Abstract
In this study, a novel deep learning-based methodology was investigated to predict breast cancer response to neo-adjuvant chemotherapy (NAC) using the quantitative ultrasound (QUS) multi-parametric imaging at pre-treatment. QUS multi-parametric images of breast tumors were generated using the data acquired from 181 patients diagnosed with locally advanced breast cancer and planned for NAC followed by surgery. The ground truth response to NAC was identified for each patient after the surgery using the standard clinical and pathological criteria. Two deep convolutional neural network (DCNN) architectures including the residual network and residual attention network (RAN) were explored for extracting optimal feature maps from the parametric images, with a fully connected network for response prediction. In different experiments, the features maps were derived from the tumor core only, as well as the core and its margin. Evaluation results on an independent test set demonstrate that the developed model with the RAN architecture to extract feature maps from the expanded parametric images of the tumor core and margin had the best performance in response prediction with an accuracy of 88% and an area under the receiver operating characteristic curve of 0.86. Ten-year survival analyses indicate statistically significant differences between the survival of the responders and non-responders identified based on the model prediction at pre-treatment and the standard criteria at post-treatment. The results of this study demonstrate the promising capability of DCNNs with attention mechanisms in predicting breast cancer response to NAC prior to the start of treatment using QUS multi-parametric images.
Similar content being viewed by others
Introduction
Breast cancer is the most common cancer type and the foremost cause of cancer-related death among women1,2. In 2020, around 2.3 million new cases of breast cancer were diagnosed worldwide and it caused around 0.7 million deaths among females1. Locally advanced breast cancer (LABC) is an aggressive subtype of breast cancer that includes up to 20% of new cases each year3. LABC is often identified with tumors greater than 5 cm in size and possibly with skin and/or chest wall involvement. Moreover, LABC includes patients diagnosed with inflammatory breast cancer or multiple positive axillary lymph nodes3,4.
Patients diagnosed with LABC suffer from high risk of relapse and metastasis with a local recurrence rate of about 48% in 5 years5. With availability of different systemic and targeted regimens, neoadjuvant chemotherapy (NAC) followed by surgery is currently considered as the standard treatment for LABC patients6,7,8,9. In some cases, the surgery is followed by adjuvant radiation and/or hormonal therapies to reduce the risk of cancer recurrence4,6. Although response to NAC has demonstrated a high correlation to the patient survival, complete pathological response is limited to less than 30% of the patients, with about 30% of the patient do not even partially respond to NAC3,4,6,10,11,12,13,14. To determine the tumor pathological response to NAC, post-surgical histopathology is considered as the standard approach6,7,8,9. However, post-surgical evaluations cannot be used for adjusting the NAC or switching to salvage treatment.
Currently, monitoring tumor response to NAC mostly relies on physical examination or standard anatomical imaging to assess the changes in tumor size. The main limitation of these methods is that detectable changes in tumor dimensions usually become apparent after several months of therapy, and in some cases a measurable change may not become evident on imaging despite a pathological response to NAC15. Early prediction of tumor response to NAC can permit therapy adjustments by modifying the regimen, dose, and/or sequence of treatment options, switching to more effective treatments or even salvage therapies before it is potentially too late for individual patients16,17. A personalized strategy for LABC treatment is anticipated to improve tumor response to neoadjuvant therapies, spare patients from unnecessary side effects of ineffective treatment, and improve their overall survival and quality of life.
Ultrasound is a portable, rapid and cost-efficient imaging modality that can be applied to characterize tissue physical properties without injection of any exogenous contrast agents. In particular, quantitative ultrasound (QUS) techniques have been introduced to derive quantitative measures of tissue biophysical properties that are independent of instrument settings, with a lower level of dependence to the operator18. Quantitative ultrasound spectral analysis techniques examine the frequency dependence of the ultrasound radiofrequency (RF) signal backscattered from the underlying tissue which can be used for tissue micro-structure characterization18. The QUS parameters derived from the analysis of normalized power spectrum of RF signal including the mid-band fit (MBF), spectral slope (SS), spectral 0-MHz intercept (SI), effective scatterer diameter (ESD) and effective acoustic concentration (EAC) have shown promises in detecting and characterizing malignancies, examination of liver tissues and detecting cardiovascular disease19,20,21,22,23,24,25.
It has been shown that QUS spectral parameters can detect tumor cell death induced by various anti-cancer-therapies26,27,28,29. Also, several studies have demonstrated that the hand-crafted features derived from the QUS parametric maps can be used to predict and monitor breast cancer response to neoadjuvant chemotherapy before or within weeks after the start of treatment, with a high correlations to clinical and pathological response identified at the end of treatment30,31,32,33. For example, it has been demonstrated that textural features of QUS spectral parametric maps have higher correlations to histological tumor cell death in response to chemotherapy in comparison with QUS mean-value parameters34. Further, A few studies have revealed the potential of the textural features of QUS parametric images in predicting LABC tumor response to NAC as early as 1 week after starting the treatment35,36,37. In a recent study, Tadayyon et al. have demonstrated that using the QUS hand-crafted features derived from both the tumor core and its margin could improve the performance of tumor response prediction before starting the treatment38.
