Abstract
Freshwater ecosystems are under multiple threats in modern times such as water extraction for human consumption, industries and agricultural activities, water contamination and habitat destruction for example. At the same time the biodiversity of these ecosystems are often poorly studied, especially in arid countries such as Iran. In this work, we study one of the ecologically important members of Iranian freshwater fauna, freshwater crab species of the genus Potamon. Here, we barcoded the different populations occurring in the country and delimited the species to allow for a better understanding of their distribution and taxonomy. In this study, we evaluated the taxonomical statues of Potamon species in Iran using genetic data. In addition, we created the first barcoding reference for Iranian freshwater crabs, which is an important resource for future environmental and conservation studies.
Similar content being viewed by others
Introduction
Freshwater crabs (Crustacea: Decapoda: Brachyura Linnaeus, 1758) are different groups of Eubrachyurans, comprising approximately 20% of the biodiversity of Brachyuran. They are classified into three superfamilies: Potamoidea (Ortmann, 1896), Gecarcinucoidea Rathbun, 1904 and Trichodactyloidea (H. Milne Edwards, 1853)1. A new superfamily Pseudothelphusoidea has been proposed recently2. More than 1300 extant species of freshwater crabs have been described to date1. Freshwater crabs have a pantropical distribution range in the inland water bodies of the continents and adjacent islands, mainly in tropical and subtropical habitats of Neotropical, Palaearctic, Oriental, Australasian, and Afrotropical biogeographic regions1. They are among the largest detritivorous macroinvertebrate species3 in freshwater ecosystems, where they play vital functional roles in ecological structure4, 5.
On the other hand, due to the rapid loss and deterioration of habitats in freshwater ecosystems, especially in the tropics, the survival of many species, including crabs, is endangered. Approximately one-sixth of all freshwater crab species are at high risk of extinction, and approximately one-third are endangered6. Most species of endangered crabs are endemic to a restricted-range habitat and survive under the pressure of habitat loss, changing drainage patterns, and water pollution. Therefore, more studies are needed to understand their diversity and to protect endangered populations6. This is especially important in Middle Eastern countries, where freshwater ecosystems are under numerous environmental pressures. Iran, in the region, has one of the most diverse varieties of freshwater habitats, but most freshwater studies focus on ichthyofauna conservation7. Freshwater studies on freshwater crabs usually focus on taxonomy and species descriptions. In Brandis et al.8, seven species of freshwater crabs of the genus Potamon have been reported from Iran: Potamon persicum Pretzmann, 1962; P. ruttneri Pretzmann, 1962; P. strouhali Pretzmann, 1962; P. transcaspicum Pretzmann, 1962; P. ibericum (Beiberstein, 1808); P. bilobatum Storch and Turkay, Brandis, 2000 and P. gedrosianum Alcock,1909. Later, Keikhosravi and Schubart described a new species, P. ilam Keikhosravi and Schubart, 20149, and revalidated P. elbursi Pretzmann, 197610. Posteriorly, P. gedrosianum was reported in Iran from Zabol (southeastern Iran)11.
The identification of the different species in the field, in collections or in labs is relatively challenging due to the similarity and high variation in the morphological characteristics of these species. The lack of taxonomic expertise and field guides makes it difficult to truly evaluate the conservation statuses and diversity of these important members of freshwater ecosystems. In recent years, with the development and proliferation of molecular techniques, one could easily identify different populations, directly or indirectly (i.e., environmental DNA approaches). Molecular approaches present their own set of challenges, one of them being the availability of easily accessible reference databases. In this study, we sampled different populations of freshwater crabs of the Potamon genus to (i) identify their taxonomic placement, (ii) evaluate the taxonomic validity of the recognized species using molecular data, and (iii) create a barcode reference for different species inhabiting Iranian freshwater ecosystems.
