Abstract
Proteins are the essential biological macromolecules required to perform nearly all biological processes, and cellular functions. Proteins rarely carry out their tasks in isolation but interact with other proteins (known as protein–protein interaction) present in their surroundings to complete biological activities. The knowledge of protein–protein interactions (PPIs) unravels the cellular behavior and its functionality. The computational methods automate the prediction of PPI and are less expensive than experimental methods in terms of resources and time. So far, most of the works on PPI have mainly focused on sequence information. Here, we use graph convolutional network (GCN) and graph attention network (GAT) to predict the interaction between proteins by utilizing protein’s structural information and sequence features. We build the graphs of proteins from their PDB files, which contain 3D coordinates of atoms. The protein graph represents the amino acid network, also known as residue contact network, where each node is a residue. Two nodes are connected if they have a pair of atoms (one from each node) within the threshold distance. To extract the node/residue features, we use the protein language model. The input to the language model is the protein sequence, and the output is the feature vector for each amino acid of the underlying sequence. We validate the predictive capability of the proposed graph-based approach on two PPI datasets: Human and S. cerevisiae. Obtained results demonstrate the effectiveness of the proposed approach as it outperforms the previous leading methods. The source code for training and data to train the model are available at https://github.com/JhaKanchan15/PPI_GNN.git.
Similar content being viewed by others
Introduction
Proteins are organic macromolecules made up of twenty standard amino acids. They are responsible for performing nearly all biological processes and cellular functions in organisms1. DNA transcription and replication, hormone regulation, metabolism, molecular cell signaling, and signal transduction are some examples of life activities that involve protein interactions2,3. Furthermore, the knowledge of PPI has proven to be helpful in new drug discovery as well as the prevention and diagnosis of diseases4. However, they rarely carry out their functions in isolation but interact with other proteins in their surroundings and accomplish their tasks. There are several high-throughput experimental methods such as Yeast two-hybrid screens (Y2H)5, Tandem affinity purification (TAP)6, and Mass spectrometric protein complex identification (MS-PCI)7 which are used to identify the interaction between proteins. These experimental methods contributed to the creation of PPI datasets for different species but at a slow speed. Moreover, as the experimental environment and device resolution influence the output, the PPI data collected by these methods have a high rate of false positives and false negatives8,9. When used in conjunction with experimental methods, high-throughput computational methods improve the accuracy and quality of PPI prediction10.
To date, there have been a plethora of works that employed traditional machine learning (ML) techniques to solve several problems in the computational biology domain, such as protein classification, structure prediction, protein interaction prediction, etc. Support vector machines (SVMs), Random forest (RF), Neural networks (NNs) are widely used ML algorithms to classify protein interactions. The inputs to these algorithms are hand-engineered features, which are mainly derived from underlying protein sequences using physicochemical properties, evolutionary information, and distribution patterns of amino acids. For example, Shen et al.11 have employed a conjoint triad feature extraction method, which divides 20 amino acids into seven groups based on their properties, and SVM with a kernel function as a learning algorithm. Guo et al.12 have used the autocovariance (AC) method to encode protein sequences and SVM as a classifier to predict PPI. AC method is intended to consider the neighboring effect by capturing the physicochemical properties of residues a certain distance apart in the sequence. Li et al.13 have developed a method incorporating a mixture of evolutionary features based on a Position-specific scoring matrix (PSSM) and physicochemical properties. The discriminative vector machine (DVM) is used as a classifier to predict protein interactions. Huang et al.14 have adopted the global encoding representation of proteins with a weighted sparse representation-based classifier to classify PPI. Li et al.15 have proposed a sequence-based method to predict self-interacting proteins. Firstly, the known protein sequences are converted into a Position-specific scoring matrix (PSSM), and then Low-Rank Approximation (LRA) is used to get feature vectors from PSSM. Finally, the extracted vectors are the inputs to the rotation forest classifier, which distinguishes self-interacting and non-self-interacting proteins. The gradient boosting decision tree algorithm has been introduced to predict PPI by Zhou et al.16. This algorithm is based on several protein descriptors such as frequency, composition, transformation, distribution, and autocovariance to encode protein sequences. Apart from sequence-based features, we have other sources of information to get input features for PPI models, including gene fusion17, protein structure18, function19,20, gene expression profile21, etc. Ding and Kihara22 reviewed and classified several computational methods based on input features mentioned above. Among all sources of proteins, sequence-derived features are the most commonly used to predict PPI.
