Abstract
Acinetobacter has been frequently detected in backwater areas of the Three Gorges Reservoir (TGR) region. We here employed Caenorhabditis elegans to perform biosafety assessment of Acinetobacter strains isolated from backwater area in the TGR region. Among 21 isolates and 5 reference strains of Acinetobacter, exposure to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii ATCC 19606T, A. junii NH88-14, and A. lwoffii DSM 2403T resulted in significant decrease in locomotion behavior and reduction in lifespan of Caenorhabditis elegans. In nematodes, exposure to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii and A. lwoffii also resulted in significant reactive oxygen species (ROS) production. Moreover, exposure to Acinetobacter isolates of AC1, AC15, AC18, and AC21 led to significant increase in expressions of both SOD-3::GFP and some antimicrobial genes (lys-1, spp-12, lys-7, dod-6, spp-1, dod-22, lys-8, and/or F55G11.4) in nematodes. The Acinetobacter isolates of AC1, AC15, AC18, and AC21 had different morphological, biochemical, phylogenetical, and virulence gene properties. Our results suggested that exposure risk of some Acinetobacter strains isolated from the TGR region exists for environmental organisms and human health. In addition, C. elegans is useful to assess biosafety of Acinetobacter isolates from the environment.
Similar content being viewed by others
The Three Gorges Reservoir (TGR), whose distance is approximately 662.9 km, is a major water source in China. Water fluctuation in the TGR region produces a water-level fluctuating zone (WLFZ) every year1. That is, a novel ecosystem is created by construction of the TGR Dam. Meanwhile, due to rapid development in industrialization and urbanization in the recent years, various organic and inorganic pollutants are potentially released into the environment in the TGR region through industrial or residential wastewater2,3,4,5. Mover, a large amount of bacterioplankton community, including waterborne pathogens, has been detected in the TGR region6,7,8.
Caenorhabditis elegans can be used to perform the toxicological study at the whole animal level9,10,11,12. C. elegans has become an ideal surrogate model to determine both pathogenesis and conserved mechanisms in host-microbe interactions of human pathogens13,14. More importantly, C. elegans is highly sensitive to various environmental exposures15,16,17,18. Considering the sensitivity to environmental exposure, it has been employed to perform biosafety evaluation of water samples in TGR region in both flood season and quiet season19,20. Based on our previous toxicity evaluation, only acute exposure to water sample in backwater area resulted in toxic effects on nematodes, such as decrease in locomotion behavior and activation of oxidative stress20. Moreover, both liquid phase and solid phase contributed to toxicity induction of water sample in backwater area20. In the liquid phase, the potential toxicants were suggested to be the organic pollutants20. Nevertheless, the toxicity contributors in the solid phase of water sample in backwater area in the TGR region are still largely unclear.
Environmental pathogens are an important component in the solid phase of surface water samples. Existence of high prevalence pathogens has been found in backwater areas of the TGR region7,8. Environmental Acinetobacter was realized as an important nosocomial pathogen during the late 1970s, and has received an increasing attention because of its potential to cause severe nosocomial infections and formation of multiple-drug and pan-drug resistance21,22. Acinetobacter occupies a considerable position in nature because it prevails in natural environments, such as soil, oceans, fresh water, and sediments23,24. Nevertheless, the biosafety properties of Acinetobacter in the TGR region remain largely unclear. We here aimed at performing biosafety assessment of Acinetobacter strains isolated from backwater areas in the TGR region in nematodes. Among the isolated 21 Acinetobacter strains, exposure to four isolates (AC1, AC15, AC18, and AC21) resulted in toxic effects in nematodes. Our data implied the possible exposure risk of some Acinetobacter strains in the TGR region for environmental organisms and human health.