The deep learning approaches have recently been investigated in different applications of medical image analysis39,40. Such methodologies can potentially remove the process of extracting carefully designed hand-crafted features from images required for conventional machine learning techniques. Instead, the deep learning frameworks optimize their data-driven feature maps during the iterative training procedure41. In this context, a few studies have been conducted on adapting deep convolutional neural networks (DCNN) for NAC response prediction in breast cancer patients using magnetic resonance imaging (MRI)42,43,44. Moreover, few studies have explored the potential of DCNNs in analyzing ultrasound images of breast tumors for cancer classification. For example, Byra et al. have demonstrated the potential of convolutional neural networks for breast lesion classification using Nakagami parametric images45. To our knowledge, no previous study has explored the efficacy of deep learning techniques with QUS multi-parametric images for therapy response prediction.
In this study, the effectiveness of DCNN methodologies on QUS spectral multi-parametric images has been investigated to predict LABC response to NAC before the start of treatment. The QUS spectral parametric images were generated using the ultrasound data acquired from 181 LABC patients at pre-treatment. The patient responses to NAC were identified after their surgery using the standard clinical and pathological criteria, and used as the ground truth to evaluate the performance of the prediction models. The dataset was randomly partitioned into a training set and an independent test set. Different DCNN architectures including RAN46, and ResNet47 were investigated for feature extraction in the developed framework. In a set of experiments, the feature maps were extracted from the tumor core and the core and its margin. After averaging the features on different tumor cross-sections, a fully connected network was utilized for response prediction. The results demonstrate that the developed model with the RAN architecture for extracting feature maps from the expanded parametric images of the tumor core with margin had superior performance with an AUC of 0.86 on the independent test set. The Kaplan–Meier survival analyses show that the patients predicted as the responders using this model demonstrated a significantly better survival rate in comparison with those predicted as non-responders.
Materials and methods
Study protocol
This study was conducted following the guidelines and regulations in accordance with institutional research ethics board approval from Sunnybrook Health Sciences Centre (SHSC), Toronto Canada. The study was open to all women aged 18–85, diagnosed with LABC and planned for NAC followed by surgery. After obtaining written informed consent, 181 eligible patients were recruited for the study. A core needle biopsy was done for each patient to confirm the cancer diagnosis and grade the tumor. The initial tumor size was determined for each patient using the magnetic resonance (MR) images of the affected breast. Ultrasound B-mode images and radiofrequency (RF) data were acquired from the patients (in supine position with arms above the head) before the start of NAC, following a standardized protocol. Three experienced sonographers were responsible for ultrasound data acquisition. For NAC, 62.9% of the patients received doxorubicin, cyclophosphamide followed by paclitaxel/docetaxel (AC-T/D), 32.6% were treated with 5-fluorouracil, epirubicin, cyclophosphamide followed by docetaxel (FEC-D), and 4.5% with paclitaxel and cyclophosphamide (TC). The patients with HER2+ tumors also received tratuzumab. Patients were followed up to 10 years after their treatment and their clinical data were recorded for survival analysis. Out of the 181 patients, about 30% (n = 50) were randomly selected through a stratified random sampling and kept unseen as an independent test set, and the remaining patients (n = 131) were considered as the training set and used to develop and optimize the predictive models.
Clinical and pathological response evaluation
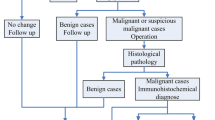
In keeping with the institutional guidelines, all patients had breast surgery after completing their neoadjuvant chemotherapy. Before surgery, the residual tumor size was determined using MRI. Standard histopathology was performed on the surgical specimens to assess the pathological response of tumor to NAC. The specimens were stained with hematoxylin and eosin (H&E) and prepared when possible on whole-mount 5″ × 7″ pathology slides. The mounted slides were digitized using a confocal scanner (TISSUEscope, Huron Technologies, Waterloo, ON). A board-certified pathologist who remained blinded to the study results examined all pathology samples. A modified response (MR) grading system based on response evaluation criteria in solid tumors (RECIST)48 and histopathological criteria38,49 was used to categorize the patients into two groups of responders and non-responders, as before50. In the MR grading system, the MR score is defined as follows: MR 1: no reduction in tumor size; MR 2: less than 30% reduction in tumor size; MR 3: between 30 and 90% reduction in tumor size or a very low residual tumor cellularity determined histopathologically; MR 4: more than 90% reduction in tumor; MR 5: no evident tumor and no malignant cells identifiable in sections from the site of the tumor (ductal carcinoma in situ may be present). In this study, patients with a MR score of 1–2 (less than 30% reduction in tumor size) were considered as non-responders, and patients with a MR score 3–5 (more than 30% reduction in tumor or very low residual tumor cellularity) were determined as responders. In keeping with this, 138 and 43 patients were identified as responders and non-responders, respectively.
Ultrasound data acquisition
Ultrasound RF data were acquired utilizing an RF-enabled Sonix RP system (Ultrasonix, Vancouver, Canada) and an L14-5/60 transducer. The transducer operated at the center frequency of ~ 6 MHz with a − 6 dB bandwidth of 3–8 MHz. The RF data were acquired with a sampling frequency of 40 MHz and digitized with a 16-bit resolution. For each tumor, the ultrasound data were acquired at four to seven image planes across the breast with approximately 1 cm intervals. The focal depth was set at the center of the tumor depending on the individual patient circumstances. The breast region for ultrasound scanning was specified by an oncologist who determined the acquisition scan planes via a physical examination of the patient. The image size along the lateral and axial directions was 6 cm and 4–6 cm, respectively.