Results
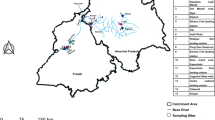
The final sampling resulted in 110 individuals from six species (Table 1), which covers all the major freshwater bodies of Iran inhibited by this genus (Fig. 1). The final dataset consisted of 923 positions (mean sequence length of 798 bp), from which 205 were parsimony informative, 36 singletons and 682 invariable sites. ModelFinder analysis did not merge any partitions; therefore, each codon position formed an independent partition. The resulting phylogenetic tree (Fig. 2) does not have enough resolution to recover the phylogenetic relationships of the genus. However, each species forms a relatively clear cluster, which helps identify the species boundaries. All species recovered highly supported monophyletics, with the exception of Potamon persicum. The only two sequences representing P. bilobatum in our study were clustered inside the P. ibericum clade, making them unidentifiable from the latter mentioned species. Sequences identified as P. gedrosianum were placed with high support as the sister group to all other species of Potamon from Iran. P. transcaspicum was only represented by a single sequence. Despite having a wide distribution and being overrepresented in this study, P. ibericum does not show a clear population structure. The genetic distances observed within each species were highest in P. ibericum, with a 2% genetic distance (Table 2). The shortest genetic distance between sister species is 3% between P. persicum and P. ilam.
The sampling map of the studied populations. Points marked with an blue pentagon represent P. ibericum, red star P. bilobatum, green circle P. elbursi, pink triangle P. persicum, light blue rectangle P. ilam, yellow asterisk P. transcaspicum, orange hexagon P. ruttneri and brown diamond P. gedrosianum. The colours used for each species correspond to the same colours used in Fig. 2. The map was created using the software ArcGIS 10.8.1.
Discussion
At present, the Iranian members of the genus Potamon are represented by 9 species in the literature: P. bilobatum; P. elbursi; P. gedrosianum; P. ibericum; P. ilam; P. persicum; P. ruttneri; P. strouhali and P. transcaspicum. Based on our results, we suggest that the taxonomic status of P. bilobatum should be studied in more detail, and our study supports the synonymy of P. bilobatum with P. ibericum. As seen in Fig. 2, both nominal species are indifferent from each other in the tree. This result relies on the sequences of samples identified in other studies12, where the paratypes of the P. bilobatum have been sequenced. Even if the COI barcode region did separate perfectly the other species studied here, a single marker might not be sufficient to confirm the taxonomy of the genus. Therefore, we believe more specific studies on the subject are needed to resolve P. bilobatum’s taxonomic status.
The result of our analyses divides the samples identified as P. persicum into two independent lineages, which could be caused by the lack of resolution and support in that part of the tree. This could be improved with a higher sampling size for the populations of this species. On the other hand, the average genetic distance within all samples identified as P. persicum was comparable to the average genetic distances within P. ibericum samples. This supports the idea that the structure observed in the tree for P. persicum corresponds to population structures observable in widespread species and is probably not due to a speciation event. The interspecific and intraspecific genetic distance gap in Iranian members of the Potamon genus seems to be a value between 2 and 3% genetic distance.
Conclusions
In this study, we present the first barcode reference for different populations of potamid crabs inhabiting Iranian freshwater bodies. We evaluated the taxonomic statuses of different described species using molecular data that showed rather high genetic diversity within species. This is a first step to improve the identification of the different species for future studies using molecular techniques. Our results offer an important molecular resource for environmental and conservation studies. We believe these results are especially important these days, as eDNA approaches are becoming an important part of all conservation and biodiversity studies, and these approaches rely strongly on molecular references. Proper species identification is the basis for future studies on the ecology and conservation of these highly susceptible species to climate change.
Methods
Taxon sampling
A total of 35 specimens from 19 localities were sampled in this project, covering the main distribution range of the genus in Iran. In addition, all available barcode sequences from Iran in GenBank, a total of 75, were downloaded and included in the study (Table 1 and Supplementary Material). Other available COI sequences (eleven in total) from Iran (accession numbers LN833869-LN833879) were omitted from the study, as they corresponded mainly to the second half of the COI gene, which overlapped very shortly with the barcode region, and the rest of our dataset. These sequences were identified as P. elbursi, which is represented in our study by other better suited sequences. To root the phylogenetic tree, the barcode sequences for two other potamid species were downloaded from GenBank, Socotra pseudocardisoma (AY803585) and Johara tiomanensis (AB290644).
