Abstract
Melibiose-derived AGE (MAGE) is an advanced glycation end-product formed in vitro in anhydrous conditions on proteins and protein-free amino acids during glycation with melibiose. Our previous studies revealed the presence of MAGE antigen in the human body and tissues of several other species, including muscles, fat, extracellular matrix, and blood. MAGE is also antigenic and induces generation of anti-MAGE antibody. The aim of this paper was to identify the proteins modified by MAGE present in human body fluids, such as serum, plasma, and peritoneal fluids. The protein-bound MAGE formed in vivo has been isolated from human blood using affinity chromatography on the resin with an immobilized anti-MAGE monoclonal antibody. Using mass spectrometry and immunochemistry it has been established that MAGE epitope is present on several human blood proteins including serum albumin, IgG, and IgA. In serum of diabetic patients, mainly the albumin and IgG were modified by MAGE, while in healthy subjects IgG and IgA carried this modification, suggesting the novel AGE can impact protein structure, contribute to auto-immunogenicity, and affect function of immunoglobulins. Some proteins in peritoneal fluid from cancer patients modified with MAGE were also observed and it indicates a potential role of MAGE in cancer.
Similar content being viewed by others
Introduction
Advanced glycation end-products (AGEs) form during glycation and accumulate in a body disturbing protein and tissue homeostasis1. AGEs constitute a group of antigens that can induce generation of the autoantibodies2,3. Recently, our group has revealed presence of the novel unconventional AGE antigen accumulating in different tissues of various species. This is a structural analog of the product called MAGE that forms from melibiose (mel) during an in vitro protein glycation in anhydrous conditions. The structure of a low-molecular MAGE product (protein-free) determined by NMR analysis showed a mixture of isomers with an open chain and a cyclic form of the fructosamine moiety. Using the model MAGE we have also shown that the serum from diabetic patients contains the autoantibody specifically binding MAGE4. Immunohistochemical analyzes with the in-house generated anti-MAGE antibody have shown that the novel AGE structural analog accumulates in a variety of human and animal tissues. An intense staining was observed on slides with sections from skeletal muscle of human, horse, pig, frog, and fish, in cardiomyocytes of pig, chicken and rat, and in smooth muscle of pig and rat. In addition, MAGE has been detected in the extracellular matrix of various animal tissues4, including various cancer tumors (manuscript in preparation). MAGE has also been associated with diabetic complications and atherosclerosis5,6. Interestingly, in other studies MAGE showed genotoxicity on human peripheral blood lymphocytes, melanoma, lung cancer, and colorectal cancer cells in vitro, while protein-free adducts prevented cells from this effect7. This observation indicates a potential role of MAGE in vivo, however the origin, detailed structure, and specific effect in different tissues remain to be elucidated. It should be noted that melibiose is produced during food fermentation by bacteria of the genus Bifidobacterium8,9, Lactobacillus, Lactococcus, Leuconostoc10, and yeast11. It is also found in cocoa beans, honey, and processed soybeans12,13. Supplied with food, melibiose is absorbed in the small intestine through para-cell junctions existing between adjacent enterocytes14. This suggests an exogenous source of mel as a substrate for further MAGE production. However, there are currently no data available on possible MAGE formation in the gut and subsequent absorption into the bloodstream.
Glycation occurs on various molecules in the body, including proteins in the circulatory system and solid tissues on the extra- and intracellular proteins. The formation of AGE adducts is favored on the long-lived proteins. This group of proteins includes lens crystallins15, collagen, blood albumin16, insulin, immunoglobulins17, and low-density lipoproteins (LDL)18,19,20,21. Glycation of insulin in pancreatic cells or blood makes it difficult to maintain homeostatic glucose levels and stimulate lipogenesis22. Glycated albumin and methylglyoxal (an intermediate glycation metabolite) can enhance insulin resistance, e.g. by blocking the PI3K/Akt signaling pathway and inducing oxidative stress, which leads to a strong inhibition of metabolic processes regulated by this hormone23,24. These factors contribute to the chronic hyperglycemia observed in type 2 diabetes, which in turn enhances protein glycation19. In addition, a high content of glycated albumin in the blood may disturb immune balance and induce oxidative stress, causing tissue inflammation25. This process can also have a detrimental effect on the immune system as glycation of immunoglobulins impacts protein function19. Glycated LDL proteins interact with scavenger receptors on the surface of macrophages causing differentiation into foam cells and atherosclerotic plaques26. Moreover, glycation provides proteins with immunogenic properties and causes their accumulation in plasma in form of immune complexes19,27. In the extracellular matrix and connective tissue, collagen, laminin, vitronectin, and elastin are targets for glycation. Such modifications result in increased stiffening of the walls of blood vessels or heart muscle, cause cardiovascular complications (especially in diabetic patients), and lead to the development of angiopathy, retinopathy, nephropathy, neuropathy, and cardiomyopathy1,28,29,30,31.
AGEs can also interact with some specific receptors and excerpt cellular effects. The best known receptor—RAGE has been identified on the surface of various types of cells and tissues, e.g. on macrophages, phagocytes, neutrophils, hepatocytes, endothelial smooth muscle, nervous system, and mesangial cells1,32. Under physiological conditions, RAGE is expressed at a low level, but elevated concentration of ligands (including AGEs) causes receptor overexpression in pathological or chronic inflammation associated with diabetes, Alzheimer's and cardiovascular disease or cancer33. RAGE interaction with the ligands initiates signaling pathways through the mitogen-activated protein kinases (MAPKs), such as p44/42 (ERK1/2), p38, and JNK34. In effect activation of NF-κB leads to upregulation of Jak/STAT and p21ras pathways, AKT kinases, GSK-3β, and rac-1 molecules35 responsible for several effects, i.e. inflammation, cell proliferation, angiogenesis, fibrosis, thrombogenesis, and apoptosis32. This creates pro-inflammatory conditions with release of cytokines Il-1 (interleukin-1), Il-6, and TNF-α (tumor necrosis factor)36. Moreover, the AGEs—RAGE interaction can induce NADPH oxidase and oxidative stress through reactive oxygen species (ROS), which also affect induction of the NFκB37,38. The increased production of ROS and the dysregulated inflammatory process largely contribute to cancer formation38. AGEs were shown to play a role in proliferation, migration, invasion, and/or increased angiogenic potential of cancer cells39. The tumor-promoting effects of AGEs have been revealed in studies on several cancer, like colon40, prostate41, lung42, liver43,44, breast45, pancreatic46,47, and melanoma48. It has been reported that AGEs derived from glucose are present in cancer tissues and blood of gastric cancer (GC) patients. Interestingly, RAGE expression is increased in in invasive cancer compared to the non-invasive tumor and it is associated with disease progression. Therefore, in invasive cancer the accumulation of glucose-derived AGEs and RAGE expression is associated with the potential risk for disease progression49. However, the effect of MAGE on cancer microenvironment and its role in RAGE-mediated cellular response remain to be discovered.
To date, only few AGEs have been characterized and confirmed in vivo, with many other perhaps crucial for a specific pathology, are yet to be described. While the structure and basic biological properties of the in vitro obtained MAGE has been elucidated4,7, there is need for determination of the natural in vivo counterpart. The goal of the presented paper was to identify the proteins modified by MAGE present in human body fluids, especially in blood and peritoneal fluid from healthy, diabetic and cancer individuals. We present the isolation of natural MAGE antigen, identification of the type of carrier protein, and characterization by means of spectrometric and immunochemical methods. We also indicate the presence of the MAGE antigen on proteins derived from blood and peritoneum in gastric cancer (GC) patients. Our data initiate an interesting perspective on MAGE in cancer microenvironment.