QUS parametric map generation
For generating the parametric images, QUS spectral analyses were performed in conjunction with a sliding window analysis (described below) to derive MBF, SI, ESD, and EAC parameters22,23. The mean power spectrum was obtained by averaging over the Fourier transform of the Hanning-gated RF data calculated for every scan line of the analyzed window. The average power spectrum was normalized using a reference phantom method to remove the effects of the system transfer function and transducer beam-forming51,52. The reference phantom was composed of 5–30 μm diameter glass beads embedded in a homogeneous background of microscopic oil droplets in gelatin (Medical Physics Department, University of Wisconsin, USA). The reference phantom had an attenuation coefficient of 0.576 dB/MHz.cm and a speed of sound parameter of 1488 m/s. The MBF and SI parameters were estimated using a linear regression analysis within the − 6 dB bandwidth of the transducer22,53,54. The ESD and EAC parameters were derived by fitting a spherical Gaussian form factor model to the estimated backscatter coefficient55,56.
To generate the QUS parametric maps for each tumor, the tumor core was manually outlined by trained staff under the supervision of expert oncologists on each scan plane using the associated B-mode image. The tumor margin contour was automatically generated with a thickness of 5 mm around the core, based on the observations of a previous study38. The parametric maps were generated for all imaging planes of the tumor using a sliding window analysis throughout the entire region of interest (tumor core and margin) with windows of size 2 mm × 2 mm and 95% overlap in both lateral and axial direction, where the calculated parameter for each window was assigned to its center. The sliding window size was selected such that it covers sufficient ultrasound wavelengths in the axial direction for spectral analysis while preserving texture in generated parametric maps, with an overlap size to obtain isotropic pixels57,58.
Deep learning model
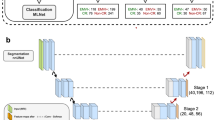
The scheme of the deep learning framework developed in this study for response prediction is shown in Fig. 1. The framework consists of two cascaded networks. The first network is a DCNN, with several convolutional layers as its backbone, adapted to extract the optimal feature maps from the QUS parametric images and is called the feature network in this paper. Two main architectures including a modified residual network version 101 (ResNet)47 and a modified residual attention network version 56 (RAN)46, were investigated in this study as the backbone of the feature network. Figure 1 demonstrates the fully connected layers after the convolutional layers (backbone) in the feature network that are applied in training this network on single parametric images to extract the optimal feature maps for response prediction. The figure also depicts the adapted ResNet and RAN architectures with their residual and attention modules. In the residual module applied in the ResNet architecture, the convolutional layers could be skipped through the identity branch. This strategy permits a more efficient training of very deep networks such as ResNet to achieve an improved performance on unseen samples. In the attention module applied in the RAN architecture, the trunk branch determines the information that can be passed through the network, whereas the mask branch determines the amount of information from the trunk branch that should be passed. Therefore, the module is able to pass the important information with higher weights and reduce the effect of less-important information in the network’s output.
As shown in the Fig. 1, the optimized feature maps obtained from the feature network are averaged over all parametric images associated with each tumor, and subsequently used in a second fully connected network (predictive network) adapted for response prediction at the patient level (described further below). The predictive network consists of two fully-connected layers with an input layer with the same size as the flatten feature vector (256), a middle layer with 100 neurons, and a softmax layer at the end with an output size of two to predict the probability of the response categories (responder vs. non-responder) for each patient. Drop-out layers have been added after the first and second layer of this network to avoid overfitting and enhance its generalization performance.
Preprocessing and model training
Before training the models, the parametric images were preprocessed and adjusted for the convolutional model. About 25% (31 patients) of the training set was randomly selected as a validation set to optimize the network hyperparameters. The parametric images were resampled to the size of 512 × 512 pixel. Then the pixel values in the parametric images of the training set were normalized to (0 1) to facilitate the training convergence. The training set normalization parameters were used for normalizing the validation and test sets. In order to improve the network training and alleviate the problem of having a relatively small training dataset, data augmentation was applied on the training set. For augmenting the training data, flipping horizontally and shifting both horizontally and vertically (maximum shift: 30% of image size) were stochastically applied.
In the first step of model training, the feature network was trained to generate the optimal feature maps for single QUS parametric images. For training the feature network, parametric images of the training set were fed into the network while different imaging planes of each tumor were considered as independent inputs with the tumor response as their output. The optimal feature maps for each imaging plane were acquired by feeding its corresponding parametric images into the trained feature network. For each patient, the optimal feature maps were calculated for all 2D imaging planes of the tumor, flatten to a 1D vector, and subsequently averaged over the entire tumor volume to obtain an averaged feature vector with size of 256 × 1 that was used in the predictive network. This strategy was applied to standardize the input size of the networks for different tumors with various sizes and, consequently, different number of QUS parametric images. The predictive network was trained using the averaged feature vectors associated with the patients in the training set, and evaluated over the independent test set for response prediction. For training the networks, the cross entropy was used as the loss function, with a cost weight ratio of C:1 (C ≥ 1; optimized as described below) for non-responders to responders to account for the unbalance in the dataset. The network hyperparameters including the dropout rate (range 0.3–0.7), width of the hidden fully connected layers (range 50–300), learning rate (range 0.1–0.00001), cost weight (range 1 ≤ C ≤ 10), and batch size (range 4–16) were optimized using the validation set. Preliminary experiments were conducted using the validation set to select the network training optimizer among the Adam and stochastic gradient descent (SGD) methods, where the Adam optimizer was selected and applied59. The optimal hyperparameters for training the models were as follows: dropout rate = 0.5, learning rate = 0.0001, cost weight = 5, batch size = 8. Early stopping was used to avoid overfitting by monitoring the network’s performance on the validation set during the training process.