Researchers have recently carried out considerable works on PPI using the latest deep learning techniques, which improve the model’s performance. These techniques allow them to use high-dimensional and complex input features. Sun et al.23 have used a stacked autoencoder to get compact and relevant representation of sequence-based input features. Du et al.24 have proposed a deep learning-based PPI model known as DeepPPI, which learns the high-level features from protein descriptors and outperforms the traditional ML algorithms. Hashemifar et al.25 have developed a model known as DPPI, which consists of three modules: siamese-like CNN, random projection, and prediction. The input to the DPPI is the sequence profiles of proteins in a pair, and the output is the binary value representing whether they will interact or not. Gonzalez-Lopez et al.26 have proposed a PPI model in which features are learned using NLP techniques such as embedding and recurrent neural networks from raw protein sequences. They have shown that the state-of-the-art results can be obtained using only raw data without relying on feature engineering to predict protein interactions. EnsDNN (Ensemble deep neural network) is a very complex PPI model proposed by Zhang et al.27. It uses autocovariance (AC), local descriptor (LD), and multi-scale continuous and discontinuous local descriptor (MCD) methods to get different feature representations of protein sequences. These feature vectors are then individually fed to the nine independent neural networks that differ in the number of layers and neurons, which produce a total of 27 neural networks. The output of each neural network is then fed to the multi-layer perceptron model having two hidden layers to predict the labels. A deep multi-modal framework, which utilizes structural and ontology-based features, is proposed by Jha et al.28 to predict the protein interactions and outperform the existing works.
Most of the above-mentioned deep-learning methods have only considered sequence-based features. The PPI models based on structural information of proteins are significantly less explored. Nowadays, the latest deep learning techniques, such as deep convolutional neural networks (CNN), have been used widely because of their potential to extract features seamlessly from structural data. For example, in our previous work29, we have used structural information with sequence-based features to predict the interactions between proteins. We have utilized a pre-trained ResNet50 model to extract structural features from the 2D volumetric representations of proteins. The obtained results suggest that techniques for image-related tasks can be extended to work for protein structures. But these approaches of analyzing molecular structure have certain issues such as high computing cost and interpretability.
Graph neural networks have made significant progress in recent years and have emerged as key tools in graph-based applications. For example, Huang et al.30 have predicted the associations between miRNA and drug resistance using graph convolution. The problem of predicting associations between them is formulated as a link prediction problem. Other applications of graph neural networks include prediction of chemical stability31, protein interface prediction32, protein solubility prediction33 and modeling the side effects of polypharmacy34. The prediction of interactions between proteins using the graph-based technique can be implemented in two ways: molecular structure-based and PPI network-based. Yang et al.35 have proposed the signed variational graph auto-encoder (S-VGAE) to predict the interactions between proteins by considering the PPI network as an undirected graph. This representation learning model can effectively use the graph structure and seamlessly assimilate protein sequence information as features. The current work explores the applicability of graph-based neural networks to predict protein interactions utilizing the graphical molecular representations of proteins. This representation of proteins allows for a natural spatial representation of molecular structures and is more interpretable than 2D volumetric representation.
In this paper, we propose a framework that combines graph-based techniques and language models (SeqVec36 and ProtBert37) to predict PPI. The molecular graph of a protein has nodes representing the amino acids of which proteins are made up of. The language model is used here to get features for each residue (node in graph) directly from protein sequences. The advantage of using the language model-based feature vectors is that it does not require domain knowledge to encode the sequences. The obtained results show its applicability in predicting protein interactions. We use graph-based methodologies, such as graph convolutional network (GCN)38 and graph attention network (GAT)39, to learn features from protein representations combining structural and sequence information. Finally, the feature vectors of proteins in pairs are concatenated and fed to the classifier having two hidden layers and an output layer. The following are the key contributions of this work:
-
1.
We use the graphical representation of proteins with residues as nodes. We believe that it will improve the model’s performance as it covers spatial structural/low-level properties.
-
2.
We use pre-trained language models (SeqVec and ProtBert) to get the feature vector for each residue/node in the constructed graph. We show that this feature vector is more beneficial than other features such as physicochemical properties of residues, one-hot encoding of amino acids, etc.
-
3.
We propose two graph-based architectures: GCN-based and GAT-based to learn features from protein representation integrating spatial structure and sequence features. The obtained results propound the superiority of the proposed method over the existing works.
Materials and methodology
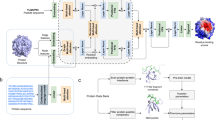
The proposed methodology to predict PPI consists of three modules: protein graph construction, feature extraction, and classifier to predict interactions between them. This section discusses each module and the deep learning techniques used to implement them in detail. In addition, we describe the datasets used to evaluate the performance of our approach. All experiments were performed in accordance with relevant guidelines and regulations.
