heating in glacial acetic acid and fused sod. acetate to yield the targeted pyrazole- thiazol-4(5H)-one hybrids 9a-p.
Spectral and elemental analyses data supported the proposed structure for target pyrazole- thiazol-4(5H)-one hybrids 9a-p herein reported. 1H NMR spectra for pyrazolines 9a-p showed three doublet of doublet signals of pyrazoline ring; one for CH and two for CH2 about δ (5.90) and (3.67 and 4.39) ppm, respectively with three reciprocal values for coupling constant (4.0, 11.2, 18.0) Hz. Moreover, 1H NMR spectra for compounds 9d-h and 9l-p confirmed their structure through the presence of additional aliphatic singlet signal for OCH3 around δ 3.80 ppm, whereas, the 1H NMR spectra of pyrazole- thiazol-4-one hybrids 9c and 9k revealed the presence of another aliphatic signal for (N(CH3)2) around δ 3.01 ppm. In turn, the 13C NMR spectra for the targeted pyrazole- thiazol-4-one hybrids 9a-p disclosed the presence of aliphatic signals of pyrazoline ring around (46.55 and 62.40) ppm, for CH2 and CH, respectively, in addition, the signal of (C = O) functional group appeared around δ 179.90 ppm. Lastly, the 13C NMR spectra of pyrazolines 9d-h and 9l-p disclosed the presence of the methoxy group around δ 56.0 ppm.
Biological evaluation
In vitro anti-proliferative activity
The cytotoxic effects of herein prepared pyrazole- thiazol-4-one hybrids 9a-p against BC MDA-MB-231 and T-47D cell lines, obtained from American Type Culture Collection (ATCC), were determined using the MTT assay protocol. It is worth to mention that the selected cancer cell lines in this study are EGFR-expressing cell lines25,26,27,28. Doxorubicin and Erolitinib were utilized as positive control (Table 1).
Generally, the obtained results disclosed that MDA-MB-231 cell line was more susceptible to the influence of the tested members than T-47D. Also, it was noticed that the 4-fluorophenyl derivative 9g (IC50 = 0.62 ± 0.03 against MDA-MB-231 and 3.14 ± 0.11 µM against T-47D) and the 4-chlorophenyl derivative 9k (IC50 = 1.14 ± 0.06 against MDA-MB-231 and 4.92 ± 0.28 µM against T-47D) were the most potent cytotoxic members against the two breast cancer cell lines, comparing to the positive control, doxorubicin (IC50 = 3.67 ± 0.2 and 2.73 ± 0.09 µM against MDA-MB-231 and T-47D, respectively), Table 1.
According to all results, it can be concluded that the 4-fluorophenyl derivatives 9a-h were slightly more advantageous than the 4-chlorophenyl derivatives 9i-p against the explor cell lines.
Regarding the activity toward MDA-MB-231 BC cell line, hybrids 9c, 9f, 9h, 9j and 9p displayed good cytotoxicity with (IC50 = 2.54 ± 0.138, 3.40 ± 0.185, 5.82 ± 0.317, 6.24 ± 0.34 and 6.55 ± 0.357 μM, respectively). In the meantime, compounds 9a, 9b, 9e, 9m, and 9p, with IC50 = 19.84 ± 1.08, 12.24 ± 0.67, 14.13 ± 0.77, 18.95 ± 1.03 and 10.81 ± 0.59 μM, respectively, exhibited moderate cytotoxicity, whereas, pyrazolines 9d, 9i, 9l, and 9n with IC50 ranging from 35.15 ± 1.916 to 58.01 ± 3.16 μM displayed the lowest cytotoxic activity.
Additionally, cytotoxicity evaluation against T-47D breast cancer cell line highlighted that compounds 9c and 9o displayed good anticancer actions with IC50 values of 8.45 ± 0.27 and 7.36 ± 0.30 μM, respectively. In turn, pyrazolines 9b, 9e, 9h, and 9p with (IC50 ranging from 11.42 ± 0.67 to 19.47 ± 0.92 μM) exhibited moderate cytotoxicity. Besides, compound 9f, 9j, 9m, and 9n with IC50 ranging from 27.1 ± 1.08 to 67.29 ± 2.51 μM demonstrated weak cytotoxic activity. Finally, pyrazolines 9a, 9d, 9i, and 9l appeared to be inactive.