Response prediction and risk assessment
In different experiments, the QUS multi-parametric images (MBF, SI, ESD, and EAC) of the tumor core, as well as the core and its margin were investigated, as the inputs to the DCNN framework and their performance were compared in response prediction. The deep learning models with different feature networks were trained and optimized using the training set. The performance of the optimized models was evaluated on the independent test set using the accuracy, sensitivity, specificity, and the ROC analysis. In this study, sensitivity refers to the ratio of the non-responses that were predicted as non-responder, and specificity refers to the ratio of the responding patients correctly predicted as responder by the model. A prediction difference analysis (PDA) was performed to visualize the importance of different regions of the input QUS parametric images to the network’s decision60. In each iteration of the modified PDA procedure applied in this study, a small patch (8 × 8 pixel with 50% overlap between adjacent patches) of one of the input parametric images was occluded (pixel values were set to zero). The absolute change in the model’s prediction (output probability) was then calculated compared to the case of inputting the original parametric images, and considered as the impact of the occluded patch on the network’s decision. The PDA maps were generated for each input parametric image by sliding the occluding patch over the image and assigning the estimated impact to its center.
The efficacy of the developed predictive models in differentiating the LABC patients with different recurrence-free survival was assessed through Kaplan–Meier survival analysis. The survival curves were generated for the responders and non-responders identified based on each model’s prediction at pre-treatment, and at post-treatment based on the clinical and histopathological criteria. A long-rank test was applied to assess for statistically significant differences between the survival curves of the two response cohorts obtained in each experiment.
Results
Table 1 presents the clinical and histopathological characteristics of the participating patients. The patients had an average initial tumor size of 5.2 cm, and an average residual tumor size of 2.5 cm at the end of treatment. Using the MR grading system, 76.2% and 23.8% of the patients were identified as responders and non-responder, respectively, at the end of treatment.
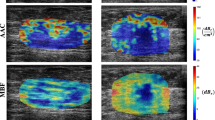
Figure 2 demonstrates QUS parametric maps of MBF, SI, ESD, and EAC overlaid on the ultrasound B-mode images obtained from representative responding and non-responding patients, respectively. As observed in these representative images, the QUS parametric maps associated with the responding and non-responding patients demonstrated different mean and spatial pattern of pixel values within the tumor core and margin. The figure also shows the PDA maps associated with these parametric images, visualizing the relative impact of different regions in each image to the network’s decision for response prediction. Figure 3 demonstrates H&E stained histopathology images of the surgical specimens acquired from representative responding and non-responding patients. In responding patients, minimal tumor cellularity usually remained within the tumor bed after chemotherapy, as evident in the histopathology slides. In contrast, histopathology images of the non-responding patients typically indicated large areas of residual disease with minimal chemotherapy effects.
Ultrasound B-mode images (A), and parametric overlays of MBF (B), SI (C), ESD (D), and EAC (E) on B-mode images acquired at pre-treatment from a representative responder and non-responder to NAC, and the associated PDA maps visualizing the level of impact of different regions in each parametric image on the network’s decision (model 4 in Table 2). The tumor core has been outlined with white dashed line.
Table 2 presents the results of response prediction in different experiments on the validation and independent test sets. The ROC curves associated with the validation and test sets for different predictive models are demonstrated in Fig. 4. Using the ResNet architecture as the model’s backbone to extract feature maps from the parametric images of the tumor core resulted in an AUC of 0.77 on the independent test set. Extending the input parametric images to include both the tumor core and its margin improved the AUC of this model to 0. 83.
ROC curves generated for responding and non-responding patients in the validation set and independent test set identified at pre-treatment using the predictive models 1–4 in Table 2 (A–D).
Applying the extracted features from the parametric images of the tumor core using the RAN architecture resulted in an accuracy of 80%, and an AUC of 0.82 on the independent test set. Similar to the model with the ResNet architecture as the feature extractor, the overall performance of this model improved by extending the input parametric images to include the tumor margin. In particular, this model resulted in the best prediction performance with an accuracy and AUC of 88%, and 0.86, respectively, on the independent test set. All models demonstrated a relatively similar performance on the validation and test sets, implying a good generalizability of the trained models on never seen samples.
Figure 5 presents the 10-year recurrence-free survival curves for the responders and non-responders identified based on the clinical and histological criteria at post treatment, and at pre-treatment using the four predictive models presented in Table 2. The survival analysis demonstrated a statistically significant difference (p value = 0.030) between the survival curves of the responders and non-responders identified at post-treatment. Among the response cohorts predicted at pretreatment, the ones identified using the predictive models with the RAN as their feature network demonstrated a statistically significant difference or approaching significance. Specifically, whereas the model that input the parametric images of the tumor core approached a significant difference (p value = 0.058), the one with the input parametric images extended to the tumor margin demonstrated a statistically significant difference between the survival of the two predicted cohorts (p value = 0.040). The response cohorts identified by the other two models at pre-treatment did not show a significant difference in survival.
Recurrence-free survival curves for the two patient cohorts in the independent test set. The responders and non-responders were identified at post treatment based on the clinical and histopathological criteria (A), and at pre-treatment using the predictive models 1–4 in Table 2 (B–E).