Datasets
In this work, we have used the PPI datasets of two organisms: Human and S. cerevisiae. The Pan’s human dataset40 is available at http://www.csbio.sjtu.edu.cn/bioinf/LR_PPI/Data.htm. The positive pairs of this dataset are collected from the human protein reference database (HPRD, 2007 version). The elimination of duplicate and self-interacting pairs gives a total of 36,545 instances. For the S. cerevisiae, there are a total of 22,975 interacting protein pairs, downloaded from the Database of Interacting Proteins (DIP; version 20160731). After performing the preprocessing, such as removing protein pairs that have a protein with fewer than 50 amino acids, we have a total of 17,257 interacting pairs. The negative pairs for both organisms are generated by randomly pairing proteins with different subcellular localization. These generated negative instances should not be in the positive PPI dataset. The information regarding the subcellular localization of proteins is collected from the Swiss-Prot database. The Pan’s human negative PPI dataset has a total of 36,323 non-interacting samples consisting of pairs generated using the method discussed above and the negative instances from the Negatome database41. For S. cerevisiae, there are a total of 48,594 negative pairs. The CD-HIT42 tool with a sequence identity level of 40% as the cutoff has been used to remove protein pairs that are considered to be homologous (having too much sequence identity).
The 3D structures of all proteins utilized in this study are downloaded in PDB format from the RCSB Protein Data Bank (https://www.rcsb.org). It limits the number of samples in both PPI datasets as the structural information is not available for all proteins. The final statistics of these datasets are reported in Table 1.
Graph representation of proteins
In this work, we have constructed the molecular graph of proteins, also known as amino-acids/residues contact network, using the PDB files. The PDB file is a text file containing structural information such as 3D atomic coordinates. Let G(V, E) be a graph representing the proteins, where each node (\(v \in V\)) is the residue and interaction between the residues is described by an edge (\(e \in E\)). Two residues are connected if they have any pair of atoms (one from each residue) having the Euclidean distance less than a threshold distance. The cutoff distance used in the literature is 6 angstroms (Å)32, and we are also using the same value for threshold distance.
Node features
Each node in the protein’s graph has some properties associated with it. These node features are mainly obtained from protein sequences and structures. In this work, the node features are extracted from protein sequences employing two pre-trained language models (LSTM-based36 and BERT-based37). In addition, we have also considered some other methods to get node features, such as one-hot encoding of 20 standard amino acids and physicochemical properties of residues. In the case of the one-hot encoding method, each node is represented as a vector of length 20. The seven physicochemical properties of amino acids provided by Meiler et al.43 are assumed to influence the interactions between proteins by creating hydrophobic forces or hydrogen bonds between them. So, these features are used as other feature vectors for each node in the graph. Table 2 reports the dimension of the node’s features obtained using the aforementioned techniques. Among these node features (based on the results obtained), the protein graph with node features extracted using the pre-trained LSTM-based language model (LM) outperforms the methods based on other feature vectors. The results are discussed in the subsection Performance of GNN Variants using Different Node Features.
Figure 1 depicts all the steps of generating a protein graph from a PDB file. The first step is to get each protein’s PDB sequence and molecular graph structure using a python script. Then the residue-level features are merged with a molecular graph, creating a final protein graph.
Graph models
CNN-based models work effectively as feature extractors. But the limitation with these models is that they can only operate on regular Euclidean data like 2D grid images and 1D sequences. To work on non-Euclidean data, such as graph data, a graph neural network (GNN) has been developed, which can directly process the graph. Over the years, there has been a rapid development in GNN and has offered several variants44. This work explores the two popular GNN variants, GCN38 and GAT39, to extract features from proteins represented as graphs.
Graph convolutional network
The GCN is used to learn features from data represented as graphs. Let \(P = (V, E)\) be a graph representation of a protein, where the number of vertices is the length of the PDB sequence (i.e., \(\vert V \vert = L\)). The connection between every pair of nodes is encoded in the adjacency matrix, \(A \in R^{L, L}\). The matrix, \(X \in R^{L, F}\), contains the residue-level features for all nodes of the given graph (F: dimension of node features). Each layer of GCN takes the adjacency matrix (A) and node embeddings from the previous layer (\(H^{(l)} \in R^{L, F_l}\)) as input and outputs the node-level embeddings for the next layer (\(H^{(l+1)} \in R^{L, F_{l+1}}\)).