Results
Acinetobacter isolates from the TGR region
In the TGR region, we isolated 21 Acinetobacter strains (one A. johnsonii, one A. haemolyticus and 19 Acinetobacter sp. strains) (Table S1). Based on phylogenetic analysis after 16S rRNA gene sequencing, these 21 isolates belong to the genus Acinetobacter, exhibiting a similarity of 95.38–99.93% with known Acinetobacter strains in GenBank (Table S1). In phylogenetic tree (N-J) constructed with both isolated and known Acinetobacter strains, these 21 isolates branched deeply with three Acinetobacter clusters consisting of important clinical Acinetobacter species, such as A. johnsonii H10 (FJ009371), A. junii NH88-14 (FJ447529), A. baumannii ATCC19606T (HE651907), A. lwoffii DSM2403T (X81665) and A. haemolyticus TTH04-1 (KF704077) (Fig. 1). Five reference Acinetobacter strains were selected and used21. Currently, the genus Acinetobacter comprises 68 species with validly-published names (https://apps.szu.cz/anemec/Classification.pdf, May 25, 2021). Among the named species, A. baumannii is the most studied species associated with clinical infections followed by the non-A. baumannii species A. haemolyticus, A. junii, A. johnsonii, and A. lwofii21.
Effect of different Acinetobacter strains isolated from the TGR region and reference strains on lifespan of nematodes
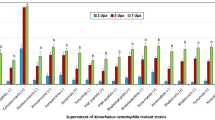
L4-larvae were exposed to different Acinetobacter strains for 24-h. Totally 21 Acinetobacter strains isolated from the TGR region and 5 reference strains of Acinetobacter species were used for the lifespan analysis. Based on the comparison of lifespan curves, exposure to Acinetobacter strains of AC2, AC3, AC4, AC5, AC6, AC7, AC8, AC9, AC10, AC11, AC12, AC13, AC14, AC16, AC17, AC19, AC20, A. johnsonii H10, and A. haemolyticus TTH0-4 could not alter lifespan curve (Fig. 2). Similarly, Acinetobacter strains of AC2, AC3, AC4, AC5, AC6, AC7, AC8, AC9, AC10, AC11, AC12, AC13, AC14, AC16, AC17, AC19, AC20, A. johnsonii, and A. haemolyticus also could not influence mean lifespan (Fig. 2). Different from these, the lifespan curves of nematodes exposed to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii ATCC 19606T, A. junii NH88-14, and A. lwoffii DSM 2403T were significantly (P < 0.01) different from that in control nematodes (Fig. 2). Additionally, exposure to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, and A. lwoffii significantly decreased the mean lifespan (Fig. 2). Thus, Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, and A. lwoffii potentially resulted in adverse effects on lifespan of nematodes.
Effect of exposure to different Acinetobacter strains isolated from the TGR region and reference strains on locomotion behavior of nematodes
Locomotion behavior is more sensitive than lifespan for assessing toxicity of environmental toxicants or stresses25. After exposure for 24-h, Acinetobacter strains of AC2, AC3, AC4, AC5, AC6, AC7, AC8, AC9, AC10, AC11, AC12, AC13, AC14, AC16, AC17, AC19, AC20, A. johnsonii, and A. haemolyticus did not obviously affect locomotion behavior (Fig. 3). In contrast, exposure to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, and A. lwoffii significantly decreased locomotion behavior (Fig. 3).
Effect of exposure to different Acinetobacter strains isolated from the TGR region and reference strains in inducing activation of oxidative stress of nematodes
Oxidative stress is one cellular contributor to toxicity of exposure to toxicants or stresses25,26,27. We further employed the ROS production to examine effect of Acinetobacter strains in inducing oxidative stress. Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, and A. lwoffii for 24-h resulted in obvious induction of ROS production (Fig. 4A).
Effect of exposure to different Acinetobacter strains isolated from the TGR region and reference strains in inducing activation of oxidative stress in nematodes. (A) Effect of exposure to different Acinetobacter strains in inducing ROS production in wild-type nematodes. (B) Effect of exposure to different Acinetobacter strains on SOD-3::GFP expression. The L4-larvae nematodes were exposed to Acinetobacter for 24-h. Control, unexposed nematodes. Bars represent means ± SD. **P < 0.01 vs control.
SOD-3/Mn-SOD provides a molecular basis for antioxidation defense response25. Moreover, we observed that exposure to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, and A. lwoffii for 24-h further led to significant increase in expression of SOD-3::GFP (Fig. 4B).