Next, we inspected the influence of the substitution on the benzylidine moiety on the cytotoxic activities. Substitution of the benzylidine moiety at C-4 with hydrophilic or lipophilic electron donating groups (like the hydroxyl, methoxy and N,N-dimethylamino groups) resulted in an enhanced cytotoxic activities in comparison to the counterparts bearing unsubstituted benzylidine motif. In details, compounds 9b, 9c and 9e bearing para-substituted benzylidine motifs showed better activity against both MDA-MB-231 (IC50 = 12.24 ± 0.67, 2.54 ± 0.138 and 14.13 ± 0.77 μM, respectively) and T-47D (IC50 = 18.61 ± 0.87, 8.45 ± 0.27 and 3.14 ± 0.11 μM, respectively) cell lines than the unsubstituted analogue 9a (IC50 = 19.84 ± 1.08 and > 100 μM, toward MDA-MB-231 and T-47D cells respectively). Similarly, pyrazolines 9j, 9k and 9m exerted more potent activity towards both MDA-MB-231 (IC50 = 6.24 ± 0.34, 1.14 ± 0.06 and 18.95 ± 1.03 μM, respectively) and T-47D (IC50 = 35.10 ± 2.04, 4.92 ± 0.28 and 27.63 ± 1.24, respectively) cell lines than the corresponding unsubstituted counterpart 9i (IC50 = 24.68 ± 1.35 and > 100 μM, toward MDA-MB-231 and T-47D cells respectively).
It’s noteworthy mentioning that moving the methoxy group from para to ortho position led to a decrease of the cytotoxic influence against both the tested cancer cell lines; hybrids 9d (IC50 = 45.02 ± 2.45 and > 100 μM) and 9l (IC50 = 58.01 ± 3.16 and > 100 μM) vs. compounds 9e (IC50 = 14.13 ± 0.77 and 12.37 ± 0.33 μM) and 9m (IC50 = 18.95 ± 1.03 and 27.63 ± 1.24 μM). Moreover, it was found that the best di-substitution pattern (hydroxyl and methoxy substitution) for the benzylidene moiety is the 3-hydroxy-4-methoxy pattern for the 4-fluorophenyl bearing derivatives 9a-h and the 4-chlorophenyl bearing derivatives 9i-p against both the examined cell lines, except the 4-hydroxy-3-methoxy substitution in compound 9p toward MDA-MB-231 cell line.
Furthermore, the cytotoxic impact for the most potent anti-proliferative thiazolyl pyrazolines reported in this work against MDA-MB-231 cell line (9c, 9f., 9g, 9h, 9j, 9k and 9p; with IC50 value less than 10 µM) was evaluated toward the normal human MCF-10A cells (Table 1). The examined pyrazolines (9c, 9f., 9g, 9h, 9j, 9k and 9p) exerted weak to non-significant activities against MCF-10A cell line (IC50 range: from 31.74 ± 3.10 to 83.62 ± 6.74), which highlights their safety and selectivity toward the cancer cells.
EGFR inhibitory activity
From the aforementioned data presented in Table 1 concerning the cytotoxicity, compounds 9g and 9k were identified as the most promising members among the series and showed superior cytotoxicity against MDA-MB-231 and T-47D cell lines. So, compounds 9g and 9k were further evaluated for their potential EGFR inhibitory action, using erlotinib as a reference EGFR inhibitor.
Table 2 shows the data as IC50 values. The results displayed that the two candidates exhibited EGFR inhibitory activity at the nanomolar level. In comparison to erlotinib (IC50 = 57 ± 3 nM), compounds 9g and 9k exhibited good activity as EGFR inhibitors in nanomolar concentrations (IC50 = 267 ± 12 and 395 ± 17 nM, respectively).
Cell cycle analysis
In this work, MDA-MB-231 cells were treated with the most cytotoxic members (compounds 9g and 9k) at a concentration of 0.62 and 1.14 µM, respectively (corresponding to their IC50 values against MDA-MB-23 cells). Investigating the results, it can be noticed that compounds 9g and 9k induced a significant increase in the cell population at Sub-G1 phase (31.05% and 27.86%, respectively), comparing to control cells (2.13%). For the S phase, the percentage of MDA-MB-23 cells was increased from 32.11% in control cells to 35.43% for compound 9g while the % was decreased to 28.45% for compound 9k.