Here, \(H^{(0)}\) = \(X \in R^{L, F}\), \(F_l\) and \(F_{l+1}\) represent the dimensions of node-level embeddings for layers l and \(l+1\), respectively. The more specific expression of Eq. (1), as defined by Kipf and Welling38 is:
Here, \(\hat{A}\) is the adjacency matrix added with identity matrix \(I_L \in R^{L,L}\) (\(\hat{A} = A+I_L\)). The addition of identity matrix to adjacency matrix enforces the self-loops in the graph, ensuring the inclusion of node’s own features in the sum during convolution operation. \(\hat{D}\) is the diagonal node degree matrix calculated as: \(\hat{D}_{ii} = \sum _{j=1}^{L}{\hat{A}_{ij}}\). It is used to normalize the adjacency matrix in a symmetric manner \((\hat{D}^{-0.5}\hat{A}\hat{D}^{-0.5})\) so that we get the normalized residue features after each convolutional layer. After the GCN layer, each residue feature vector is updated as a weighted sum of the features of neighboring nodes in the graph, including residue’s own feature (Eq. 2). \(W^{(l+1)}\) is the trainable weight matrix.
Graph attention network
GAT39 is an attention-based architecture that operates on graph-structured data. The idea is to use a self-attention method to compute the hidden representation of each node in the graph by paying attention to its neighbors. In GAT, for input feature matrix \(X \in R^{L, F}\), we will get the learned feature matrix \(H \in R^{L, F'}\) as an output. Here, L is the number of nodes in the graph, and F and \(F'\) are the input and output dimensions, respectively. The expression to calculate the learned feature of node i is:
Here, \(\sigma\) is the LeakyReLU activation function used to apply non-linearity to aggregated features of neighboring nodes, \(N_i\). W is the weight matrix used to apply a linear transformation to the input feature matrix, X. The normalized attention coefficient between node i and j, represented as \(\alpha _{ij}\), is calculated as:
where \(a \in R^{2F'}\) is a weight vector. Let \(H \in R^{L, F'}\) be the learned feature matrix of protein graph P after one GNN layer. Here, \(F'\), the dimension of a node’s feature, is fixed for all graphs’ nodes. But, L, which represents the number of nodes in a graph, may vary as each protein has different number of residues. To ensure the fixed-size representations of all proteins, we have added a global pooling layer after the GNN layer. The pooling layer is calculated by performing some operation (mean, max, sum) on the GNN layer’s output (\(H \in R^{L, F'}\)) across the number of nodes. In our case, we use the mean pooling to get a fixed size \((1, F')\) representation of all proteins, independent of the number of nodes L in a graph.
After the pooling layer, we add a fully connected (FC) layer with a LeakyReLU activation function to get the final representation of a protein from its pooled representation. Similarly, we get the learned feature vector of another protein in a pair. The concatenated feature vectors of proteins in a pair are then fed to the classifier having two FC layers and an output layer. To avoid over-fitting, we add a dropout layer with a dropout rate of 0.2 after each FC layer. The output layer is the sigmoid layer, which determines whether the given pairs are interacting (≥ 0.5) or non-interacting \((<0.5)\). The LeakyReLU is used as an activation function to add non-linearity to the output of a fully connected layer. Figure 2 depicts the overall steps of the proposed methodology.
Language models
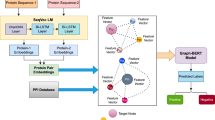
Proteins are the long chain of amino acids, where each amino acid (residue) can be considered as a word and each sequence as a sentence. Recently, researchers have started using language models (LMs) from natural language processing (NLP) to encode the protein sequence. They have trained many protein LMs to generate embeddings for proteins, can be used as input features to deep learning models. These input features have been proven to be more valuable than the earlier approaches (substitution matrices, capturing biophysical properties) to perform tasks like protein subcellular localization and structure prediction. In this work, we explore two protein LMs models: SeqVec36 (LSTM-based) and ProtBert37 (BERT-based). The procedure of getting feature vectors using these language models is depicted in Figure 3.
The SeqVec is the adaption of the bi-directional language model, ELMo (Embeddings from language models)45 and is context-dependent. The SeqVec model has the same configuration as the standard ELMo architecture with some minor changes such as the reduction of tokens to 28 (20: standard amino acids, 2: rare amino acids, 3: unknown or ambiguous amino acids, 2: tokens to represent the start and end of the sequence, 1: masking). To handle the increased length of protein sequences, the number of unrolling steps has been increased. This language model consists of one character convolution (CharCNN)46 layer and two bi-directional LSTM layers. The CharCNN layer maps each amino acid to the fixed-length (1024) latent space. This layer does not consider information from neighbouring words. The output of this layer is the input of the first bi-LSTM layer. Each bi-LSTM unit has a dimension of 1024 (first 512 for a forward pass and the last 512 for a backward pass). The ELMo-based SeqVec model is trained on the UniRef50, a dataset of 33 million protein sequences. For a protein sequence with a length L, this pre-trained embedder generates an embedding of size (3, L, 1024). We get the feature vectors of this size by concatenating the outputs of 3 layers (1 CharCNN and 2 bi-LSTMs), each layer having the 1024-dimensional embedding for each word or amino acid. The per-residue embedding is generated by taking the summation of the embeddings of the 3 layers.