Results
Identification of blood proteins reacting with anti-MAGE antibody
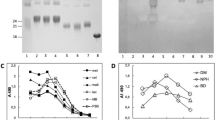
The proteins present in serum sample obtained from a healthy volunteer were at first separated by 2D electrophoresis (Fig. 1A) and subjected to a Western Blotting (WB) analysis to identify MAGE modifications. Staining with the specific anti-MAGE monoclonal antibody (mAb) revealed several proteins caring the MAGE antigen (Fig. 1B). Some of the spots, i.e. proteins with ~ 25 kDa (spot 1–4) and ~ 75 kDa (5–6) were selected for protein identification using mass spectrometry analysis.
Identification of proteins glycated with MAGE. Protein from human serum were subjected to isoelectric focusing between pH 3–7, separated on 8% SDS-PAGE gel and stained with Coomassie Safe (A). Additional gels were subjected to WB analysis with anti-MAGE mAb followed by secondary Ab anti-mouse IgE-HRP (B) or by incubation with the secondary Ab only (C). The indicated spots 1–6 were excised from the gel. Molecular mass markers on the right side of each panel indicate mass in kDa. The original gel and blots are presented in Supplementary Fig. S4.
Several proteins have been identified in each of the excised samples (spot 1–6), although the employed criteria were very stringent, included hits with score > 1000 and high sequence coverage (Table 1). The spots 1–4 that in 2D gel migrated as the proteins around 25 kDa, revealed presence of the light chain of immunoglobulin kappa and lambda, while the spot 5 and 6 migrating as proteins around 75 kDa showed several proteins including serum albumin, heavy chain of immunoglobulin mu and gamma. These results suggest that the common and highly abundant blood proteins can be modified with MAGE. Presence of other proteins, especially with unrelated molecular mass (spot 1–3—albumin, spot 3—serotransferrin, spot 5—complement C3) needs further explanation and verification.
Isolation of MAGE analog from human blood
After an initial 2D electrophoresis tracking of the blood proteins modified by MAGE the antigen has been extracted by employing an immunoprecipitation. The purified anti-MAGE monoclonal antibody (Supplementary Figs. S1, S2) and Pierce Direct IP Kit (Thermo Scientific, Waltham, MA, USA) were used in accordance with the manufacturer's instructions. Serum samples from the diabetic patients was applied to columns with the immobilized anti-MAGE antibody. The same experiment was performed with a human blood plasma from the healthy donors. The material bound to the Sepharose-anti-MAGE resin was analyzed by SDS-PAGE and showed number of proteins (Fig. 2A, lane 1 and 2). Some of the proteins were also eluted from the control resin without anti-MAGE antibody (Fig. 2A, lane 3 and 4). However, in this case only few proteins showed WB staining with anti-MAGE antibody, i.e. 59.4 kDa (1S) and 50.0 kDa (2S) (Fig. 2B; lane 1) in a blood of the diabetic patients, and proteins of 61.7 kDa (1P) and 49.3 kDa (2P) (Fig. 2B; lane 2) in a blood of the healthy donor. Some weak reaction was evident in the sample eluted from the control resin (1PN, Fig. 2B; lane 4), however the mass of this protein (56.7 kDa) was different than the protein eluted from Sepharose-anti-MAGE resin (Fig. 2B, lane 1, 2), suggesting the non-specific binding of this blood protein to the resin not present in sample 2. The non-specific secondary antibody reaction in WB was excluded by staining of the membrane with the secondary antibody only (Fig. 2C). The gel bands (marked in red in Fig. 2A) corresponding to the proteins labeled with the anti-MAGE antibody (Fig. 2B, lane 1, 2) were excised and subjected to the mass spectrometry protein identification.
Extraction of blood proteins glycated with MAGE. Proteins from serum of the diabetic patient and plasma of healthy donor were immunoprecipitated with the Sepharose-anti-MAGE resin and the same volume of each sample was analyzed by SDS-PAGE (A). The separated in the gel proteins that bound to the Sepharose-anti-MAGE antibody resin (lane 1, 2) and the control resin (lane 3, 4) were stained with Coomassie Brilliant Blue or were subjected to WB with the anti-MAGE antibody (B). As the negative control, the additional membrane was probed with the secondary anti-mouse IgE-HRP antibody (C). The proteins indicated in panel A by red boxes corresponding to bands 1S, 2S (diabetic serum) and 1P, 2P (plasma from healthy donor) showed specific binding with the anti-MAGE antibody and were excised from gel for mass spectrometry analysis. M—molecular mass standard; MAGE (MB-mel). The original gel and blots are presented in Supplementary Fig. S5.
Mass spectrometry analysis of the excised protein bands revealed several proteins in the analyzed samples (Table 2). In the sample from diabetic patients (1S), the strongest signal (score) and sequence coverage of 38.8% was shown for albumin, α-1-antitrypsin, and heavy chain of immunoglobulin A (IgA) and G (IgG). In the 2S sample, IgG1 heavy chain (30% sequence coverage) and the constant regions of the IgG3 and IgG2 heavy chains were identified. The sample 1P from the healthy donor plasma contained fibrinogen α-chain as the protein with the strongest signal, although sequence coverage was slightly greater for the heavy chain of IgA (20.4%). In the 2P sample, the strongest signal was obtained for the heavy chain of IgG1 (28.7% sequence coverage). In this sample the presence of IgG2 heavy chain as well as β and γ fibrinogen was also found.
Based on the identification score, sequence coverage (Table 2), and theoretical protein mass the most common proteins in human blood, i.e. albumin (HSA), IgG, and IgA were selected for further consideration. To verify the mass spectrometry results we performed a series of additional WB experiments with specific anti-HSA, anti-human IgG, and anti-human IgA antibodies (Fig. 3). The molecular mass of the identified protein bands was determined using the LabImage software (Kapelan Bio-Imaging, Leipzig, Germany) and were summarized in Table 3.
Verification of proteins extracted from human blood by WB analysis. The proteins extracted from diabetic patient serum (S), plasma of healthy donor (P) were transferred on the PVDF membrane along with protein marker (M), HSA, IgG, IgA (2 µg/well) and were probed with anti-HSA (A), anti-mouse IgG-HRP (B), anti-human IgG-HRP (C), and anti-human IgA-HRP (D) antibodies. The original blots are presented in Supplementary Fig. S6.
The samples obtained from serum and plasma were probed by WB with the anti-HSA antibody showed the presence of proteins with a mass of ~ 59 kDa (Fig. 3A; S-band 2, P-band 3), that migrated like HSA protein loaded as a control (Fig. 3A; HAS-band 1). On the membrane probed with the anti-mouse IgG-HRP antibody (negative control), the weak band of about 49 kDa was observed (Fig. 3B; bands 4, 5), that presents a non-specific cross-reactivity of the secondary antibody to human IgG present in the samples and proves the HSA identity in the studied samples.
To verify presence of human IgG in the extracted from human blood MAGE protein, the membrane was incubated with the anti-human IgG-HRP antibodies. It showed the proteins with molecular mass of about 49–50 kDa (Fig. 3C; S-band 7, P-band 8) that migrated like IgG loaded as a control (Fig. 3C; IgG-band 6).
Similarly, the reactivity of the extracted proteins with anti-human IgA-HRP antibody showed the protein band of about 61 kDa (Fig. 3D; S-band 10, P-band 11) that migrated similarly to IgA loaded as a control (Fig. 3D; IgA-band 9). The results suggest that the in vivo MAGE antigen extracted from human blood is present on several proteins including HSA (59 kDa), IgG (50 kDa), and IgA (61 kDa).