Moreover, compounds 9g and 9k exhibited insignificant decrease in the cell population (31.0217% and 40.61%, respectively) at G0-G1 phase, comparing to the control cells (53.45%). Additionally, a marked decrease in the cell population for compounds 9g (2%) and 9k (3.08%) was observed at the G2/M phase, comparing to control cells (12.31%). These findings verify that the cytotoxicity of compounds 9g and 9k against MDA-MB-231cells was due to arresting the cell growth at G2/M phase (Fig. 2, Table 3).
Apoptosis assay
The most potent anticancer agents in the current work 9g and 9k were chosen for the assessment of apoptosis in MDA-MB-231cell line using Annexin V/propidium iodide (PI) double staining assay method. In this method, MDA-MB-231 cells were incubated with compounds 9g and 9k at the IC50 concentrations (0.62 and 1.14 µM, respectively) for 48 h.
The results revealed that compounds 9g and 9k triggered more apoptotic cells in comparison to control cells. In particular, pyrazoline 9g induced apoptosis by 32.32% (early apoptosis = 8.39% & late apoptosis = 23.93%) while pyrazoline 9k induced apoptosis by 34.22% (early apoptosis = 14.22% & late apoptosis = 20%) compared to 0.94% in the control cells (0.73% and 0.21% for early and late apoptosis, respectively). From these findings, we concluded that compounds 9g and 9k could induce apoptosis in MDA-MB-231 cells (Fig. 3 and Table 4).
Modeling studies
Docking study
Elucidating the potential binding mode between new compounds and their target became inevitable in lead discovery studies to provide futuristic insights for further optimization. Accordingly, both compounds 9k and 9g were docked into the predetermined active site of EGFR using MOE 2019.02.
Exploring the binding mode of the crystal reference erlotinib with the EGFR has revealed the essential hydrogen bond interactions with the key residues Met 769 and Leu 768. Besides, the phenyl and quinazoline rings of erlotinib participated in carbon hydrogen bond interactions with Glu 738 and Gln 767, respectively. The importance of solvent molecules in EGFR active site was unequivocal, acting as a bridge for two interactions between the nitrogen of quinazoline ring with Cys 751 and Thr 766, Fig. 4.
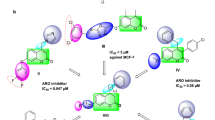
Interestingly, the docking of both compounds 9k and 9g induced a favorable binding upon interacting with EGFR through a binding pattern perfectly aligned with that of erlotinib. The synthesized compounds 9g and 9k achieved docking scores of −11.8 and −11.4 kcal/mol respectively, that nearly match the docking score of erlotinib (− 12.0 kcal/mol). As depicted from Fig. 5, the carbonyl group of the thiazol-4(5H)-one ring in compound 9g acted as a hydrogen bond acceptor for 2 H-bonds with the key residues Met 769 and Leu 768, in addition the para methoxy group of compound 9g was engaged in a carbon hydrogen bond interaction with Glu738. Once again, the water molecules played a crucial role in linking compound 9g with Thr766 and Gln767.
In a similar manner, compound 9k engaged in two hydrogen bonds via its carbonyl group with the essential residues Met 769 and Leu 768. The N,N-dimethyl aniline ring of compound 9k involved in one hydrogen bond with Lys 721 and three carbon hydrogen bonds with Glu738, Asp831 and a water molecule. Unlike compound 9g, the water effect was less significant in case of compound 9k, which explains the superiority of compound 9g in docking score and biological activity. Noteworthy, the difference in the kinase activity of compounds 9g and 9k could be attributed to the grafting of different substituents (para-fluoro in 9g and para-chloro in 9k) to 4,5-dihydro-1H-pyrazolyl ring. The large size of the chlorine atom hinders the ability of compound 9k to get closer enough with the solvent molecules unlike the smaller fluorine atom that allows compound 9g to get closer enough to the water molecule attached to the Thr766 and Gln767, Fig. 5–6.