The second embedder that we use to get per-residue embedding (node features in a protein graph) is ProtBert, trained on the BFD-100 dataset. The BFD (Big Fat Database) has 2.1 billion protein sequences and more than 393 billion tokens (amino acids). The ProtBert LM employed the BERT model to generate descriptive features for each residue. The BERT47 is a deep learning model based on transformers and has been extensively used for transfer learning in NLP. For any protein sequence of length L, ProtBert LM first performs the tokenization and adds positional encoding to each token. The output of this layer is then passed through the stacked self-attention layers, which generate context-aware embeddings. Each self-attention-based encoding layer outputs an embedding of length 1024 for each residue of a protein sequence. The last hidden state of the attention stack having the dimension of (L, 1024) is used as a feature matrix. Since the length of the natural language sentences is much smaller when compared to that of protein sequences, some changes are made in the protein LMs, such as increasing the number of layers and unrolling steps.
Results and analysis
This section analyzes the performance of the proposed method to predict PPI in terms of various evaluation metrics. These metrics include accuracy (acc), sensitivity (Sn), specificity (Sp), precision (Pr), F-score, Matthew correlation coefficient (MCC), the area under the precision-recall curve (AUPRC), and area under the receiver operating characteristic curve (AUROC).
Experimental setup
We use PyTorch, an open-source deep learning framework, to build the proposed PPI models. To build GNN model variants that we explore in this work, we use PyTorch geometric. For current PPI work, the models’ performance with more than one GNN layer (e.g., 2 and 3) is either comparable to or less than that of a one-layer GNN model. So, in this work, the number of GNN layers to encode the protein graph representation is chosen to be one. The classifier module of the proposed work has two fully connected (FC) layers having 256 and 64 units, respectively. The Adam optimizer with a learning rate of 0.001 is used to minimize the MSE loss while training the model. After every FC layer in the proposed PPI model, we use a dropout layer with a dropout rate of 0.2. The values of these parameters, such as dropout and learning rate, number of FC layers, number of units in each layer, are set following the previous works28,33
Performance of GNN variants using different node features
In this work, we have represented each protein as a graph and employed different graph neural networks (GCN and GAT), which operate on these graph-structured data to get protein-level representation. The selection of input features greatly influences the performance of any model. Here, we have explored different methods to extract features for nodes of protein graphs (see Table 2). Tables 3 and 4 illustrate the performance of GNN models utilizing different node features. As visible, the residue embedding generated using LSTM-based LM yields the best results for both GCN and GAT models. For the human PPI dataset, the GAT model obtains an accuracy of 98.13%, F-score of 98.73%, MCC of 95.20% and these values are slightly better than those obtained by its GCN counterpart in the respective performance metric. The GAT model for the S. cerevisiae dataset improves the results moderately than its GCN counterpart in terms of all evaluation metrics. From the results reported in these tables, we can observe that the performance of GNN models utilizing BERT-based LM embeddings is superior to those utilizing one-hot encoding and physicochemical properties to obtain node features. It shows the potential of language models to capture the hidden associations directly from raw sequences. The proposed model takes advantage of both graph neural networks and language models. The GNN models compute the hidden representation of a node by considering its neighboring nodes that are distant in sequence but nearer to each other in 3D space. Also, the per-residue feature extraction from sequences directly does not require background knowledge. Along with results on test data (20% of the dataset), we have also reported the average results of 5-fold cross-validation with the standard deviation of well-performing models using LSTM-based LM embeddings in Table 5 for both PPI datasets. As evident, the GAT-based classifiers performed a bit better than the GCN-based classifiers for most of the evaluation metrics.
Effect of number of GNN layers on the performance of PPI models
One of the parameters that affects the performance of deep learning models is the number of layers48. In general, it is assumed that the more GNN layers there are, the more information may be gathered from the edge and node features. However, in reality, the excessive layers would cause a decrease in performance due to vanishing gradients and over-smoothing49. Smoothing is defined as the similarity between nodes in a graph. It is considered to be the essential nature of GNN as the nodes in the graph exchange messages with each other. When stacking multiple layers, this message interaction between them would make the representations of the nodes similar and cause over-smoothing. To demonstrate the effect of the number of GNN layers for PPI tasks, we experimented with up to 3 layers of GNN in the proposed PPI framework. The results are reported in Table 6. As evident, when we increase the number of GNN layers from one to two, the performance of PPI models (GCN/GAT-based) in terms of evaluation metrics is comparable. With three layers, the values of performance metrics are lower than those with one or two layers, except for sensitivity. Since the performance of PPI models with one and two GNN layers is comparable, we choose a model with one GNN layer.