The results of WB analysis and the molecular mass estimation, based on comparison with the marker proteins, have been summarized in Table 3. Protein band 1S with mass of 59.4 kDa labeled with anti-MAGE antibody (Fig. 2B, lane 1) corresponds to the mass of band 2 in Fig. 3A, lane S, suggesting it is HSA. This has also been confirmed by mass spectrometry analysis of the sample 1S that we identified to be a human albumin protein (Table 2). Bands 2S and 2P (Fig. 3B) with mass of 50.0 and 49.3 kDa, respectively, correspond to bands 7 and 8 (Fig. 3C) detected by the WB with anti-IgG antibodies. These results have been also confirmed by mass spectrometry that identified human IgG in samples 2S and 2P (Table 2). Band 1P (Fig. 2B, lane 2) of 61.7 kDa labeled in WB with anti-MAGE antibody corresponds to band 11 (61.5 kDa) in Fig. 3D and confirms it is IgA. Mass spectrometry analysis also indicated the presence of IgA in sample 1P (Table 2).
Our results suggest the in vivo MAGE antigen extracted from human blood is present on several proteins including HSA (59 kDa), IgG (50 kDa), and IgA (61 kDa). Interestingly, in these preliminary experiments we noticed different proteins modified by MAGE in the tested samples from the diabetic patients comparing to the healthy people. Serum albumin and IgG were the major proteins found to contain MAGE antigen in diabetic serum in contrast to healthy people that plasma showed mostly immunoglobulins G and A to be modified. In addition, the sample after immunoprecipitation with the Sepharose-anti-MAGE resin contained other proteins identified by mass spectrometry (α-1-antitrypsin, vitronectin, vimentin, antithrombin or fibrinogen; Table 2), however they need further verification.
Analysis of body fluid samples for presence of MAGE-modified proteins
The presence of MAGE antigen was next verified in body fluid samples from bigger number of subjects. Our previous observation suggested that the sections from different cancer tissues show positive reactivity with the anti-MAGE antibody (manuscript in preparation). The peritoneal fluid is often collected during the diagnostic laparoscopy. It presents a good target specimen for identification of the cancer markers released from the growing tumor and it is used for determination of some diagnostic markers50. Therefore, inhere we aimed to study the presence of MAGE antigen in body fluids—blood and peritoneum of the GC patients. For this purpose, WB analysis with the anti-MAGE antibody was performed on 10 serum samples (Fig. 4). Several bands were marked in patients’ serum, including proteins around 60 kDa and 25 kDa (Fig. 4A, lane 2–11), suggesting a reactivity with MAGE antigen, similar like for MB-mel used as a control (Fig. 4A, lane 12). At the same time, the secondary anti-mouse IgE-HRP antibody staining was weaker for some samples (lane 3, 4, 5, 7, 9, 10, Fig. 4B) that suggests the specific binding of the monoclonal anti-MAGE antibody to the serum proteins from patients with GC.
WB analysis of human body fluids with anti-MAGE monoclonal antibody. Ten samples of serum (A, B—lane 2–11) and corresponding peritoneal fluid (C, D—lane 2–11) from patients with GC were subjected to WB with the anti-MAGE antibody (A, C) or as a control with secondary antibody anti-mouse IgE-HRP (B, D). Molecular marker indicated in kDa (lane 1) and MB-mel (lane 12) was run as the controls. The original blots are presented in Supplementary Fig. S7.
Further, we tested the samples of peritoneal fluids that have been previously collected along the blood from the same patients with GC. The samples transferred on the membrane were exposed to the anti-MAGE monoclonal antibody (Fig. 4C) and interestingly they also showed presence of MAGE antigen on some proteins of mass about 60 kDa. The control staining with the secondary anti-mouse IgE-HRP antibody showed weaker staining for the corresponding bands (lane 2, 3, 5, 6, 7, 8, 10, 11, Fig. 4D), suggesting that MAGE antigen in addition to blood is also present on some proteins in peritoneal fluids of GC patients.
Discussion
Antibodies can be used as powerful tools to identify trace amount of an analyte in a sample. The antigen-binding properties determine both the sensitivity and selectivity of the immunoassay. The high specificity of antibody is fundamental to their natural function51. The generated earlier anti-MAGE antibody specifically binds MAGE antigen synthesized on various carrier proteins (Supplementary Fig. S1). We have previously reported no cross-reactivity of the anti-MAGE antibody with other known AGEs4. The specificity was also confirmed in competitive ELISA, with model MAGE generated on MB and protein-fee lysine (Supplementary Fig. S2). Using the anti-MAGE monoclonal antibody, we have identified some proteins (~ 25 and 75 kDa) in human blood from the healthy donor that were clearly separated on a 2D electrophoresis and contained MAGE antigen (Fig. 1). This encouraged us to attempt extraction of the in vivo MAGE antigen from human blood. MAGE was isolated from the diabetic serum (pooled from several patients) and from the donors’ plasma (healthy individuals). The extracted proteins were analyzed by WB (Fig. 2B) and mass spectrometry showing the presence of 59.4 kDa (1S) and 50.0 kDa (2S) proteins in the blood of diabetic patients in contrast to 61.7 kDa (1P) and 49.3 kDa (2P) proteins in the blood of healthy donors. In both samples the proteins with mass over 25 kDa were identified. We suspect these are the light chains of the secreted immunoglobulins52 however it needs further confirmation.
In the next step, several types of antibodies specific for different proteins and mass spectrometry analysis were employed for identity verification of the proteins (extracted from human blood) modified with MAGE antigen. In sample 1S (59.4 kDa) the human serum albumin (HSA) showed the best score and 38.8% sequence coverage. This protein was confirmed by WB with the specific anti-HSA antibody (Fig. 3A). Albumin is the most abundant plasma protein and accounts for over 50% of serum proteins in healthy subjects. It plays a key role in maintaining oncotic pressure and is known to be a versatile protein carrier for the transport of various endogenous and exogenous ligands53. HSA has a half-life of approximate 3 weeks, sufficient for significant glycation progressing to AGE formation54. The structure of HSA consists of a single polypeptide chain of 585 amino acids with a theoretical molecular mass of 66.438 kDa55. The molecular mass calculated from spectrometric analysis shows 71.317 kDa (Table 1) and the difference might result from the protein in vivo modifications56. It has been proven that this protein is subject to several chemical modifications, including acetylation, oxidation, glycosylation, glycation, carbonylation, and phosphorylation affecting its binding and antigenic properties57. After sample separation by SDS-PAGE, the protein molecular mass was calculated to be 59.4 kDa (Fig. 3). Migration of proteins in a polyacrylamide gel in the presence of SDS, which does not correlate with the molecular mass formula, is called “gel shift” and appears to be fairly common but has not yet been fully clarified58. This effect is explained by differences in the secondary and tertiary structure of the protein, the intrinsic net charge of the protein, and the amount of bound SDS58,59,60,61. The literature shows that post-translational modifications of albumin cause changes in the secondary and tertiary structure of HSA, causing e.g. reduced content of α-helices in the structure of polypeptide62. It may affect the degree of detergent loading and result in a difference between the actual mass of the protein and the mass calculated from the electrophoretic migration in a gel. The difference in theoretical and observed molecular mass however does not challenge the identity of HSA as the in vivo MAGE-carrier, since it has been confirmed by mass spectrometry (Table 2).