Molecular dynamics
RMSD and RMSF analysis
In the current study, further in silico investigations were achieved through molecular dynamic simulations. Molecular dynamics (MD) simulation provides many valuable information and parameters to study the dynamicity of biological systems. Amongst these information, MD could provide insights into precise estimation of the binding strength of a docked complex of a ligand and a target. Accordingly, the predicted binding co-ordinates retrieved from the docking of EGFR with 9g, 9k and erlotinib were moved forward to MD simulation. To provide a comparative mean for the effect of each ligand on the stability of the EGFR enzyme, the later was subjected to MDS using the Apo form.
As demonstrated by Fig. 7, all the three inhibitors were able to stabilize the EGFR enzyme as indicated by their lower RMSD values comparing to the RMSD value of Apo EGFR. The EGFR-9g and EGFR-9k complexes had RMSD values of 1.8 and 2.1 Å, respectively, that are very comparable to that of EGFR-erlotinib (1.75 Å), while the RMSD of the Apo EGFR reached 4.5 Å. In cancer cells, EGFR serves as the switch-on for the intracellular signaling pathways to trigger cell division and uncontrolled growth. In this respect, the high dynamicity seen in the Apo EGFR as discerned from the high RMSD values is perfectly aligned with its intended oncogenic function. The capability of compounds 9g and 9k to restrict the dynamic nature of the EGFR via the formation of stable complexes as indicated by the lower RMSD values is a valid indicator for their inhibitory impact on EGFR (Fig. 7).
To further endorse the outputs of the RMSD calculations, the RMSF values for all the residues in the four systems have been computed. As expected the RMSF values lead to the same conclusion retrieved from the RMSD calculations in which the average RMSF of the Apo EGFR residues reached 4.2 Å, while the average RMSF values for the EGFR residues in complex with 9g, 9k and erlotinib reached an average 1.8, 2.1, 1.7 Å respectively (Fig. 8). To this end, both RMSD and RMSF values validated the proposed binding mode between 9g and 9k with the EGFR active site and attributed the inhibitory activity of both molecules to their ability to form a stable complex with the EGFR.
Binding free energy calculations using MM-PBSA approach
Attempting to further evaluate the strength of binding between the EGFR enzyme and the newly prepared compound 9g and 9k, the g_mmpbsa package generated by Kumari et al. was brought in action to calculate the binding free energies between the EGFR enzyme and the proposed molecules 9g and 9k. The generated trajectories from the production stage were used to calculate all the forms of binding free energy. These energy types include Electrostatic energy, van der Waal energy, Polar solvation energy and SASA energy. All the previous types of energy were calculated to the three complexes containing EGFR bound to either 9g, 9k or erlotinib (Table 5).
Interestingly, the calculated binding free energy for the three small molecules were comparable to each other in which compound 9g and 9k achieved binding free energies of − 288.1 ± 4.9 and − 251.9 ± 3.8 (kJ/mol) respectively, whereas the crystal reference erlotinib achieved − 286.8 ± 4.4 (kJ/mol). These results augmented all the in silico calculations giving credit to the predicted binding mode of both 9g and 9k within EGFR. Moreover, all the MD and energy calculations favored compound 9g over compound 9k as consistent with the early results of enzyme assay.
In terms of binding energy the energy contribution for each residue was calculated to provide deep insights into the effect of compounds 9g and 9k upon engaging in the binding pocket of EGFR. This contribution was calculated through decomposing the total binding free energy of each complex into per residue contribution energy. As demonstrated by Fig. 9, the binding of compound 9g to the EGFR active site resulted in favor energy contribution in all the Key residues especially those of close contact to compound 9g such as Met769 and Leu768 Glu738 Thr766 and Gln767. In a similar fashion, the binding of compound 9k contributed favorably to the free energy of the surrounding residues significantly to Met769, Leu768 and Glu738 and in lesser extent to Thr766 and Gln767. Consistent with the results of docking, MD and MMPBSA analyses, the per residue decomposition results supported the predicted binding mode of compounds 9g and 9k with EGFR kinase.