Discussion and conclusion
In this study, a deep learning methodology was proposed and investigated for predicting breast tumor response to NAC using the QUS parametric images acquired at pre-treatment. In the proposed framework, a DCNN architecture is utilized to extract optimal feature maps from QUS multi-parametric images. The flatten feature vectors associated with different cross-sections of a tumor are averaged over the entire tumor volume. A fully-connected network is subsequently applied for response prediction, using the average feature vectors. The performance of the framework was investigated in a set of experiments using two different DCNN architectures including the ResNet and RAN as the backbone feature extractor, and the QUS parametric images of the tumor core, and the tumor core with margin as the input. The best performance in response prediction was achieved with the RAN architecture and the extended parametric images of the tumor core and margin, with an AUC of 0.86 on the unseen test set. Long-term survival analyses demonstrated statistically significant differences between the survival of the responders and non-responders identified at pre-treatment based on this model’s prediction, and at post-treatment based on the standard clinical and pathological criteria.
A number of previous studies have investigated the potential of QUS multi-parametric imaging in predicting and monitoring response of breast cancer tumors to NAC38,61. In those studies, handcrafted features such as the statistical and textural features of the QUS parametric maps were analyzed and applied for response prediction using traditional machine learning methods. With predefined hand-crafted features, the feature extraction process is easier to understand and analyze. But these features only carry limited information based on their definition. Further, carefully determining and defining optimal discriminatory features for each specific application is crucial in this approach but is not always feasible. In this study, we investigated the potential of DCNN architectures for extracting optimal features from QUS multi-parametric images automatically through multiple convolutional layers. The advantage of using deep learning model for feature extraction is that the model learns during the training process to extract the important information while considering the whole image. As the PDA maps generated for the QUS parametric images implied, different regions of these parametric maps contribute substantially to the model’s prediction, albeit with varying levels.
It has been shown that intratumor heterogeneity is an important factor in responsiveness of tumors to cancer therapy, with several studies demonstrated the value of imaging-based quantitative features of tumor heterogeneity for response prediction23,62,63,64,65. The results obtained in the study here using the DCNN feature extractors are in agreement with the observations of those studies. Specifically, the features maps extracted from the QUS multi-parametric images are computed using multiple layers of convolutional filters throughout a DCNN architecture and can quantify the spatial heterogeneity within multi-channels of the QUS images simultaneously. The results of different experiments performed in this study demonstrated that extending the input QUS parametric images to include the tumor margin in addition to the core enhances the performance of the model in response prediction. Further, whereas the PDA maps demonstrated considerable contribution of various regions of the QUS parametric images to the network’s decision, the impact of the margin areas on the model’s prediction was highlighted particularly in the MBF and EAC parametric images. These results are in agreement with observations of the previous studies in which the imaging-based characteristics of tumor margin were shown important in different diagnostic and prognostic applications38,65,66,67.
The RAN architecture demonstrated a better performance compared to the ResNet in deriving optimal feature maps from QUS parametric images for response prediction. Both architectures use residual modules to overcome the overfitting throughout their very deep layers. The RAN architecture also utilizes attention modules that potentially facilitate feature optimization. Specifically, the attention module enables the network to focus on significant and influential regions of images while extracting the feature maps. The observations of this study imply that the attention modules in the RAN architecture have been successful in identifying the important regions of the QUS parametric maps for response prediction.
A potential limitation associated with this study is the relatively small dataset that was available to train and optimize the models. Training large DCNN models on small datasets may result in overfitting and lack of generalizability. Effective approaches including network regularization through drop-out layers and early training stopping was applied in this study to reduce the chance of model overfitting and improve its generalizability. An independent unseen test set was applied to evaluate the model’s performance, where the results demonstrated a good generalizability on never seen samples. Availability of larger datasets in future would permit more comprehensive training of the models and more rigorous evaluation of their prediction performance for new patients. In terms of NAC, the majority of the patients in this study (95.5%) were treated with either AC-T/D or FEC-D. The performance of the response prediction models in this study was relatively similar for the patients treated with different NAC regimens with the number of incorrect predictions proportional to the number of patients receiving each regimen. Nevertheless, response to different NAC regimens may be different for a patient. Future stratified analysis on larger datasets with separate models for different NAC regimens and possibly various molecular subtypes may result in more robust predictive models. Such models can also facilitate future randomize clinical trials to investigate the adjustment of NAC regimen for patients with low likelihood of response to the standard treatment. In conclusion, this study demonstrated that DCNN models can be adapted in the context of quantitative imaging for therapy response prediction. The results indicated a better performance of the attention-guided convolutional networks in deriving optimal quantitative features form QUS multi-parametric images. The deep learning models developed in this study could predict the survival-linked response of LABC patients to NAC before starting the treatment with a high accuracy. Predicting the outcome of NAC at pre-treatment would help the clinicians to adjust ineffective treatment regimens for individual patients. The results obtained in this study are promising and encourage further investigations using other DCNN architectures with the data acquired from larger (multi-institutional) cohorts of patients to evaluate the robustness of the methodologies in clinic.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. https://doi.org/10.3322/caac.21660 (2021).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68(6), 394–424. https://doi.org/10.3322/caac.21492 (2018).
Giordano, S. H. Update on locally advanced breast cancer. Oncologist 8(6), 521–530. https://doi.org/10.1634/theoncologist.8-6-521 (2003).
Hortobagyi, G. N. Comprehensive management of locally advanced breast cancer. Cancer 66(6 Suppl), 1387–1391. https://doi.org/10.1002/1097-0142(19900915)66:14+%3c1387::aid-cncr2820661414%3e3.0.co;2-i (1990).