In the sample 1S (Fig. 2B; Table 1), apart from albumin, the α-1-antitrypsin and heavy chains of IgA and IgG were also identified with high scores. Water-soluble and tissue-diffusing α-1-antitrypsin is a circulating glycoprotein with a molecular mass of 52 kDa63. Its primary function is to protect tissues from enzymes, such as neutrophil elastase and proteinase 3, released during inflammation64. The half-life in the blood of α-1-antitrypsin is 4–5 days, thus is not sufficient for generation of AGEs, and only allows the formation of Amadori products54,63. However, the presence of other MAGE-analogous structures on this protein that result from other modifications cannot be ruled out, especially that the origin of the in vivo MAGE has not been elucidated so far. The presence of immunoglobulins G and A in the 1S sample was also excluded by the WB analysis (Fig. 3; Table 2), based on the reaction with specific antibodies showing bands of different molecular mass in comparison to the result of WB with the anti-MAGE antibodies (Fig. 2B).
The sample 1P corresponding to the proteins with a mass of about 61.7 kDa (Fig. 3B) showed presence of fibrinogen α (highest score) and IgA heavy chain (highest sequence coverage) (Table 2). Fibrinogen is a multifunctional acute-phase plasma protein, playing a key role in hemostasis and the coagulation cascade. It occurs in the blood plasma of healthy people at concentrations of 1.5–4 mg/ml. This protein consists of two identical subunits containing three polypeptide chains referred to as Aα, Bβ, and γ chains with a total mass of approximately 340 kDa65. According to the literature, the molecular mass of the fibrinogen α chain is ~ 95 kDa66, which significantly exceeds the value of the mass determined by SDS-PAGE and WB (Fig. 3). Moreover, fibrinogen has a short blood half-life of 3–5 days67 and its glycation seems unlikely. Thus it was not considered in our further studies, but other MAGE analogous structures (different in vivo origin) might mediate binding to the anti-MAGE antibody. The mass spectrometry (Table 2) and immunoblotting analysis provided evidence that sample 1P consists of immunoglobulins (Fig. 3D). The protein band with a mass of 61.5 kDa, evidenced with the anti-IgA antibody, correlated with the mass of the band evidenced with the anti-MAGE antibody (Fig. 3B, band 1P). Theoretical mass of the IgA heavy chain constant region (38.49 kDa) reflects the specific peptide identified by mass spectrometry without considering the variable domain of the whole heavy chain molecule. In electrophoresis, we observed the entire heavy chain of IgA migrating around 60 kDa as it is found in literature68,69. Immunoglobulin A is the second most abundant antibody in human blood. It neutralizes toxins and prevents from microbial invasion through the mucosa epithelial barrier. Although this protein has a fairly short half-life of 5–7 days 70, there are disease states in which the elimination of IgA is impaired and the protein is deposited in the tissues71, allowing for its glycation.
In the sample 2S (serum of diabetic patients) and in the sample 2P (plasma of healthy donors) heavy chain IgG1 (sequence coverage 30 and 28.7%, respectively) was identified by mass spectrometry as the protein with the highest identification score (Table 2). The theoretical molecular mass of this protein is 49.93 kDa and it is consistent with the protein band observed in WB with anti-MAGE antibody (50.0 and 49.3 kDa for 2S and 2P bands, respectively) (Fig. 3, Table 2). In addition, the same size proteins were revealed upon membrane staining with anti-IgG antibody (50.0 kDa for band 7 and 49.2 kDa for band 8 (Fig. 3C, Table 2). Literature describes the human IgG heavy chain as a protein of about 50 kDa72, which is in agreement with our observation in WB for both the positive control and test samples (Fig. 3). Slightly lower mass of IgG in the sample 2P (healthy donor) may indicate a lower degree of modification of this protein compared to the IgG in diabetic patient sera. Constant regions of IgG3 and IgG2 heavy chains were also identified in sample 2S, and constant regions of IgG2 and IgG4 heavy chains in sample 2P. The antibodies used in immunoblotting to verify the presence of IgG in the sample are directed to the Fc region of human IgG, which did not allow for the precise determination of the subclass of the identified protein. IgG accounts for about 75% of human serum immunoglobulins and is the primary antibody of the secondary immune response. It has a half-life of 23 days, which is sufficient to generate AGEs through the glycation process73.
In the same sample 2P the other proteins, i.e. β and γ chains of fibrinogen have been identified (Table 2) that theoretical mass was calculated as 56.58 kDa and 52.11 kDa. In the studies by Repetto et al. fibrinogen β chain migrated at about 50, 40 and 37 kDa74, while in the studies of Luo et al. the mass of the β chain was determined at 52 kDa and the γ chain at 46 kDa75. The observed masses only slightly differ from the mass of the band determined by us (49.3 kDa, Fig. 3B) on a membrane probed with the anti-MAGE antibody. The short half-life of this protein makes it less attractive target for glycation, therefore we focused on IgG identified with a higher score.
Our data suggest that MAGE antigen is present in vivo on several proteins, mainly albumin, IgG and IgA—the main components of blood plasma, accounting for over 70% of all plasma proteins (mainly HSA and IgG)54. This is the first report showing specific proteins modified with MAGE found in serum of the diabetic patients (albumin and IgG) and in plasma of the healthy people (IgG and IgA). Results suggest that the MAGE antigen is physiologically present on immunoglobulins for which glycation is a common post-translational modification76, but in pathology and oxidative stress formation of MAGE antigen can also occur on other proteins, like albumin. These observations require further confirmation in a larger sample cohort. We assume that IgA modified with MAGE identified in the plasma sample from the healthy donor and not isolated from the serum sample of the diabetic patients, may be explained by abundance of MAGE-modified albumin and IgG under hyperglycemic conditions that saturated the Sepharose-anti-MAGE resin, preventing from binding the proteins of a lower abundance. Further studies to identify additional MAGE-modified proteins in human blood should consider the changes in the immunoprecipitation protocol, i.e. higher concentration of the anti-MAGE antibody and sample re-precipitation.
Significance of our study refers to the glycation-induced change in function of the circulating natural antibodies and potentially some therapeutic antibodies used recently more often in clinics. AGEs increase immunogenicity of immunoglobulins and cause production of antibodies against this modified protein77, as reported in type 2 diabetes78, atherosclerosis79, and rheumatoid arthritis (RA)80,81,82. Glycation is also a suspected cause of decreased elimination of IgA by the liver and the protein accumulation in blood83. Serum IgA levels are significantly higher in diabetic patients than in healthy subjects84,85,86. High levels of IgA is a sign of diabetic nephropathy86, the most commonly recognized type of glomerular disease in the world and manifested by the formation of mesangial IgA deposits87. Glycation of HSA have also serious impact on its function in defense against oxidative stress88, transport of various ligands, and regulating of an oncotic pressure due to changes in the secondary and tertiary structure89. HSA glycation status is associated with development of autoantibodies in diabetic patients90,91 and progression of cardiovascular disease92. In this paper we present for the first time that several MAGE-modified proteins are present not only in blood but also in a peritoneal fluid collected from the GC patients (Fig. 4). Similarly, Deng et al. reported an accumulation of the glucose-derived AGEs in tumor and blood of the GC patients49, suggesting a potential role of MAGE in cancer. Therefore, some proteins with molecular mass about 25 kDa and 60 kDa modified by MAGE found in our study in peritoneal fluid (suggesting immunoglobulin protein and HSA) will be further investigated to determine the protein identity and relevance to disease pathology and progression. The interesting direction will be also further studies on plasma from diabetic patients to verify the presence of MAGE on fibrinogen in this pathological state.
In summary, the presented data add to the knowledge on protein modifications in vivo and their potential role in diabetes, cancer, and adaptive immune response associated with modification of the circulating antibodies.