Conclusions
In the current work, new series of pyrazole-thiazol-4(5H)-one hybrids (9a-p) were synthesized as potential anti-breast cancer molecules. The anti-proliferative activities for the synthesized molecules (9a-p) were evaluated against MDA-MB-231 and T-47D BC cell lines. The obtained results showed that MDA-MB-231 cell line was more sensitive to the influence of the tested members than T-47D. Also, it was noticed that the 4-fluorophenyl derivative 9g (IC50 = 0.62 ± 0.03 against MDA-MB-231 and 3.14 ± 0.11 µM against T-47D) and the 4-chlorophenyl derivative 9k (IC50 = 1.14 ± 0.06 against MDA-MB-231 and 4.92 ± 0.28 µM against T-47D) were the most potent cytotoxic members against the two BC cell lines, comparing to the positive control, doxorubicin (IC50 = 3.67 ± 0.2 and 2.73 ± 0.09 µM against MDA-MB-231 and T-47D, respectively). Accordingly, both compounds 9g and 9k were tested for their activity against EGFR, where their achieved nanomolar inhibitory activity; IC50 = 267 ± 12 and 395 ± 17 nM for 9g and 9k, respectively. Further investigations unveiled their ability to arrest cell cycle at G2/M phase in addition to apoptosis induction in the MDA-MB-231 cell line as evidenced by Annexin V analysis. Interestingly, the docking of both compounds 9g and 9k induced a favorable binding upon interacting with EGFR through a binding pattern perfectly aligned with the crystal reference erlotinib. Compound 9g achieved docking score (− 11.8 kcal/mol) favorable than 9k (− 11.4 kcal/mol) and both scores are close to the docking score of erlotinib (− 12.0 kcal/mol). The docking results were further endorsed by 200 ns of molecular dynamic simulations alongside with MM-PBSA calculations and both revealed that compound 9g had more stable complex with EGFR than 9k with RMSD of 1.8, Å and binding free energy of − 288.1 ± 4.9 kJ/mol. The results of per residue decomposition supported predicted binding mode of compounds 9g and 9k with EGFR enzyme and showed favorable energy contribution of the key residues in the active site especially upon the binding compound 9g. The overall molecular modeling results attributed the superiority of compound 9g to the smaller size of the fluorine substituent that enables perfect fitting of compound 9g in EGFR active pocket.
Experimental
Chemistry
General
The IR spectra were recorded on Schimadzu FT-IR 8400S spectrophotometer. The NMR spectra were recorded via Bruker spectrometer at 400 MHz. 13C NMR spectra were run at 100 MHz in DMSO-d6. Chemical shifts (δH) were reported relative to the solvent (DMSO-d6). Elemental analyses were performed by FLASH 2000 CHNS/O analyzer, Thermo Scientific. Chalcones 3a-b were reported previously29,30.
Synthesis of 3-(naphthalen-1-yl)-4,5-dihydropyrazole-1-carbothioamide derivatives 5a-b
To hot stirred suspension of 1-(naphthalen-1-yl)propen-1-one derivatives 3a-b (4.5 mmol) in 30 mL of abs. EtOH with sodium hydroxide (0.55 g, 13.5 mmol), equivalent amount of thiosemicarbazide 4 (0.40 g, 4.5 mmol) was added. The mixture was heated under reflux for 2 h with TLC monitoring. After full consumption of all starting, the produced precipitate was collected, washed with hot water (4 × 5 mL) and recrystallized form acetonitrile to furnish the intermediates 3-(naphthalen-1-yl)-4,5-dihydro-1H-pyrazole-1-carbothioamides 5a-b, which utilized in the next reaction without more purification.
Synthesis of 3-(naphthalen-1-yl)-4,5-dihydropyrazol-1-yl)thiazol-4(5H)-one derivatives 7a-b
The previously prepared thioamide intermediates 5a-b (2 mmol) and sod. acetate (0.34 g, 4 mmol) were added to a solution of ethyl bromoacetate 6 (0.33 g, 2 mmol) in absolute ethanol (12 mL). The reaction mixture was refluxed for 4 h with TLC monitoring. After completion of the reaction, it was cooled to r.t. The formed solid was filtrated, washed with hexane (3 × 4 mL) and recrystallized from glacial acetic acid to produce the key intermediates 3-(naphthalen-1-yl)-4,5-dihydro-1H-pyrazol-1-yl)thiazol-4(5H)-one derivatives 7a-b, which exploited in the next step without further purification.