Gardin, G. et al. Locally advanced non-metastatic breast cancer: Analysis of prognostic factors in 125 patients homogeneously treated with a combined modality approach. Eur. J. Cancer 31(9), 1428–1433. https://doi.org/10.1016/0959-8049(95)00199-S (1995).
Cleator, S. J., Makris, A., Ashley, S. E., Lal, R. & Powles, T. J. Good clinical response of breast cancers to neoadjuvant chemoendocrine therapy is associated with improved overall survival. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 16(2), 267–272. https://doi.org/10.1093/annonc/mdi049 (2005).
Guarneri, V. et al. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J. Clin. Oncol. 24(7), 1037–1044. https://doi.org/10.1200/JCO.2005.02.6914 (2006).
Hennessy, B. T. et al. Outcome after pathologic complete eradication of cytologically proven breast cancer axillary node metastases following primary chemotherapy. J. Clin. Oncol. 23(36), 9304–9311. https://doi.org/10.1200/JCO.2005.02.5023 (2005).
Nikas, J. B., Low, W. C. & Burgio, P. A. Prognosis of treatment response (pathological complete response) in breast cancer. Biomark. Insights 7, 59–70. https://doi.org/10.4137/BMI.S9387 (2012).
Fisher, B. et al. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J. Clin. Oncol. 16(8), 2672–2685. https://doi.org/10.1200/JCO.1998.16.8.2672 (1998).
Sethi, D. et al. Histopathologic changes following neoadjuvant chemotherapy in various malignancies. Int. J. Appl. Basic Med. Res. 2(2), 111. https://doi.org/10.4103/2229-516x.106353 (2012).
Chuthapisith, S., Eremin, J. M., El-Sheemy, M. & Eremin, O. Neoadjuvant chemotherapy in women with large and locally advanced breast cancer: Chemoresistance and prediction of response to drug therapy. Surgeon 4(4), 211–219. https://doi.org/10.1016/S1479-666X(06)80062-4 (2006).
Haque, W. et al. Response rates and pathologic complete response by breast cancer molecular subtype following neoadjuvant chemotherapy. Breast Cancer Res. Treat. 170(3), 559–567. https://doi.org/10.1007/s10549-018-4801-3 (2018).
Byrski, T. et al. Pathologic complete response rates in young women with BRCA1-positive breast cancers after neoadjuvant chemotherapy. J. Clin. Oncol. 28(3), 375–379. https://doi.org/10.1200/JCO.2008.20.7019 (2010).
Brindle, K. New approaches for imaging tumour responses to treatment. Nat. Rev. Cancer 8(2), 94–107. https://doi.org/10.1038/nrc2289 (2008).
von Minckwitz, G. et al. S3–2: Neoadjuvant chemotherapy adapted by interim response improves overall survival of primary breast cancer patients—Results of the gepartrio trial. Cancer Res. https://doi.org/10.1158/0008-5472.sabcs11-s3-2 (2011).
Von Minckwitz, G. et al. Neoadjuvant vinorelbine-capecitabine versus docetaxel-doxorubicin-cyclophosphamide in early nonresponsive breast cancer: Phase III randomized gepartrio trial. J. Natl. Cancer Inst. 100(80), 542–551. https://doi.org/10.1093/jnci/djn085 (2008).
Mamou, J. & Oelze, M. L. Quantitative Ultrasound in Soft Tissues (Springer, 2013).
Feleppa, E. J. et al. Typing of prostate tissue by ultrasonic spectrum analysis. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 43(4), 609–619. https://doi.org/10.1109/58.503779 (1996).
Yang, M., Krueger, T. M., Miller, J. G. & Holland, M. R. Characterization of anisotropic myocardial backscatter using spectral slope, intercept and midband fit parameters. Ultrason. Imaging 29(2), 122–134. https://doi.org/10.1177/016173460702900204 (2007).
Coleman, D. J. et al. A model for acoustic characterization of intraocular tumors. Investig. Ophthalmol. Vis. Sci. 26(4), 545–550 (1985).
Tadayyon, H., Sadeghi-Naini, A., Wirtzfeld, L., Wright, F. C. & Czarnota, G. Quantitative ultrasound characterization of locally advanced breast cancer by estimation of its scatterer properties. Med. Phys. https://doi.org/10.1118/1.4852875 (2014).
Tadayyon, H., Sadeghi-Naini, A. & Czarnota, G. J. Noninvasive characterization of locally advanced breast cancer using textural analysis of quantitative ultrasound parametric images. Transl. Oncol. 7(6), 759–767. https://doi.org/10.1016/j.tranon.2014.10.007 (2014).
Sadeghi-Naini, A. et al. Quantitative ultrasound spectroscopic imaging for characterization of disease extent in prostate cancer patients. Transl. Oncol. 8(1), 25–34. https://doi.org/10.1016/j.tranon.2014.11.005 (2015).
Guimond, A. et al. Quantitative ultrasonic tissue characterization as a new tool for continuous monitoring of chronic liver remodelling in mice. Liver Int. 27(6), 854–864. https://doi.org/10.1111/j.1478-3231.2007.01493.x (2007).
Banihashemi, B. et al. Ultrasound imaging of apoptosis in tumor response: Novel preclinical monitoring of photodynamic therapy effects. Cancer Res. 68(20), 8590–8596. https://doi.org/10.1158/0008-5472.CAN-08-0006 (2008).