Methods
All chemicals were from Sigma-Aldrich (Saint Louis, MO, USA), unless otherwise stated. All methods were performed in accordance with the relevant guidelines and regulations.
Human body fluids
In this study we have used plasma or serum collected from the healthy donors (Local Bioethics Committee at Medical University of Wroclaw, KB-770/2018 and Local Bioethics Committee at Medical University of Lublin, KE-0254/157/2016) and serum from the patients with diabetes (Local Bioethics Committee at Medical University of Wroclaw, KB-384/2012, donated by dr Agnieszka Bronowicka-Szydełko). Additionally, we used serum and the corresponding peritoneal fluid collected from the patients with GC described in our earlier studies50 (Local Bioethics Committee at Medical University of Lublin, KE–0254/182/2018). The study was approved by the indicated ethics committee and the informed consent was obtained from all subjects.
MAGE antigen synthesis
The high molecular mass glycation products (HMW-MAGE) used in this study were generated in dry conditions (MWG) as described earlier4. Briefly, the miliQ water solution mixtures of melibiose (mel) and one of a model proteins: horse myoglobin (MB), bovine serum albumin (BSA) or rabbit immunoglobulin G (rIgG), in 100:1 molar ratio (carbohydrate:protein) were prepared and subjected to MWG reaction. Next, the samples were dissolved in milliQ water and centrifuged at 5000 × g, 15 min to remove any insoluble precipitates. The obtained supernatants were washed several times with milliQ water on Amicon Ultra-15 centrifuge filters (Merck Millipore, Burlington, MA, USA) with a 30 kDa cut-off to remove the unreacted substrates. The resulting glycated proteins were frozen at − 80 °C, lyophilized, and stored at − 20 °C until further studies.
SDS-PAGE and Western blotting
The model protein and samples immunoprecipitated with the Sepharose-anti-MAGE resin were diluted 1:1 with Non-Reducing Lane Marker Sample Buffer (Thermo Scientific, Waltham, MA, USA), supplemented with 100 mM β-mercaptoethanol and denatured by incubation at 100 °C for 5 min. The samples were separated on 12% polyacrylamide gel using a Mini-Protean Tetra Cell apparatus (Bio-Rad, Hercules, CA, USA) for 90 min under constant voltage of 100 V. The gel was next stained with Quick Coomassie Protein Stain (ProteinArk, Sheffield, UK). The results were visualized on a PXi Touch gels and blots detection and analysis system (SYNGENE, Cambridge, UK). The molecular mass of the selected bands was determined with LabImage gel analysis software (Kapelan Bio-Imaging, Leipzig, Germany). For WB the proteins separated on a polyacrylamide gel were next transferred to a PVDF membrane (0.45 μm, Immobilon-P Transfer Membrane PVDF, Merck-Millipore, Burlington, MA, USA) using a Mini Trans-Blot Cell apparatus (Bio-Rad, Hercules, CA, USA). The transfer was carried out for 1 h at constant voltage of 100 V (for model protein). Then the membrane was blocked with 5% solution of skimmed milk powder (SM Gostyń, Poland) in TBS-T (Tris with 0.05% Tween 20) for 1 h, at room temp or overnight at 4 °C. After washing 3 times with TBS-T, the membrane was incubated for 2 h, at room temp with one of the antibodies: goat anti-human IgG-HRP (Abcam, Cambridge, UK), goat anti-human IgA-HRP (Abcam, Cambridge, UK) diluted 1:15 000 in phosphate-buffered saline (PBS) pH 7.4 (single step, no secondary antibodies), rabbit anti-HSA (human serum albumin, Thermo Scientific, Waltham, MA, USA) diluted 1:5 000 or with culture medium from the hybridoma cells producing mouse anti-MAGE monoclonal antibodies, with incubation carried overnight at 4 °C, followed by 3 times washing with TBS-T solution. Next, when appropriate, the membranes were incubated for 1.5 h, at room temp with anti-rabbit IgG-HRP antibody (Abcam, Cambridge, UK) diluted 1:2 000 or goat anti-mouse IgE-HRP antibody (Origene, Rockville, MD, USA) diluted 1:5 000 in PBS. After washing 5 times with TBS-T, antibody binding was visualized by incubation with SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Scientific, Waltham, MA, USA). The results were photographed using PXi Touch gels and blots detection and analysis system (SYNGENE, Cambridge, UK).
Electrophoresis of the human body fluids (blood and peritoneal fluid) from GC patients was performed on miliQ water-diluted samples (1:1) subjected to protein precipitation with cold methanol (chilled to − 20 °C) by mixing 1 volume of sample with 4 parts of 100% methanol. The protein pellet in each sample obtained after centrifugation (14 000 × g, 5 min, 4 °C) was dissolved in the original sample volume of RIPA buffer (EMD Milipore, Billerica, MA, USA), mixed with a Laemmli sample buffer containing 20% β-mercaptoethanol, and denatured by incubation at 97 °C for 10 min. Equal volume of each sample was separated on 8% polyacrylamide gel at constant voltage of 200 V for 50 min. Next the proteins were transferred onto the PVDF membrane with constant voltage of 200 V (for human body fluids). After incubation with a blocking solution (5% solution of skim milk powder from Carl Roth, Karlsruhe, Germany dissolved in PBS containing 0.05% Tween (PBS-T)) for 1 h at room temp the membrane was incubated overnight at 4 °C with antibodies anti-MAGE diluted in blocking solution. Next, the excess of antibodies was washed out 3 times with PBS-T and the membrane was incubated for 4 h with secondary antibody anti-mouse IgE-HRP (Origene, Rockville, MD, USA) diluted 1:8 000 in PBS-T. After 3 times washing with PBS-T, antibody binding was visualized and photographed using ChemiDoc MP V3 imaging system (BioRad, Hercules, CA, USA) after incubation with a Clarity Western ECL Substrate (BioRad, Hercules, CA, USA).
2-D Electrophoresis and identification of the proteins for mass spectrometry analysis
Sample (120 µl) of human serum from the healthy donor, previously diluted 1:1 in miliQ water, was mixed with cold methanol (480 µl) and incubated for 20 min at − 20 °C to precipitate proteins. After centrifugation at 13 000 rpm for 5 min, the pellet was retained and subjected to the sample preparation procedure using the Cleanup solution according to the manufacture protocol (BioRad, Hercules, CA, USA). The sample was finally reconstituted in 150 µl of Rehydration solution (8 M urea, 2% CHAPS, 50 mM DTT, 0,2% (w/v) Bio-Lyte 3/10 ampholytes, trace of Bromophenol Blue). Before application on the gel the sample was vortexed, sonicated for 5 min and centrifuged at 13 000 rpm for 3 min to remove any residual insoluble material. Isoelectric focusing was performed on a strip within pH 3–10 using a BioRad PROTEAN i12 IEF Cell (BioRad, Hercules, CA, USA) with gradient parameters: 250 V linear for 20 min, 4000 V linear for 2 h, rapid 4000–10 000 VHr, hold at 500 V and rehydration for 14 h. Next day the strip was dried on a paper towel and equilibrated in a tray with solution I (6 M Urea, 2% SDS, 0.375 M Tris–HCl pH 8.8, 20% glycerol, 2%, DTT) and solution II (6 M Urea, 2% SDS, 0.375 M Tris–HCl pH 8.8, 20% glycerol, iodoacetamide) for 10 min each. The second-dimension electrophoresis was carried on 8% polyacrylamide gel at 200 V for 42 min. Next the gel was rinsed 3 × for 10 min with miliQ water and stained with Bio-Safe Coomassie Stain (BioRad, Hercules, CA, USA) or transferred to the PVDF membrane at 150 V for 90 min. The membrane was subjected to WB with anti-MAGE mAb diluted in bocking solution (as above for peritoneal fluids WB) followed by anti-mouse IgE-HRP (Origene Rockville, MD, USA) diluted 1:6 000 in PBS-T. The membrane serving as the negative control was incubated only with secondary antibody without the anti-MAGE antibody exposure. The results were photographed using an ChemiDoc MP V3 imaging system (BioRad, Hercules, CA, USA) after incubation with a Clarity Western ECL Substrate (BioRad, Hercules, CA, USA).