Synthesis of target molecules 9a-p
To hot stirred solution of the appropriate aldehydes 8a-h (0.5 mmol) in glacial AcOH (15 mL) with two equivalent amount of fused sod. acetate (0.08 g, 1 mmol) at round flask, an equivalent amount of the previously prepared key intermediates 7a-b (1 mmol) was added. The reaction solution was left under reflux for 6 h, the formed solid was collected while hot, washed with MeOH, and recrystallized from DMF to get the targeted 9a-p.
The spectral (IR and NMR) and elemental analysis for all the newly prepared pyrazolines 9a-p were provided in the Supporting Materials.
Biological evaluation
All the biological assays conducted in this study were performed as reported earlier; MTT cytotoxicity31,32 cell cycle33, Annexin V-FITC Apoptosis34,35,36 and EGFR kinase37 assays, (Supplementary Materials).
Molecular modeling studies
Docking study
In this work, all the docking studies were conducted using Molecular operating environment (MOE 2019.02) Software38,39. The X-ray crystal structures of EGFR in complex with erlotinib were downloaded from the protein databank PDB IDs 1M17. At the beginning, the hydrogens and charges of the receptors were optimized using AMBER10: EHT embedded in MOE software. The binding site of the EGFR enzyme was constructed where the co-crystalized erlotinib is bound. As a part of docking validation, the x-ray coordinates of the co-crystalized ligand was retrieved through re-docking into the pre-determined active site. The previous step resulted in RMSD value of 0.77 Å between the co-crystalized pose and the docking pose indicating a valid docking protocol. After that, compounds 9g and 9k were docked into the EGFR binding domain using triangular matcher and London dg as a placement and scoring methods, respectively. At last, 2D and 3D interaction diagrams were generated by MOE to analyze the socking results.
Molecular dynamics
In this work, 4 molecular dynamic simulations (MDS) were conducted for 200 ns using GROMACS 2.1.1 software40. The free EGFR enzyme and the retrieved docking coordinates of the same enzyme bound Erlotinib, 9k and 9g were used as input structures for the molecular dynamics. The typical work scheme of Gromacs simulation was applied to conduct the four MDS41,42,43,44,45 (Supplementary Materials).
MM-PBSA calculation and per residue contribution
The MM-PBSA package of Kumari et al.46 was contrived to calculate the binding free energy between the ligands and the EGFR enzyme using the following equation.
\(\Delta {\text{G}}_{{({\text{Binding}})}} = {\text{ G}}_{{({\text{Complex}})}} - {\text{ G}}_{{({\text{Receptor}})}} - {\text{ G}}_{{\left( {{\text{Ligand}}} \right) }}\) (Supplementary Materials)
Both the complexes of EGFR-9g and EGFR-9k complexes were subjected to such calculations and compared to the binding energy values of EGFR-Erlotinib.
To evaluate the contribution of each residue to the binding of 9g and 9k, the total free energy of each complex was decomposed per residues.
Data availability
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
WHO. Breast cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 1 December 2021).
Waks, A. G. & Winer, E. P. Breast cancer treatment: A review. JAMA 321, 288–300 (2019).
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Hsu, J. L., Hung, M.-C.J.C. & Reviews, M. The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev. 35, 575–588 (2016).
Butti, R. et al. Receptor tyrosine kinases (RTKs) in breast cancer: Signaling, therapeutic implications and challenges. Mol. Cancer 17, 1–18 (2018).
Tomiguchi, M. et al. Fibroblast growth factor receptor-1 protein expression is associated with prognosis in estrogen receptor-positive/human epidermal growth factor receptor-2-negative primary breast cancer. Cancer Sci. 107, 491–498 (2016).
Palmieri, D. et al. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 67, 4190–4198 (2007).
Templeton, A. J. et al. Prognostic relevance of receptor tyrosine kinase expression in breast cancer: A meta-analysis. Cancer Treat. Rev. 40, 1048–1055 (2014).
Normanno, N., Bianco, C., De Luca, A. & Salomon, D. S. The role of EGF-related peptides in tumor growth. Front. Biosci. Landmark 6, 685–707 (2001).