To analyze the contribution of sequence embeddings generated using a language model (LSTM-based), we designed a baseline, depicted in Figure 4. The language model used in this work generates both per-residue and per-protein embeddings. The per-residue embedding is used as node features in a graphical representation of a protein. The per-protein embedding is generated by taking the mean across the protein sequence length. The generated embeddings for protein sequences are then fed to an MLP classifier for protein interaction prediction. The results are presented in Table 7.
Effect of sample size on the performance of PPI models
Our method to predict PPI is based on structural information, which reduces the number of samples compared to sequence-based methods. So, to check the robustness of the current work, we train the models for different sample sizes and report the results in Table 8. In Table 8, we have reported the values of the performance metrics such as accuracy, sensitivity, specificity, and MCC, which we think are sufficient to analyze the effect of sample size on PPI models’ performance. As seen from the table, the performance of GNN models for both datasets increases as the number of samples increases. Initially, the GCN model outperforms the GAT model for both datasets, but gradually it improves the performance with more samples and yields the best results for final datasets. It is to be noted that all samples for the human PPI dataset are the same as reported in Table 1. But for the second dataset, following previous works of literature, we made this skewed dataset to a balanced dataset by randomly reinserting the positive samples into the original one. It gives us the S. cerevisiae dataset with a total number of samples of 8854, in which the ratio of positive samples to the negative samples is 1:1. For other sample sizes (Human: 4k, 8k, 12k; S. cerevisiae: 2k, 4k, 6k), we randomly select protein pairs from the final benchmark datasets having the fair distribution of interacting and non-interacting pairs. If we observe the values of sensitivity (Sn) and specificity (Sp) for all samples of the human PPI dataset, we can infer that the proposed work performs well on the skewed dataset as well. The number of positive samples is almost three times the number of negative samples for this dataset. After analyzing the patterns of the results with the sample sizes, we can say that in the future, if we have more structural data than now, we may get better results for PPI datasets.
Performance comparisons with previous work
We have considered recent works for performance comparison, including sequence-based methods23,24,25,26,50, multi-modal methods28,29, and the PPI network graph-based method35. Most works have reported training accuracy for human dataset (Sun’s work23: 97.19%, Yang’s work35: 99.15%). We achieve a training accuracy of more than 99.50%, which is higher than previous recent work. However, the training results are not sufficient to compare the PPI models. Therefore, we use the test set to check the predictive capability of PPI models on unseen data. Table 9 summarizes the performance comparisons between the proposed approach and previous works for the human dataset on the test set. As seen from the table, our method to predict PPI outperforms the previous methods in terms of most of the evaluation metrics, thus, indicating the proposed approach’s efficacy utilizing the GNN and LM. For the S. cerevisiae dataset, we have also performed the comparative study as summarized in Table 10, which also demonstrates our work’s effectiveness. Following the previous works, in Table 10 we have reported the values, which are the average of 5-fold cross-validation results. It should be noted that the current work outperforms our previous works that used a multi-modal framework in terms of the majority of the performance metrics. However, the performance improvement is not very large, but significant enough.
Conclusion
In this work, we propose a method that combines graph neural network (GNN) and language model (LM) to predict the interaction between proteins. First, we build a molecular protein graph (amino acids/residues as nodes) from a PDB file containing structural information. Then we use LM to generate per-residue embedding from the PDB sequence, which is used as the node’s feature of the protein graph. The GNN-based model then extracts features from the protein’s graphical representation (combining structural and sequence information). Finally, we concatenate the outputs of the GNN-based model for each protein pair, and the resulting vectors are then fed to the PPI classifier. This classifier has two fully connected layers and an output layer. We have evaluated the performance of our method on two popular datasets, and the obtained results demonstrate its effectiveness. The proposed work outperforms the previous approaches, including PPI network-based graph auto-encoder model. However, PPI network-based model has the advantage over its protein structure-based counterpart in terms of the number of samples as the structural information is not available for all existing proteins. In the future, we will explore other deep learning-based approaches to learn features from protein representations (sequences and structures) such as multi-scale representation learning51 and intrinsic-extrinsic convolution and pooling for learning on 3D protein structures52.