The protein spots visualized on the gel stained with Coomassie were photographed and manually correlated using the PhotoShop software with the spots of proteins reacting with the anti-MAGE mAb on the PVDF membrane and were excised from the gel. For mass spectrometry analysis the proteins that did not show reactivity with the anti-mouse-IgE-HRP antibody (negative control) were selected. The excised gel pieces were stored at − 20 °C until analysis.
Isolation of MAGE-modified proteins from human blood
The analogue of MAGE present naturally in human blood was extracted from the samples collected in the Military Center for Blood Donation and Haemotherapy in Wroclaw (honorary blood donors) and the Department of Angiology, Diabetology and Hypertension in Wroclaw (diabetic patients). The presence of the MAGE antigen in the tested diabetic sera was previously confirmed by competitive ELISA5. MAGE extraction was performed using the purified murine anti-MAGE monoclonal antibody (described in Supplementary data) immobilized on Sepharose resin using the Pierce Direct IP Kit (Thermo Scientific, Waltham, MA, USA). The procedure was carried out in accordance with the manufacturer's instruction. Briefly, for each sample, 8 µg of anti-MAGE antibody was mixed with 20 µl of Sepharose resin suspension and incubated for 2 h, at room temp on a shaker (BioSan, Riga, Latvia) at low speed. The Sepharose-anti-MAGE resin was next washed and incubated with serum sample (mixed from several patients) or blood plasma containing 1 mg of protein (for 2 h, at room temp and then overnight at 4 °C with gentle rocking), followed by incubation with the blocking solution (1 M Tris–HCl). The resin incubated only with a blocking solution, without anti-MAGE antibody was used as the negative control. After the resin was appropriately washed, the resin-bound material was eluted with the elution solution (25–75 µl). The collected samples were stored at − 20 °C until further analysis.
Identification of MAGE-modified proteins present in human blood by mass spectrometry
Proteins extracted from human blood on Sepharose-anti-MAGE resin were separated on a polyacrylamide gel (SDS-PAGE) and stained. The pieces of gel with protein bands and the spots from 2-D gel (corresponding to the proteins identified in WB with anti-MAGE Ab) were excised and after drying, the proteins were reduced with 10 mM DTT (dithiothreitol), alkylated with 50 mM iodoacetamide, and digested with trypsin at 37 °C overnight. The obtained peptides were extracted from the gel with 0.1% TFA (trifluoroacetic acid) containing 2% acetonitrile. Mass spectrometry analysis was performed at the Environmental Mass Spectrometry Laboratory, Institute of Biochemistry and Biophysics, Polish Academy of Sciences in Warsaw. The analysis employed a liquid chromatography coupled with an Orbitrap mass spectrometry (Thermo Scientific, Waltham, MA, USA). Protein identification was performed using the MASCOT software (Matrix Science, Boston, MA, USA) by comparison with the UniProt protein sequence database.
References
Pietkiewicz, J., Seweryn, E., Bartys̈, A. & Gamian, A. Receptory końcowych produktów zaawansowanej glikacji-znaczenie fizjologiczne i kliniczne. Postepy Higieny i Medycyny Doswiadczalnej 62, 511–523 (2008).
Nagai, R. et al. Antibody-based detection of advanced glycation end-products: Promises vs limitations. Glycoconj. J. 33, 545–552 (2016).
Ashraf, J. M. et al. Recent advances in detection of AGEs: Immunochemical, bioanalytical and biochemical approaches. IUBMB Life 67, 897–913 (2015).
Staniszewska, M. et al. The melibiose-derived glycation product mimics a unique epitope present in human and animal tissues. Sci. Rep. 11, 2940 (2021).
Bronowicka-szydełko, A., Krzystek-korpacka, M. & Gacka, M. Association of novel advanced glycation end-product ( AGE10) with complications of diabetes as measured by enzyme-linked immunosorbent assay. J. Clin. Med. 10, 4499 (2021).
Indyk, D., Bronowicka-Szydełko, A., Gamian, A. & Kuzan, A. Advanced glycation end products and their receptors in serum of patients with type 2 diabetes. Sci. Rep. https://doi.org/10.1038/s41598-021-92630-0 (2021).
Czech, M. et al. The genotoxic and pro-apoptotic activities of advanced glycation end-products (MAGE) measured with micronuclei assay are inhibited by their low molecular mass counterparts. Genes 12, 729 (2021).
O’Connell, K. J. et al. Metabolism of four α-glycosidic linkage-containing oligosaccharides by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 79, 6280–6292 (2013).
O’Callaghan, A. & van Sinderen, D. Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 7, 925 (2016).
Yoon, M. Y. & Hwang, H. J. Reduction of soybean oligosaccharides and properties of α-d-galactosidase from Lactobacillus curvatus R08 and Leuconostoc mesenteriodes JK55. Food Microbiol. 25, 815–823 (2008).
Baú, T. R., Garcia, S. & Ida, E. I. Changes in soymilk during fermentation with kefir culture: Oligosaccharides hydrolysis and isoflavone aglycone production. Int. J. Food Sci. Nutr. 66, 845–850 (2015).
Schievano, E., Tonoli, M. & Rastrelli, F. NMR quantification of carbohydrates in complex mixtures: A challenge on honey. Anal. Chem. 89, 13405–13414 (2017).
Mao, B. et al. In vitro fermentation of raffinose by the human gut bacteria. Food Funct. 9, 5824–5831 (2018).
Sandek, A. et al. Studies on bacterial endotoxin and intestinal absorption function in patients with chronic heart failure. Int. J. Cardiol. 157, 80–85 (2012).
Fan, X. & Monnier, V. M. Protein posttranslational modification (PTM) by glycation: Role in lens aging and age-related cataractogenesis. Exp. Eye Res. 210, 108705 (2021).
Ahmad, S. et al. Inhibitory effect of metformin and pyridoxamine in the formation of early, intermediate and advanced glycation end-products. PLoS ONE 8, e72128 (2013).
Rehman, S., Faisal, M., Alatar, A. A. & Ahmad, S. Physico-chemical changes induced in the serum proteins immunoglobulin g and fibrinogen mediated by methylglyoxal. Curr. Protein Pept. Sci. 21, 916–923 (2020).
DeGroot, J. et al. Accumulation of advanced glycation end products as a molecular mechanism for aging as a risk factor in osteoarthritis. Arth. Rheum. 50, 1207–1215 (2004).
Fournet, M., Bonté, F. & Desmoulière, A. Glycation damage: A possible hub for major pathophysiological disorders and aging. Aging Dis. 9, 880–900 (2018).
Akhter, F., Khan, M. S., Alatar, A. A., Faisal, M. & Ahmad, S. Antigenic role of the adaptive immune response to d-ribose glycated LDL in diabetes, atherosclerosis and diabetes atherosclerotic patients. Life Sci 151, 139–146 (2016).
Akhter, F., Salman Khan, M., Faisal, M., Alatar, A. A. & Ahmad, S. Detection of circulating auto-antibodies against ribosylated-LDL in diabetes patients. J. Clin. Lab. Anal. 31, 22039 (2017).