Normanno, N. et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366, 2–16 (2006).
Maennling, A. E. et al. Molecular targeting therapy against EGFR family in breast cancer: Progress and future potentials. Cancers 2019, 11 (1826).
Yanase, K. et al. Gefitinib reverses breast cancer resistance protein-mediated drug resistance. Mol. Cancer Therap. 3, 1119–1125 (2004).
Tiseo, M. et al. Predictors of gefitinib outcomes in advanced non-small cell lung cancer (NSCLC): Study of a comprehensive panel of molecular markers. Lung Cancer 67, 355–360 (2010).
Bareschino, M. A. et al. Erlotinib in cancer treatment. Ann. Oncol. 18, vi35–vi41 (2007).
Kumar, A., Petri, E. T., Halmos, B. & Boggon, T. J. The structure and clinical relevance of the EGF receptor in human cancer. J. Clin. Oncol. 26, 1742 (2008).
Shien, K. et al. Acquired resistance to EGFR inhibitors is associated with a manifestation of stem cell–like properties in cancer cells. Cancer Res. 73, 3051–3061 (2013).
Zhao, Z. et al. Exploration of type II binding mode: A privileged approach for kinase inhibitor focused drug discovery?. ACS Chem. Biol. 9, 1230–1241 (2014).
Furet, P. et al. Modelling study of protein kinase inhibitors: Binding mode of staurosporine and origin of the selectivity of CGP 52411. J. Comput. Aided Mol. Des. 9, 465–472 (1995).
Gandin, V. et al. Targeting kinases with anilinopyrimidines: discovery of N-phenyl-N’-[4-(pyrimidin-4-ylamino) phenyl] urea derivatives as selective inhibitors of class III receptor tyrosine kinase subfamily. Sci. Rep. 5, 16750 (2015).
Liu, Y. & Gray, N. S. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2, 358–364 (2006).
Zhang, J., Yang, P. L. & Gray, N. S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28 (2009).
Qiu, K.-M. et al. Design, synthesis and biological evaluation of pyrazolyl-thiazolinone derivatives as potential EGFR and HER-2 kinase inhibitors. Bioorg. Med. Chem. 20, 2010–2018 (2012).
Saleh, N. M., El-Gazzar, M. G., Aly, H. M. & Othman, R. A. Novel anticancer fused Pyrazole derivatives as EGFR and VEGFR-2 dual TK inhibitors. Front. Chem. 7, 917 (2020).
Ghode, P., Tripathi, R., Jain, K. & S.,. 2-(2-Arylidenehydrazinyl) thiazol-4 (5H)-ones as epidermal growth factor receptor inhibitors: A combined quantitative structure activity relationship and pharmacophore study. Curr. Enzyme Inhib. 12, 137–144 (2016).
Fitzpatrick, S. L., LaChance, M. P. & Schultz, G. S. Characterization of epidermal growth factor receptor and action on human breast cancer cells in culture. Cancer Res. 44(8), 3442–3447 (1984).
Hossein-Nejad-Ariani, H., Althagafi, E. & Kaur, K. Small peptide ligands for targeting EGFR in triple negative breast cancer cells. Sci. Rep. 9(1), 1–10 (2019).
Yarden, R. I., Wilson, M. A. & Chrysogelos, S. A. Estrogen suppression of EGFR expression in breast cancer cells: A possible mechanism to modulate growth. J. Cell. Biochem. 81(S36), 232–246 (2001).
Liao, W.-S. et al. Targeting EGFR of triple-negative breast cancer enhances the therapeutic efficacy of paclitaxel-and cetuximab-conjugated nanodiamond nanocomposite. Acta Biomater. 86, 395–405 (2019).
Ingarsal, N., Saravanan, G., Amutha, P. & Nagarajan, S. Synthesis, in vitro antibacterial and antifungal evaluations of 2-amino-4-(1-naphthyl)-6-arylpyrimidines. Eur. J. Med. Chem. 42(4), 517–520 (2007).
Jacques, A. V. et al. Synthesis of chalcones derived from 1-naphthylacetophenone and evaluation of their cytotoxic and apoptotic effects in acute leukemia cell lines. Bioorg. Chem. 1(116), 105315 (2021).
Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983).