Data availibility
The dataset used in this study is available at http://www.csbio.sjtu.edu.cn/bioinf/LR_PPI/Data.htm.
References
Alberts, B. The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell 92, 291–294 (1998).
Zhang, Q. C. et al. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature 490, 556–560 (2012).
Wang, L. et al. Advancing the prediction accuracy of protein–protein interactions by utilizing evolutionary information from position-specific scoring matrix and ensemble classifier. J. Theor. Biol. 418, 105–110 (2017).
You, Z.-H., Lei, Y.-K., Gui, J., Huang, D.-S. & Zhou, X. Using manifold embedding for assessing and predicting protein interactions from high-throughput experimental data. Bioinformatics 26, 2744–2751 (2010).
Ito, T. et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98, 4569–4574 (2001).
Gavin, A.-C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002).
Ho, Y. et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 (2002).
Mrowka, R., Patzak, A. & Herzel, H. Is there a bias in proteome research?. Genome Res. 11, 1971–1973 (2001).
Melo, R. et al. A machine learning approach for hot-spot detection at protein–protein interfaces. Int. J. Mol. Sci. 17, 1215 (2016).
You, Z.-H., Zhou, M., Luo, X. & Li, S. Highly efficient framework for predicting interactions between proteins. IEEE Trans. Cybern. 47, 731–743 (2016).
Shen, J. et al. Predicting protein–protein interactions based only on sequences information. Proc. Natl. Acad. Sci. 104, 4337–4341 (2007).
Guo, Y., Yu, L., Wen, Z. & Li, M. Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucl. Acids Res. 36, 3025–3030 (2008).
Li, Z.-W., You, Z.-H., Chen, X., Gui, J. & Nie, R. Highly accurate prediction of protein–protein interactions via incorporating evolutionary information and physicochemical characteristics. Int. J. Mol. Sci. 17, 1396 (2016).
Huang, Y.-A., You, Z.-H., Chen, X., Chan, K. & Luo, X. Sequence-based prediction of protein–protein interactions using weighted sparse representation model combined with global encoding. BMC Bioinform. 17, 1–11 (2016).
Li, J.-Q., You, Z.-H., Li, X., Ming, Z. & Chen, X. Pspel: In silico prediction of self-interacting proteins from amino acids sequences using ensemble learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 14, 1165–1172 (2017).
Zhou, C., Yu, H., Ding, Y., Guo, F. & Gong, X.-J. Multi-scale encoding of amino acid sequences for predicting protein interactions using gradient boosting decision tree. PLoS ONE 12, e0181426 (2017).
Enright, A. J., Iliopoulos, I., Kyrpides, N. C. & Ouzounis, C. A. Protein interaction maps for complete genomes based on gene fusion events. Nature 402, 86–90 (1999).
Singh, R., Xu, J. & Berger, B. Struct2net: integrating structure into protein–protein interaction prediction. In Biocomputing 2006, 403–414 (World Scientific, 2006).
Ben-Hur, A. & Noble, W. S. Kernel methods for predicting protein–protein interactions. Bioinformatics 21, i38–i46 (2005).
Bandyopadhyay, S. & Mallick, K. A new feature vector based on gene ontology terms for protein–protein interaction prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 14, 762–770 (2016).
Von Mering, C. et al. Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417, 399–403 (2002).
Ding, Z. & Kihara, D. Computational methods for predicting protein–protein interactions using various protein features. Curr. Protocols Protein Sci. 93, e62 (2018).
Sun, T., Zhou, B., Lai, L. & Pei, J. Sequence-based prediction of protein protein interaction using a deep-learning algorithm. BMC Bioinform. 18, 1–8 (2017).
Du, X. et al. Deepppi: Boosting prediction of protein–protein interactions with deep neural networks. J. Chem. Inf. Model. 57, 1499–1510 (2017).
Hashemifar, S., Neyshabur, B., Khan, A. A. & Xu, J. Predicting protein–protein interactions through sequence-based deep learning. Bioinformatics 34, i802–i810 (2018).
Gonzalez-Lopez, F., Morales-Cordovilla, J. A., Villegas-Morcillo, A., Gomez, A. M. & Sanchez, V. End-to-end prediction of protein–protein interaction based on embedding and recurrent neural networks. In 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2344–2350 (IEEE, 2018).
Zhang, L., Yu, G., Xia, D. & Wang, J. Protein–protein interactions prediction based on ensemble deep neural networks. Neurocomputing 324, 10–19 (2019).