Effect of exposure to different Acinetobacter strains isolated from the TGR region on expressions of antimicrobial genes in nematodes
In nematodes, intestine is the important organ to activate innate immune response to pathogen infection9. F55G11.4, dod-22, lys-8, lys-1, spp-12, lys-7, dod-6, and spp-1 are most studied intestinal anti-microbial genes28,29,30,31,32,33,34. We next selected these 8 intestinal antimicrobial genes to determine effect of different Acinetobacter strains isolated from the TGR region on innate immune response. The increase in these 8 intestinal antimicrobial genes function to be against pathogen infection and environmental stress28,29,30,31,32,33,34. After exposure to Acinetobacter strains of AC1, AC15, AC18, or AC21 for 24-h, expressions of some of these antimicrobial genes could be noticeably increased. Among these 8 antimicrobial genes, exposure to strain AC1 significantly increased the expressions of spp-1, lys-8, lys-7, lys-1, spp-12, dod-6, dod-22, and F55G11.4, exposure to strain AC15 significantly increased the expressions of F55G11.4, lys-8, dod-6, and lys-7, exposure to strain AC18 significantly increased the expressions of lys-8, lys-7, and spp-12, and exposure to strain AC21 significantly increased the expressions of dod-6, lys-7, spp-12, lys-1, dod-22, spp-1, and F55G11.4 (Fig. 5). In nematodes, LYS-8, LYS-7, and LYS-1 are lysozymes, SPP-12 is a saposin-like protein, DOD-6 and DOD-22 are proteins downstream of DAF-16, SPP-1 is a caenopore, and F55G11.4 is a protein containing CUB-like domain.
Morphological and biochemical properties of Acinetobacter strains of AC1, AC15, AC18, and AC21
For the Acinetobacter strains of AC1, AC15, AC18, and AC21, they did not show obvious difference in morphological properties of cell shape, arrangement of cell, Gram staining, and colony morphology (Table 1). The Acinetobacter strains of AC1, AC15, AC18, and AC21 also did not exhibit the obvious difference in biochemical properties of hydrothion, phenylalanine, gluconate, oxidase, nitrate reduction, catalase, peptone water, semi-solid agar, glucose, ornithine, raffinose, sorbitol, side calendula, and xylose (Table 1). Different from this, the Acinetobacter strains of AC1 and AC21 showed the negative reactions for the biochemical properties of l-arginine, l-lactic acid, d-fucose, l-histidine, l-malic acid, and d-serine (Table 1). The Acinetobacter strains of AC15 and AC18 exhibited the positive reactions for the biochemical properties of l-arginine, l-lactic acid, d-fucose, l-histidine, l-malic acid, and d-serine (Table 1). Additionally, the Acinetobacter strains of AC1 and AC21 showed the negative reactions for the biochemical properties of glucopeptone water, citrate, and gelation, whereas the Acinetobacter strain of AC15 exhibited the positive reactions for the biochemical properties of glucopeptone water, citrate, and gelation (Table 1).
Differences of main virulence genes among Acinetobacter strains
For understanding of differences of virulence genes from these pathogenic Acinetobacter strains, we checked for the presence of 14 main virulence genes (Table S4) in pathogenic strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii and A. lwoffii and nonpathogenic strains of AC2, AC12, AC14, AC17, A. haemolyticus, and A. johnsonii by PCR. Distribution of virulence genes in tested Acinetobacter strains was different and pathogenic Acinetobacter strains generally had more virulence genes than nonpathogenic strains (Table 2). 10 or more virulence genes were detected from pathogenic strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii and A. lwoffii (Table 2).
Discussion
Acinetobacter has attracted significant attention because it is ubiquitous in nature and commonly found in soil, water and hospital35. Many Acinetobacter species can cause serious nosocomial infections in medicine and actively participate the nutrient cycle in the ecosystem36. Due to the clinical and ecological importance of Acinetobacter, it is proposed as a model microorganism for environmental microbiological studies, pathogenicity tests, and industrial production of chemicals18. Nevertheless, many research areas including biosafety, natural transformation, biodegradation, and important physiological characteristics have been limitedly investigated or neglected. We here performed a biosafety evaluation of Acinetobacter strains isolated from backwater area in the TGR region and 5 reference strains of Acinetobacter species in nematode C. elegans.