Alavi, P., Yousefi, R., Amirghofran, S., Karbalaei-Heidari, H. R. & Moosavi-Movahedi, A. A. Structural analysis and aggregation propensity of reduced and nonreduced glycated insulin adducts. Appl. Biochem. Biotechnol. 170, 623–638 (2013).
Miele, C. et al. Human glycated albumin affects glucose metabolism in l6 skeletal muscle cells by impairing insulin-induced insulin receptor substrate (IRS) Signaling through a protein Kinase Cα-mediated mechanism. J. Biol. Chem. 278, 47376–47387 (2003).
Guo, Q. et al. Methylglyoxal contributes to the development of insulin resistance and salt sensitivity in Sprague-Dawley rats. J. Hypertens. 27, 1664–1671 (2009).
Lin, J.-A., Wu, C.-H. & Yen, G.-C. Perspective of advanced glycation end products on human health. J. Agric. Food Chem. 66, 2065–2070 (2018).
Younis, N. et al. Glycation as an atherogenic modification of LDL. Curr. Opin. Lipidol. 19, 378–384 (2008).
Gonen, B., Baenziger, J., Schonfeld, G., Jacobson, D. & Farrar, P. Nonenzymatic glycosylation of low density lipoproteins in vitro: Effects Cell-Interactive properties. Diabetes 30, 875–878 (1981).
Yamagishi, S., Ueda, S., Matsui, T., Nakamura, K. & Okuda, S. Role of ADVANCED GLYCATION END PRODUCTS (AGEs) and oxidative stress in diabetic retinopathy. Curr. Pharm. Des. 14, 962–968 (2008).
Yamagishi, S. I. & Matsui, T. Advanced glycation end products, oxidative stress and diabetic nephropathy. Oxid. Med. Cell. Longev. 3, 101–108 (2010).
Aronson, D. Cross-linking of glycated collagen in the pathogenesis of arterial and myocardial stiffening of aging and diabetes. J. Hypertens. 21, 3–12 (2003).
Bell, D. S. H. Heart failure: The frequent, forgotten, and often fatal complication of diabetes. Diab. Care 26, 2433–2441 (2003).
Perrone, A., Giovino, A., Benny, J. & Martinelli, F. Advanced glycation end products (AGEs): Biochemistry, signaling, analytical methods, and epigenetic effects. Oxidat. Med. Cell. Longev. https://doi.org/10.1155/2020/3818196 (2020).
Sorci, G., Riuzzi, F., Giambanco, I. & Donato, R. RAGE in tissue homeostasis, repair and regeneration. Biochim. Biophys. Acta Mol. Cell Res. 1833, 101–109 (2013).
Schmidt, A. M. RAGE and Implications for the pathogenesis and treatment of cardiometabolic disorders: Spotlight on the Macrophage. Arterioscler. Thromb. Vasc. Biol. 37, 613–621 (2017).
Ramasamy, R., Yan, S. F. & Schmidt, A. M. The diverse ligand repertoire of the receptor for advanced glycation endproducts and pathways to the complications of diabetes. Vascul. Pharmacol. 57, 160–167 (2012).
Liu, T., Zhang, L., Joo, D. & Sun, S.-C. NF-κB signaling in inflammation. (2017) https://doi.org/10.1038/sigtrans.2017.23
Ott, C. et al. Role of advanced glycation end products in cellular signaling. Redox Biol. 2, 411–429 (2014).
Dariya, B. & Nagaraju, G. P. Advanced glycation end products in diabetes, cancer and phytochemical therapy. Drug Discov. Today 25, 1614–1623 (2020).
Lin, J. A., Wu, C. H., Lu, C. C., Hsia, S. M. & Yen, G. C. Glycative stress from advanced glycation end products (AGEs) and dicarbonyls: An emerging biological factor in cancer onset and progression. Mol. Nutr. Food Res. 60, 1850–1864 (2016).
Liang, H. Advanced glycation end products induce proliferation, invasion and epithelial-mesenchymal transition of human SW480 colon cancer cells through the PI3K/AKT signaling pathway. Oncol. Lett. 19, 3215–3222 (2020).
Bao, J. M. et al. AGE/RAGE/Akt pathway contributes to prostate cancer cell proliferation by promoting Rb phosphorylation and degradation. Am. J. Cancer Res. 5, 1741 (2015).
Ahmad, S. et al. Oxidation, glycation and glycoxidation: The vicious cycle and lung cancer. Semin. Cancer Biol. 49, 29–36 (2018).
Chen, H. et al. Advanced glycation end products promote ChREBP expression and cell proliferation in liver cancer cells by increasing reactive oxygen species. Medicine 96, e7456 (2017).
Jabir, N. R., Ahmad, S. & Tabrez, S. An insight on the association of glycation with hepatocellular carcinoma. Semin. Cancer Biol. 49, 56–63 (2018).
Omofuma, O. O. et al. Dietary advanced glycation end-products (AGE) and risk of breast cancer in the prostate, lung, colorectal and ovarian cancer screening trial (PLCO). Cancer Prev. Res. 13, 601–610 (2020).
Wang, X., Zhang, R., Zhang, L. & Tian, Z. Glycated serum proteins: High in pancreatic cancer and low in preeclampsia. Prog. Mol. Biol. Transl. Sci. 162, 321–333 (2019).
Shahab, U. et al. The receptor for advanced glycation end products: A fuel to pancreatic cancer. Semin. Cancer Biol. 49, 37–43 (2018).
Abe, R. et al. Regulation of human melanoma growth and metastasis by AGE-AGE receptor interactions. J. Inv. Dermatol. 122, 461–467 (2004).
Deng, R. et al. Glucose-derived AGEs enhance human gastric cancer metastasis through RAGE/ERK/Sp1/MMP2 cascade. Oncotarget 8, 104216 (2017).
Gęca, K. et al. Kynurenine and anthranilic acid in the peritoneum correlate with the stage of gastric cancer disease Int. J. Tryptophan Res. https://doi.org/10.1177/1178646921106562015 (2022).
Hnasko, R. M. The biochemical properties of antibodies and their fragments. Methods Mol. Biol. 1318, 1–14 (2015).
Schroeder, H. W. & Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 125, S41–S52 (2010).
Raoufinia, R. et al. Overview of albumin and its purification methods. Adv. Pharm. Bull. 6, 495–507 (2016).
Vetter, S. W. Glycated Serum Albumin and AGE Receptors. In Advances in Clinical Chemistry (Elsevier, 2015).
More, J. & Bulmer, M. Human Serum Albumin: A Multifunctional Plasma Protein. In Production of Plasma Proteins for Therapeutic Use 159–183 (Wiley, 2012). https://doi.org/10.1002/9781118356807.ch12.
Schuhmacher, M., Glocker, M. O., Wunderlin, M. & Przybylski, M. Direct isolation of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analysis by electrospray-ionization mass spectrometry. Electrophoresis 17, 848–854 (1996).
Fanali, G. et al. Human serum albumin: From bench to bedside. Mol. Aspects Med. 33, 209–290 (2012).
Rath, A., Glibowicka, M., Nadeau, V. G., Chen, G. & Deber, C. M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. PNAS 106, 1760–1765 (2008).
Pitt-Rivers, R. & Impiombato, F. S. The binding of sodium dodecyl sulphate to various proteins. Biochem. J. 109, 825–830 (1968).
Dunker, A. K. & Kenyon, A. J. Mobility of sodium dodecyl sulphate: Protein complexes. Biochem. J. 153, 191–197 (1976).