We fixed the specimens sampled directly in absolute or 95% ethanol by injecting them into their body and covering them in jars. The diluted ethanol in the jars was changed multiple times in the first days as it absorbs the water of the samples while dehydrating and, therefore, preserving them. We observed that ethanol injection and multiple changes are crucial to obtain well-preserved DNA quality samples, as other specimens sampled not following this procedure did not amplify successfully in the majority of cases. The samples were deposited in the collections of the National Museum of Natural Sciences of Madrid (MNCN-CSIC).
DNA extraction and sequencing
Genomic DNA was extracted from a small sample (less than 2 mm in size) of muscle tissue of an ambulatory leg using the DNeasy® Blood & Tissue Kit (QIAGEN, Hilden, Germany). DNA purification was carried out using BioSprint 15 and one 5-tube strip per sample. The DNA was eluted in 200 μl of AE buffer and transferred into a 1.5 ml microtube for long-term storage.
The barcode region of the cytochrome c oxidase subunit I (COI) gene was amplified using LCO1-1490/HCO1-2198 forward and reverse primers13. Amplification was carried out in a total volume of 12 μl per reaction (1–2 μl template DNA, 1 μl of each primer, 2.75–1.75 μl Milli-Q H2O and 6.25 μl of DreamTaq Green PCR Master Mix). After confirmation of successful amplification by electrophoresis, PCR products were purified using Exo-SAP-IT® and sequenced using an external commercial company (Macrogen, Seoul, South Korea) with the same corresponding forward and reverse primers. The obtained sequences were quality checked, trimmed and assembled in Geneious software (Geneious® 10.2.6; Biomatters http://www.geneious.com)14. They were aligned with MAFFT15, 16 implemented in Geneious using the auto algorithm option. Each alignment was trimmed, manually adjusted, and visually verified to maximize positional homology, taking into account the genetic codes and the translation frames of the protein-coding gene. All the sequences have been deposited in GenBank (Table 1).
Alignment, phylogenetic inference and species delimitation
The final dataset was aligned using MAFFT implemented in Geneious and screened for sequencing errors. Such poor-quality sequencing errors were found in data downloaded from GenBank and corrected using IUPAC general degenerate nucleotide codes (ex. Gaps resulting in frameshift were replaced with Ns where possible). The maximum likelihood approach was used to construct a phylogenetic tree in IQ-Tree v 2.1.217. The best partitioning scheme and substitution model were found using ModelFinder18 as implemented in IQ-Tree (-m MFP + MERGE). For the tree reconstruction, 500 nonparametric bootstraps19 were used to evaluate the nodal support (-b 500). To delimit species within our dataset, we used bPTP20. For the bPTP approach, the phylogenetic tree was analysed using the online portal (https://species.h-its.org/). The “rooted tree” and “delete outgroup” options were selected, and the number of MCMC iterations was increased to 5 * 105. All other parameters were left in default. Alignment statistics and uncorrected distance matrices were obtained using MEGA1121. Species delimitation statistics were obtained using the Species Delimitation plugin22 on Geneious.
Data availability
The genetic sequences produced in this study are deposited in GenBank.
References
Yeo, D. C. J. et al. Global diversity of crabs (Crustacea: Decapoda: Brachyura). In Freshwater Animal Diversity Assessment (eds Balian, E. V. et al.). Hydrobiologia Vol. 595, 275–286 (2008).
Álvarez, F. et al. Revision of the higher taxonomy of Neotropical freshwater crabs of the family Pseudothelphusidae, based on multigene and morphological analyses. Zool. J. Linn. Soc. 193, 973–1001 (2021).
Ng, D. J. J. & Yeo, D. C. J. Terrestrial scavenging behaviour of the Singapore freshwater crab, Johora singaporensis (Crustacea: Brachyura: Potamidae). Nat. Singap. 6, 207–210 (2013).