The high prevalence pathogens exist in the backwater area of the TGR region7,8. The reason to carry out the biosafety assessment of Acinetobacter strains is that the Acinetobacter has been one of dominant microorganisms in the TGR region6, and Acinetobacter isolated form the water in the TGR arises most frequently in our study. The reasons to use C. elegans are that it is very sensitive to various environmental exposures, and can be employed as an ideal model for the study on the pathogenesis of human pathogens, and the mechanisms in host-microbe interactions9,13,14,16,37. More importantly, we previously have systematically performed the biosafety evaluation of water samples from the TGR region in both flood season and quiet season19,20. The reasons to select 5 reference strains of A. baumannii, A. lwoffii, A. junii, A. haemolyticus, and A. johnsonii to expose C. elegans are that the genus of Acinetobacter comprises 68 species with validly-published names (https://apps.szu.cz/anemec/Classification.pdf, May 25, 2021) and these 5 reference speices are important clinical microorganisms21,38, and A. baumannii ATCC 19606T is a model strain of pathogenic bacteria causing nosocomial infection39, followed by the non-A. baumannii species A. haemolyticus, A. junii, A. johnsonii, and A. lwofii21.
Our previous studies have suggested that both solid phase and liquid phase could contribute to toxicity induction of surface water sample collected from backwater areas in the TGR region19,20. In the liquid phase, the potential toxicants were suggested as the organic pollutants20. In this study, using lifespan as the toxicity assessment endpoint, we found that four (AC1, AC15, AC18, and AC21) of the isolated and examined Acinetobacter strains and tree reference strains of A. baumannii, A. junii, and A. lwoffii significantly reduced lifespan (Fig. 2). Using a more sensitive endpoint of locomotion behavior, we also observed the significant decrease in locomotion behavior after exposure to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, or A. lwoffii (Fig. 2), which further confirmed the detected toxic effect of exposure to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, and A. lwoffii on nematodes. These observations suggested that some of the Acinetobacter strains at the backwater area in the TGR region have the exposure risk to environmental organisms and human health. Nevertheless, not all the Acinetobacter strains at the backwater area in the TGR region potentially induced toxicity on environmental organisms. Our data indicated a crucial role of environmental pathogens in contributing to toxicity induction in the solid phase of water sample in backwater area in TGR region.
We further observed the significant ROS production in nematodes exposed to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, or A. lwoffii (Fig. 4A), which suggested the oxidative stress activated by exposure to these Acinetobacter strains. Meanwhile, we also detected the significant increase in SOD-3::GFP expression after exposure to Acinetobacter strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii, or A. lwoffii (Fig. 4B), which further confirmed the oxidative stress activated by exposure to these Acinetobacter strains. These results suggested the close association of the toxic effects of exposure to Acinetobacter strains (AC1, AC15, AC18, AC21, A. baumannii, A. junii, and A. lwoffii) with oxidative stress activation. Nevertheless, we did not detect the decrease in SOD-3::GFP expression after exposure to the above pathogenic Acinetobacter strains. This may be largely due to the short exposure duration (24-h) for these pathogenic Acinetobacter strains. Usually, long-term exposure to toxicants at high concentrations causes decrease in SOD-3::GFP expression9. Exposure to nanopolystyrene (1–10 μg/L) caused increase in SOD-3::GFP expression, whereas exposure to nanopolystyrene (1000 μg/L) resulted in decrease in SOD-3::GFP expression40.