Vlad, R. M., Brand, S., Giles, A., Kolios, M. C. & Czarnota, G. J. Quantitative ultrasound characterization of responses to radiotherapy in cancer mouse models. Clin. Cancer Res. 15(6), 2067–2075. https://doi.org/10.1158/1078-0432.CCR-08-1970 (2009).
Czarnota, G. J. et al. Tumor radiation response enhancement by acoustical stimulation of the vasculature. Proc. Natl. Acad. Sci. U.S.A. 109(30), E2033–E2041. https://doi.org/10.1073/pnas.1200053109 (2012).
Sadeghi-Naini, A. et al. Low-frequency quantitative ultrasound imaging of cell death in vivo. Med. Phys. 40(8), 082901. https://doi.org/10.1118/1.4812683 (2013).
Tadayyon, H. et al. Quantitative ultrasound assessment of breast tumor response to chemotherapy using a multi-parameter approach. Oncotarget 7(29), 45094–45111. https://doi.org/10.18632/oncotarget.8862 (2016).
Sannachi, L. et al. Non-invasive evaluation of breast cancer response to chemotherapy using quantitative ultrasonic backscatter parameters. Med. Image Anal. 20(10), 224–236. https://doi.org/10.1016/j.media.2014.11.009 (2015).
Sadeghi-Naini, A. et al. Quantitative ultrasound evaluation of tumor cell death response in locally advanced breast cancer patients receiving chemotherapy. Clin. Cancer Res. 19(8), 2163–2173. https://doi.org/10.1158/1078-0432.CCR-12-2965 (2013).
DiCenzo, D. et al. Quantitative ultrasound radiomics in predicting response to neoadjuvant chemotherapy in patients with locally advanced breast cancer: Results from multi-institutional study. Cancer Med. 9(16), 5798–5806. https://doi.org/10.1002/cam4.3255 (2020).
Sadeghi-Naini, A. et al. Conventional frequency ultrasonic biomarkers of cancer treatment response in vivo. Transl. Oncol. 6(3), 234–243. https://doi.org/10.1593/tlo.12385 (2013).
Sadeghi-Naini, A. et al. Chemotherapy-response monitoring of breast cancer patients using quantitative ultrasound-based intra-tumour heterogeneities. Sci. Rep. https://doi.org/10.1038/s41598-017-09678-0 (2017).
Sadeghi-Naini, A. et al. Early prediction of therapy responses and outcomes in breast cancer patients using quantitative ultrasound spectral texture. Oncotarget 5(11), 3497–3511. https://doi.org/10.18632/oncotarget.1950 (2014).
Dasgupta, A. et al. Quantitative ultrasound radiomics using texture derivatives in prediction of treatment response to neo-adjuvant chemotherapy for locally advanced breast cancer. Oncotarget 11(420), 3782–3792. https://doi.org/10.18632/oncotarget.27742 (2020).
Tadayyon, H. et al. A priori prediction of neoadjuvant chemotherapy response and survival in breast cancer patients using quantitative ultrasound. Sci. Rep. https://doi.org/10.1038/srep45733 (2017).
Shen, D., Wu, G. & Suk, H.-I. Deep Learning in Medical Image Analysis (Elsevier, 2017).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88. https://doi.org/10.1016/j.media.2017.07.005 (2017).
Goodfellow, I., Bengio, Y., Courville, A. & Bengio, Y. Deep learning. Healthc. Inform. Res. https://doi.org/10.4258/hir.2016.22.4.351 (2016).
Ravichandran, K., Braman, N., Janowczyk, A. & Madabhushi, A. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE-MRI. In Medical Imaging 2018: Computer-Aided Diagnosis Vol. 10575 (ed. Anu, A.) 11 (Springer, 2018). https://doi.org/10.1117/12.2294056.
Qu, Y. et al. Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (DL) method. Thorac. Cancer 11(3), 651–658. https://doi.org/10.1111/1759-7714.13309 (2020).
Ha, R. et al. Prior to initiation of chemotherapy, Can we predict breast tumor response? Deep learning convolutional neural networks approach using a breast MRI tumor dataset. J. Digit. Imaging 32(5), 693–701. https://doi.org/10.1007/s10278-018-0144-1 (2019).
Byra, M., Piotrzkowska-Wroblewska, H., Dobruch-Sobczak, K. & Nowicki, A. Combining Nakagami imaging and convolutional neural network for breast lesion classification. In IEEE International Ultrasonics Symposium, IUS (ed. Anu, A.) 1–4 (Springer, 2017). https://doi.org/10.1109/ULTSYM.2017.8092154.
Wang, F. et al. Residual attention network for image classification. In Proceedings of the 30th IEEE Conference on Computer Vision Pattern Recognition, CVPR 2017 (ed. Anu, A.) 6450–6458 (Springer, 2017).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Vol. 2016 (ed. Anu, A.) 770–778 (Springer, 2016). https://doi.org/10.1109/CVPR.2016.90.
Eisenhauer, E. A. A. et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 14865, 228–247. https://doi.org/10.1016/j.ejca.2008.10.026 (2009).
Ogston, K. N. et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: Prognostic significance and survival. Breast 12(5), 320–327. https://doi.org/10.1016/S0960-9776(03)00106-1 (2003).
Taleghamar, H., Moghadas-Dastjerdi, H., Czarnota, G. J. & Sadeghi-Naini, A. Characterizing intra-tumor regions on quantitative ultrasound parametric images to predict breast cancer response to chemotherapy at pre-treatment. Sci. Rep. https://doi.org/10.1038/s41598-021-94004-y (2021).