Shi, Y. et al. Abnormal SDS-PAGE migration of cytosolic proteins can identify domains and mechanisms that control surfactant binding. Protein Sci. 21, 1197–1209 (2012).
Neelofar, K. M., Ahmad, J., Arif, Z. & Alam, K. Elucidating the impact of glucosylation on human serum albumin: A multi-technique approach. Int. J. Biol. Macromol. 92, 881–891 (2016).
de Serres, F. & Blanco, I. Role of alpha-1 antitrypsin in human health and disease. J. Intern. Med. 276, 311–335 (2014).
Lechowicz, U., Rudzinski, S., Jezela-Stanek, A., Janciauskiene, S. & Chorostowska-Wynimko, J. Post-translational modifications of circulating alpha-1-antitrypsin protein. Int. J. Mol. Sci. 21, 1–18 (2020).
Perween, S., Abidi, M., Faizy, A. F. & Moinuddin,. Post-translational modifications on glycated plasma fibrinogen: A physicochemical insight. Int. J. Biol. Macromol. 126, 1201–1212 (2019).
Fu, Y. & Grieninger, G. Fib420: A normal human variant of fibrinogen with two extended α chains. Proc. Natl. Acad. Sci. USA 91, 2625–2628 (1994).
Vilar, R., Fish, R. J., Casini, A. & Neerman-Arbez, M. Fibrin(ogen) in human disease: Both friend and foe. Haematologica 105, 284–296 (2020).
Kerr, M. A. The structure and function of human IgA. Biochem. J. 271, 285–296 (1990).
Suzuki, T. et al. Relationship of the quaternary structure of human secretory IgA to neutralization of influenza virus. Proc. Natl. Acad. Sci. USA 112, 7809–7814 (2015).
Tomasz Kowalik, J. S. Salivary secretory IgA and its impact on dental caries. Nowa Stomatologia 211–214 (2013).
Perše, M. & Večeri večeri c-Haler, Ž. The role of IgA in the pathogenesis of IgA nephropathy. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20246199 (2019).
Charles A Janeway, J., Travers, P., Walport, M. & Shlomchik, M. J. The structure of a typical antibody molecule. in Immunobiology: The Immune System in Health and Disease. 5th edition. (Garland Science, 2001).
NapiórkowsKa-Baran, K. et al. Oznaczanie przeciwciał w codziennej praktyce. Część I-właściwości przeciwciał Determination of antibodies in everyday practice: Part I-properties of antibodies. Alergia Astma Immunol. 24, 51–58 (2019).
Repetto, O. et al. Quantitative proteomic approach targeted to fibrinogen β chain in tissue gastric carcinoma. Int. J. Mole. Sci. https://doi.org/10.3390/ijms19030759 (2018).
Luo, Y., Shi, J. & Li, J. Peroxynitrite induced fibrinogen site identification. Bio-Med. Mater. Eng. 26, 2241–2248 (2015).
Wei, B., Berning, K., Quan, C. & Zhang, Y. T. Glycation of antibodies: Modification, methods and potential effects on biological functions. MAbs 9, 586–594 (2017).
Ahmad, S., Moinuddin, & Ali, A. Immunological studies on glycated human IgG. Life Sci. 90, 980–987 (2012).
Islam, S. et al. Neo-epitopes generated on hydroxyl radical modified glycatedigg have role in immunopathology of diabetes type 2. PLoS ONE 12, e0169099 (2017).
Korça, E., Piskovatska, V., Börgermann, J., Navarrete Santos, A. & Simm, A. Circulating antibodies against age-modified proteins in patients with coronary atherosclerosis. Sci. Reports https://doi.org/10.1038/s41598-020-73877-5 (2020).
Newkirk, M. M. et al. Advanced glycation end-product (AGE)-damaged IgG and IgM autoantibodies to IgG-AGE in patients with early synovitis. Arth. Res. Ther. 5, 1 (2003).
Tai, A. W. H. & Newkirk, M. M. An autoantibody targeting glycated IgG is associated with elevated serum immune complexes in rheumatoid arthritis (RA). Clin. Exp. Immunol. 120, 188–193 (2000).
Ryan, B. J., Nissim, A. & Winyard, P. G. Oxidative post-translational modifications and their involvement in the pathogenesis of autoimmune diseases. Redox Biol. 2, 715–724 (2014).
Kawata, Y. et al. Detailed structure and pathophysiological roles of the IgA-albumin complex in multiple myeloma. Int. J. Mol. Sci. 22, 1–11 (2021).
Kawasaki, I. et al. Renal dysfunction worsened by superimposition of IgA glomerulonephritis in a patient with overt diabetic nephropathy [6]. Nephron 78, 232–234 (1998).
Nasr, S. H., Share, D. S., Vargas, M. T., D’Agati, V. D. & Markowitz, G. S. Acute poststaphylococcal glomerulonephritis superimposed on diabetic glomerulosclerosis. Kidney Int. 71, 1317–1321 (2007).
Kanauchi, M., Kawano, T. & Dohi, K. Serum IgA levels in patients with diabetic nephropathy and IgA nephropathy superimposed on diabetes mellitus. Diab. Res. Clin. Pract. 48, 113–118 (2000).
Perše, M. & Večeri-Večeric-Haler, Ž. The role of IgA in the Pathogenesis of IgA nephropathy. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20246199 (2019).
Matsuda, R., Anguizola, J., Joseph, K. S. & Hage, D. S. High-performance affinity chromatography and the analysis of drug interactions with modified proteins: Binding of gliclazide with glycated human serum albumin. Anal. Bioanal. Chem. 401, 2811–2819 (2011).
Mendez, D. L., Jensen, R. A., McElroy, L. A., Pena, J. M. & Esquerra, R. M. The effect of non-enzymatic glycation on the unfolding of human serum albumin. Arch. Biochem. Biophys. 444, 92–99 (2005).
Arif, B. et al. Structural and immunological characterization of Amadori-rich human serum albumin: Role in diabetes mellitus. Arch. Biochem. Biophys. 522, 17–25 (2012).
Neelofar, K. & Ahmad, J. An overview of in vitro and in vivo glycation of albumin: A potential disease marker in diabetes mellitus. Glycoconj. J. 34, 575–584 (2017).
Paradela-Dobarro, B. et al. Inflammatory effects of in vivo glycated albumin from cardiovascular patients. Biomed. Pharmacother. 113, 108763 (2019).
Acknowledgements
We would like to thank dr Agnieszka Bronowicka-Szydełko for providing the samples of serum and Marzena Ciesielska for collecting blood plasma. The study was supported by a doctoral scholarship from the Polish National Science Centre ETIUDA 7 Grant No. 2019/32/T/NZ4/00492 for K.G.P and the OPUS 13 Grant No. 2017/25/B/NZ4/01198 for M.S. The equipment used for LC-MS/MS analysis was sponsored in part by the Centre for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland.
Author information
Authors and Affiliations
Contributions
M.S., K.G-P. Conceived the experiments; K.G-P., I.J., K.O. conducted the experiments; K.R-P., K.G., J.T-W. collected human samples; M.S, K.G-P., J.T-W., A.G. analyzed the results; M.S., K.G-P. wrote the manuscript; M.S, K.G-P., A.G., K.R-P. edited the final version of manuscript; All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gostomska-Pampuch, K., Gamian, A., Rawicz-Pruszyński, K. et al. Proteins in human body fluids contain in vivo antigen analog of the melibiose-derived glycation product: MAGE. Sci Rep 12, 7520 (2022). https://doi.org/10.1038/s41598-022-11638-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-11638-2
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.