In nematodes, we further found that exposure to Acinetobacter strains of AC1, AC15, AC18, and AC21 induced increase in expressions of some antimicrobial genes (spp-1, dod-22, lys-8, lys-7, spp-12, dod-6, lys-1, and/or F55G11.4) (Fig. 5). Meanwhile, a pronounced increase in Acinetobacter colony-forming unit (CFU) was observed in nematodes infected with AC1, AC15, AC18, and AC21 (Fig. S1). The increase in these antimicrobial genes mediated a protective response to pathogen infection and environmental toxicants9,28,29,30,31,32,33,34. These antimicrobial genes can be expressed in the intestine (https://wormbase.org). The reason to select these intestinal antimicrobial genes is that the ROS production is mainly activated in the intestine25. Similarly, we also did not observe the suppression in expressions of these antimicrobial genes in nematodes exposed to Acinetobacter strains of AC1, AC15, AC18, or AC21, which is also largely due to the performed short exposure duration (24-h) in nematodes. Moreover, we found that exposure to Acinetobacter strains of AC1, AC15, AC18, and AC21 induced the different dysregulation of examined antimicrobial genes (Fig. 5). Exposure to AC1 could cause the increase in expressions of all 8 examined antimicrobial genes, and exposure to AC21 resulted in the increase in expressions of 7 examined antimicrobial genes (Fig. 5). In contrast, exposure to AC15 could cause the increase in expressions of only 4 examined antimicrobial genes, and exposure to AC18 could result in the increase in expressions of only 3 examined antimicrobial genes (Fig. 5). These results implied that Acinetobacter strains of AC1 and AC21 might cause the more severe toxicity at least at some aspects than Acinetobacter strains of AC15 and AC18.
In this study, we provide some lines of evidence to show the important value of C. elegans for assessing biosafety of Acinetobacter strains isolated from the TGR region. Nevertheless, C. elegans only has simple developmental structures, and dose not have some organs (such as heart, liver, lung, and kidney) observed in mammals. Therefore, the further biosafety assessment experiments in mammals for the identified four Acinetobacter strains are still needed.
We also examined morphological and biochemical properties of Acinetobacter strains of AC1, AC15, AC18, and AC21. However, we did not observe the obvious difference in morphological properties of cell shape, arrangement of cell, Gram staining, and colony morphology among the examined Acinetobacter strains of AC1, AC15, AC18, and AC21 (Table 1). In contrast, the observed difference in toxicity of Acinetobacter strains of AC1, AC15, AC18, and AC21 on nematodes might be related to the difference in some biochemical properties among the examined Acinetobacter strains of AC1, AC15, AC18, and AC21. For example, we observed the obvious difference in biochemical properties of l-arginine, l-lactic acid, d-fucose, l-histidine, l-malic acid, and d-serine in the Acinetobacter strains of AC1 and AC21 from those in the Acinetobacter strains of AC15 and AC18 (Table 1). To clarify if they share virulence factors that better induce to the nematode intestinal antimicrobial response, 14 main virulence genes (Table S4) of pathogenic strains of AC1, AC15, AC18, AC21, A. baumannii, A. junii and A. lwoffii and nonpathogenic strains of AC2, AC12, AC14, AC17, A. haemolyticus, and A. johnsonii were detected by PCR. The results showed that pathogenic Acinetobacter strains generally had more virulence genes than nonpathogenic strains (Table 2), and AC1 and AC21, AC15 and AC18 shared more of the same virulence genes, but nonpathogenic strains of AC2 and AC12 also had 11 and 14 virulence genes, respectively. Nevertheless, the exact underlying mechanism still needs the further careful examination.
Together, we performed a biosafety assessment of Acinetobacter strains isolated from backwater area in TGR region in nematodes. Among the isolated Acinetobacter strains, we identified four Acinetobacter strains with the potential to cause toxic effects on nematodes, such as the reduction in lifespan and the decrease in locomotion behavior. The observed toxic effects of Acinetobacter strains were associated with activation of oxidative stress. Moreover, exposure to toxic Acinetobacter strains caused the increase in some antimicrobial genes, suggesting the activation of innate immune response of animals against the Acinetobacter exposure. Considering the fact that we know little about the environmental Acinetobacter pathogens in the TGR region, our data provide important suggestion for exposure risk of certain Acinetobacter strains in the TGR region to environmental animals and human health. Our data has further implied that, after the long-term exposure, the Acinetobacter pathogens are potentially enriched in intestine and cause toxic effects by affecting immune response in environmental animals and human. In the future, we will further identify virulence and resistance factors and perform the sequencing for the identified four Acinetobacter strains isolated from the TGR region.