Labyed, Y., Bigelow, T. A. & McFarlin, B. L. Estimate of the attenuation coefficient using a clinical array transducer for the detection of cervical ripening in human pregnancy. Ultrasonics 51(1), 34–39. https://doi.org/10.1016/j.ultras.2010.05.005 (2011).
Yao, L. X., Zagzebski, J. A. & Madsen, E. L. Backscatter coefficient measurements using a reference phantom to extract depth-dependent instrumentation factors. Ultrason. Imaging 12(10), 58–70. https://doi.org/10.1016/0161-7346(90)90221-i (1990).
Lizzi, F. L. et al. Comparison of theoretical scattering results and ultrasonic data from clinical liver examinations. Ultrasound Med. Biol. 14(5), 377–385 (1988).
Insana, M. F., Wagner, R. F., Brown, D. G. & Hall, T. J. Describing small-scale structure in random media using pulse-echo ultrasound. J. Acoust. Soc. Am. 87(1), 179–192 (1990).
Oelze, M. L., O’Brien, W. D., Blue, J. P. & Zachary, J. F. Differentiation and characterization of rat mammary fibroadenomas and 4T1 mouse carcinomas using quantitative ultrasound imaging. IEEE Trans. Med. Imaging 23(6), 764–771. https://doi.org/10.1109/TMI.2004.826953 (2004).
Insana, M. F. & Hall, T. J. Parametric ultrasound imaging from backscatter coefficient measurements: Image formation and interpretation. Ultrason. Imaging 12(4), 245–267 (1990).
Sadeghi-Naini, A. et al. Breast-lesion characterization using textural features of quantitative ultrasound parametric maps. Sci. Rep. https://doi.org/10.1038/S41598-017-13977-X (2017).
Topp, K. A., Zachary, J. F. & O’Brien, J. Quantifying B-mode images of in vivo rat mammary tumors by the frequency dependence of backscatter. J. Ultrasound Med. 20(6), 605–612. https://doi.org/10.7863/JUM.2001.20.6.605 (2001).
Kingma, D. P. & Ba, J. L. Adam: A method for stochastic optimization. In 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings (ed. Anu, A.) (Springer, 2015).
Zintgraf, L. M., Cohen, T. S., Adel, T. & Welling, M. Visualizing deep neural network decisions: Prediction difference analysis. In 5th International Conference on Learning Representations ICLR 2017—Conference on Track Proceedings (ed. Anu, A.) (Springer, 2017).
Sannachi, L. et al. Response monitoring of breast cancer patients receiving neoadjuvant chemotherapy using quantitative ultrasound, texture, and molecular features. PLoS ONE 13(10), e0189634. https://doi.org/10.1371/journal.pone.0189634 (2018).
Byra, M., Nowicki, A., Wróblewska-Piotrzkowska, H. & Dobruch-Sobczak, K. Classification of breast lesions using segmented quantitative ultrasound maps of homodyned K distribution parameters. Med. Phys. 43(10), 5561–5569. https://doi.org/10.1118/1.4962928 (2016).
Wu, J., Gong, G., Cui, Y. & Li, R. Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J. Magn. Reson. Imaging 44(5), 1107–1115. https://doi.org/10.1002/jmri.25279 (2016).
Sadeghi-Naini, A. et al. Early detection of chemotherapy-refractory patients by monitoring textural alterations in diffuse optical spectroscopic images. Med. Phys. 42(11), 6130–6146. https://doi.org/10.1118/1.4931603 (2015).
Karami, E. et al. Quantitative MRI biomarkers of stereotactic radiotherapy outcome in brain metastasis. Sci. Rep. https://doi.org/10.1038/s41598-019-56185-5 (2019).
Lamb, P. M., Perry, N. M., Vinnicombe, S. J. & Wells, C. A. Correlation between ultrasound characteristics, mammographic findings and histological grade in patients with invasive ductal carcinoma of the breast. Clin. Radiol. 55(1), 40–44. https://doi.org/10.1053/crad.1999.0333 (2000).
Fowler, A. M., Mankoff, D. A. & Joe, B. N. Imaging neoadjuvant therapy response in breast cancer. Radiology 285(2), 358–375. https://doi.org/10.1148/radiol.2017170180 (2017).
Acknowledgements
This Research was supported by Natural Sciences and Engineering Research Council (NSERC) of Canada, Canadian Institutes for Health Research (CIHR), and Terry Fox Foundation through a New Frontiers Program Project Grant with Funds from the Lotte and John Hecht Memorial Foundation. A.S.N. holds a York Research Chair in Quantitative Imaging and Smart Biomarkers.
Author information
Authors and Affiliations
Contributions
A.S.N. and G.J.C. conceived and designed the project; A.S.N. supervised the project; H.T., S.A.J and A.S.N. developed the methodology; H.T. analyzed the data; H.T. and A.S.N. wrote and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.T., G.J.C and A.S.N. are inventors of a patent (Application No. 63/302492, pending) on deep learning of quantitative ultrasound multi-parametric images at pre-treatment to predict breast cancer response to chemotherapy.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Taleghamar, H., Jalalifar, S.A., Czarnota, G.J. et al. Deep learning of quantitative ultrasound multi-parametric images at pre-treatment to predict breast cancer response to chemotherapy. Sci Rep 12, 2244 (2022). https://doi.org/10.1038/s41598-022-06100-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-06100-2
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.