Jha, K., Saha, S. & Saha, S. Prediction of protein–protein interactions using deep multi-modal representations. In 2021 International Joint Conference on Neural Networks (IJCNN), 1–8, https://doi.org/10.1109/IJCNN52387.2021.9533478 (2021).
Jha, K. & Saha, S. Amalgamation of 3d structure and sequence information for protein–protein interaction prediction. Sci. Rep. 10, 1–14 (2020).
Huang, Y.-A., Hu, P., Chan, K. C. & You, Z.-H. Graph convolution for predicting associations between miRNA and drug resistance. Bioinformatics 36, 851–858 (2020).
Li, X. et al. Deepchemstable: Chemical stability prediction with an attention-based graph convolution network. J. Chem. Inf. Model. 59, 1044–1049 (2019).
Fout, A. M. Protein interface prediction using graph convolutional networks. Ph.D. thesis, Colorado State University (2017).
Chen, J., Zheng, S., Zhao, H. & Yang, Y. Structure-aware protein solubility prediction from sequence through graph convolutional network and predicted contact map. J. Cheminform. 13, 1–10 (2021).
Zitnik, M., Agrawal, M. & Leskovec, J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics 34, i457–i466 (2018).
Yang, F., Fan, K., Song, D. & Lin, H. Graph-based prediction of protein–protein interactions with attributed signed graph embedding. BMC Bioinform. 21, 1–16 (2020).
Heinzinger, M. et al. Modeling aspects of the language of life through transfer-learning protein sequences. BMC Bioinform. 20, 1–17 (2019).
Elnaggar, A. et al. Prottrans: towards cracking the language of life’s code through self-supervised deep learning and high performance computing. arXiv preprint arXiv:2007.06225 (2020).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907 (2016).
Veličković, P. et al. Graph attention networks. arXiv preprint arXiv:1710.10903 (2017).
Pan, X.-Y., Zhang, Y.-N. & Shen, H.-B. Large-scale prediction of human protein–protein interactions from amino acid sequence based on latent topic features. J. Proteome Res. 9, 4992–5001 (2010).
Smialowski, P. et al. The negatome database: A reference set of non-interacting protein pairs. Nucl. Acids Res. 38, D540–D544 (2010).
Li, W. & Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Meiler, J., Müller, M., Zeidler, A. & Schmäschke, F. Generation and evaluation of dimension-reduced amino acid parameter representations by artificial neural networks. Mol. Model. Ann. 7, 360–369 (2001).
Zhou, J. et al. Graph neural networks: A review of methods and applications. AI Open 1, 57–81 (2020).
Peters, M. E. et al. Deep contextualized word representations. arXiv preprint arXiv:1802.05365 (2018).
Kim, Y., Jernite, Y., Sontag, D. & Rush, A. M. Character-aware neural language models. In Thirtieth AAAI Conference on Artificial Intelligence (2016).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805 (2018).
Uzair, M. & Jamil, N. Effects of hidden layers on the efficiency of neural networks. In 2020 IEEE 23rd International Multitopic Conference (INMIC), 1–6 (IEEE, 2020).
Chen, D. et al. Measuring and relieving the over-smoothing problem for graph neural networks from the topological view. In Proceedings of the AAAI Conference on Artificial Intelligence Vol. 34, 3438–3445 (2020).
Wong, L. et al. Detection of interactions between proteins through rotation forest and local phase quantization descriptors. Int. J. Mol. Sci. 17, 21 (2016).
Somnath, V. R., Bunne, C. & Krause, A. Multi-scale representation learning on proteins. Adv. Neural Inf. Process. Syst. 34 (2021).
Hermosilla Casajus, P. et al. Intrinsic-extrinsic convolution and pooling for learning on 3d protein structures. In International Conference on Learning Representations, ICLR 2021: Vienna, Austria, May 04 2021, 1–16 (OpenReview. net, 2021).
Author information
Authors and Affiliations
Contributions
K.J. and S.S. conceived the idea, K.J. and H.S. conducted the experiments, K.J. and S.S. analysed the results, K.J. wrote the manuscript with valuable input from S.S. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jha, K., Saha, S. & Singh, H. Prediction of protein–protein interaction using graph neural networks. Sci Rep 12, 8360 (2022). https://doi.org/10.1038/s41598-022-12201-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-12201-9
This article is cited by
-
GNNMF: a multi-view graph neural network for ATAC-seq motif finding
BMC Genomics (2024)
-
A multi-source molecular network representation model for protein–protein interactions prediction
Scientific Reports (2024)
-
Exploring the SDE index: a novel approach using eccentricity in graph analysis
Journal of Applied Mathematics and Computing (2024)
-
Graph-BERT and language model-based framework for protein–protein interaction identification
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.