Dobson, M. Freshwater crabs in Africa. Freshw. Forum 21, 3–26 (2004).
Dobson, M., Magana, A. M., Mathooko, J. M. & Ndegwa, F. K. Distribution and abundance of freshwater crabs (Potamonautes spp.) in rivers draining Mt Kenya, East Africa. Fundam. Appl. Limnol. 168, 271–279 (2007).
Cumberlidge, N. et al. Freshwater crabs and the biodiversity crisis: Importance, threats, status, and conservation challenges. Biol. Conserv. 142, 1665–1673 (2009).
Jouladeh-Roudbar, A., Ghanavi, H. R. & Doadrio, I. Ichthyofauna from Iranian freshwater: Annotated checklist, diagnosis, taxonomy, distribution and conservation. Assessment 21, 1–303 (2020).
Brandis, D., Storch, V. & Türkay, M. Taxonomy and zoogeography of the freshwater crabs of Europe, North Africa, and the Middle East (Crustacea, Decapoda, Potamidae). Senckenberg. Biol. 80, 5–56 (2000).
Keikhosravi, A. & Schubart, C. D. Description of a new freshwater crab species of the genus Potamon (Decapoda, Brachyura, Potamidae) from Iran, based on morphological and genetic characters. In Advances in Freshwater Decapod Systematics and Biology 115–133 (BRILL, 2014). https://doi.org/10.1163/9789004207615_008.
Keikhosravi, A. & Schubart, C. D. Revalidation and redescription of Potamon elbursi Pretzmann, 1976 (Brachyura, Potamidae) from Iran, based on morphology and genetics. Open Life Sci. 9, 114–123 (2014).
Keikhosravi, A., Naderloo, R. & Schubart, C. D. Morphological and molecular diversity in the freshwater crab Potamon ruttneri-P. gedrosianum species complex (Decapoda, Brachyura) indicate the need for taxonomic revision. Crustaceana 89, 129–139 (2016).
Parvizi, E., Naderloo, R., Keikhosravi, A., Solhjouy-Fard, S. & Schubart, C. D. Multiple Pleistocene refugia and repeated phylogeographic breaks in the southern Caspian Sea region: Insights from the freshwater crab Potamon ibericum. J. Biogeogr. 45, 1234–1245 (2018).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Aust. J. Zool. https://doi.org/10.1071/ZO9660275 (1994).
Kearse, M. et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783 (1985).
Zhang, J., Kapli, P., Pavlidis, P. & Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876 (2013).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Masters, B. C., Fan, V. & Ross, H. A. Species delimitation—A geneious plugin for the exploration of species boundaries. Mol. Ecol. Resour. 11, 154–157 (2011).
Acknowledgements
This study was funded through funding number 20193M674 of the Spanish Ministerio de Ciencia e Innovación received by ID.
Funding
Open access funding provided by Lund University.
Author information
Authors and Affiliations
Contributions
H.R.G. and I.D. designed the experiment. H.R.G., I.D., A.J.R. and M.T. sampled the individuals. PR performed the wet laboratory experiments. H.R.G., P.R. and M.T. wrote the manuscript with inputs from all other authors. K.R.T., M.T., A.J.R. and A.R.M. helped obtain permits for sampling and finalizing the manuscript. H.R.G. and A.J.R. responded the comments provided by the reviewers and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rezaei Tavabe, K., Tavana, M., Mirvaghefi, A.R. et al. Barcoding and species delimitation of Iranian freshwater crabs of the Potamidae family (Decapoda: Brachyura). Sci Rep 12, 8288 (2022). https://doi.org/10.1038/s41598-022-12335-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-12335-w
This article is cited by
-
A new species of freshwater crab of the genus Himalayapotamon Pretzmann 1966 (Decapoda: Brachyura: Potamidae) from Jammu and Kashmir, Northern India
Proceedings of the Indian National Science Academy (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.