El-Naggar, M., Almahli, H., Ibrahim, H. S., Eldehna, W. M. & Abdel-Aziz, H. A. Pyridine-ureas as potential anticancer agents: Synthesis and in vitro biological evaluation. Molecules 23(6), 1459 (2018).
Mohammed, E. R. & Elmasry, G. F. Development of newly synthesised quinazolinone-based CDK2 inhibitors with potent efficacy against melanoma. J. Enzyme Inhib. Med. Chem. 37(2021), 686–700 (2022).
El-Naggar, M. et al. Novel thiazolidinone/thiazolo [3,2-a] benzimidazolone-isatin conjugates as apoptotic anti-proliferative agents towards breast cancer: One-pot synthesis and in vitro biological evaluation. Molecules 23(6), 1420 (2018).
Hagras, M. et al. Discovery of new quinolines as potent colchicine binding site inhibitors: Design, synthesis, docking studies, and anti-proliferative evaluation. J. Enzyme Inhib. Med. Chem. 36(1), 640–658 (2021).
Mahdy, H. A. et al. Design, synthesis, molecular modeling, in vivo studies and anticancer evaluation of quinazolin-4(3H)-one derivatives as potential VEGFR-2 inhibitors and apoptosis inducers. Bioorg. Chem. 94, 103422 (2020).
Elmetwally, S. A., Saied, K. F., Eissa, I. H. & Elkaeed, E. B. Design, synthesis and anticancer evaluation of thieno[2,3-d]pyrimidine derivatives as dual EGFR/HER2 inhibitors and apoptosis inducers. Bioorg. Chem. 88, 102944 (2019).
Scholz, C., Knorr, S., Hamacher, K. & Schmidt, B. DOCKTITE a highly versatile step-by-step workflow for covalent docking and virtual screening in the molecular operating environment. J. Chem. Inf. Model. 23, 398–406 (2015).
Cozza, G. & Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 8, 1555–1572. https://doi.org/10.1002/jcc.21256 (2008).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Schüttelkopf, A. W. & van Aalten, D. M. F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D 60, 1355–1363 (2004).
Hassab, M. A. E. et al. Toward the identification of potential α-ketoamide covalent inhibitors for SARS-CoV-2 main protease: Fragment-based drug design and MM-PBSA calculations. Processes 9(6), 1004 (2021).
El Hassab, M. A. et al. Identification of a new potential SARS-COV-2 RNA-dependent RNA polymerase inhibitor via combining fragment-based drug design, docking, molecular dynamics, and MM-PBSA calculations. Front. Chem. 8, 915 (2020).
El Hassab, M. A. et al. In silico identification of novel SARS-COV-2 2′-O-methyltransferase (nsp16) inhibitors: Structure-based virtual screening, molecular dynamics simulation and MM-PBSA approaches. J. Enzyme Inhib. Med. Chem. 36(1), 727–736 (2021).
El Hassab, M. A. et al. In silico identification of potential SARS COV-2 2′-O-methyltransferase inhibitor: Fragment-based screening approach and MM-PBSA calculations. RSC Adv. 11(26), 16026–16033 (2021).
Kumari, R., Kumar, R. & Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 54(7), 1951–1962 (2014).
Acknowledgements
The authors extend their appreciation to the Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R25), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code (22UQU4290565DSR29), maabourehab@uqu.edu.sa.
Author information
Authors and Affiliations
Contributions
Most of work was done at Kafrelsheikh University and all the authors contributed equally to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Eldehna, W.M., El Hassab, M.A., Elsayed, Z.M. et al. Design, synthesis, in vitro biological assessment and molecular modeling insights for novel 3-(naphthalen-1-yl)-4,5-dihydropyrazoles as anticancer agents with potential EGFR inhibitory activity. Sci Rep 12, 12821 (2022). https://doi.org/10.1038/s41598-022-15050-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-15050-8
This article is cited by
-
Design, synthesis, computational study and cytotoxic evaluation of some new quinazoline derivatives containing pyrimidine moiety
Scientific Reports (2023)
-
Bioinspired thiazolo-[2,3-b] quinazolin-6-one derivatives as potent anti-cancer agents targeting EGFR: their biological evaluations and in silico assessment
Molecular Diversity (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.