Methods
Water sampling
The water sample was collected in backwater area (N108° 23′ 25″, E30° 47′ 45″) in Wanzhou, Chongqing in the flood season20. The reason to select this season is that the bacterioplankton community is generally higher in this season than that in the impoundment season6. The detailed properties of collected surface water sample have been described previously20. Water sample was collected and stored as described41. In brief, the equal volumes (10 L) were collected from the depths of 0.5, 5, 10 m in the backwater area site. Water samples were used for the isolation of Acinetobacter after mixing fully in the sterile bucket, and water samples were stored at 0 °C after collection.
Acinetobacter isolation, identification, and preservation
The mixed water sample was diluted serially (1, 10−1, and 10−2), inoculated into LB medium, and incubated at 37 ± 0.5 °C for 24 h. Subculture and purification of bacterial colonies were carried out by the streak plate method. For the purified bacterial isolates, genomic DNA of different bacterial isolates was extracted using bacterial genomic DNA extraction kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. The complete bacterial 16S rRNA gene was amplified with the primer set 27 F and 1492R. PCR products were visualized using 1% agarose gels stained with ethidium bromide. Positive amplicons were quantified using a PicoGreen dsDNA Assay kit (Invitrogen, CA, USA). Purified products were sequenced and analyzed by Magigen (Guangzhou Magigen Biotechnology Co., Ltd., China). Phylogenetic tree was constructed using the Mega 5.0 program using the neighbor-joining (N-J) method with a 1000-bootstrap.
All identified Acinetobacter strains were preserved by freeze drying42. The exponential phase cells of Acinetobacter strains grown in LB medium for 18 h were suspended in aseptic no-fat skimmed milk with an initial cell concentration of 108–109 CFU/mL. The bacterial mixture within ampoules vials was frozen at − 20 °C for 2 h, followed by − 80 °C for 12 h. After that, they were loaded onto the freeze dryer. Both primary drying and secondary drying for 25 h after the freezing were performed. The freeze-dried products were packaged in blister packs and stored in the refrigerator at − 80 °C.
When needed, freeze-dried powders were diluted with sterilized water, and then the suspensions were streak-inoculated onto a LB medium using an inoculation loop. A single colony was inoculated into sterilized LB broth and the bacteria grew to the log phase in a constant temperature oscillator at 37 °C for the use43,44.
Reference strains of Acinetobacter species
A. baumannii (ATCC 19606T), A. lwoffii (DSM 2403T), A. junii (NH88-14), A. haemolyticus (TTH0-4), and A. johnsonii (H10) from China General Microbiological Culture Collection Center (CGMCC) were used to expose the Caenorhabditis elegans in this study. The information of these reference strains is listed in the Table S2.
Analysis of Acinetobacter properties
Different Acinetobacter strains inoculated on broth agar medium were incubated for 24 h at 37 °C45. Primary identification and characterization of different Acinetobacter strains were performed to determine cell shape, arrangement of cell, gram staining, and colony morphology using UVsolo 2 touch (Analytik Jena AG, Germany)46. After the growth at 37 °C for 24 h, the biochemical properties of different Acinetobacter strains were further determined using standard Enterobacteriaceae biochemical identification tube (HANGWEI, Hangzhou Microbiology Reagent Co., Ltd)47.
Maintenance of C. elegans
CF1553/muIs84[SOD-3:GFP] and wild-type N2 were used. Normal nematode growth media (NGM) plates were used to maintain nematodes48. To prepare synchronized L4-larvae, gravid worms were first treated with bleaching solution (0.45 M NaOH and 2% HOCl)49. The released eggs were let to further develop into the L4-larvae population.
Acinetobacter pathogenesis assay
The L4-larvae population was exposed to different Acinetobacter strains. Different Acinetobacter strains were seeded on modified NGM containing 0.35% peptone. Exposure to different Acinetobacter strains was started by transferring nematodes onto each assay plate by adding 60 animals to each assay NGM plate. Full-lawn assay plate was used for Acinetobacter pathogenesis assay as described50. That is, the surface of assay NGM plate was all seeded Acinetobacter strains. The aim of using full lawn assay was to exclude the possibility of effect from the avoidance behavior of nematodes to Acinetobacter strains.
Acinetobacter CFU analysis
The method was performed basically as described51. After the infection, the treated nematodes were transferred to M9 solution containing 25 mM levamisole to paralyze the nematodes. Nematodes were first transferred to a NGM plate containing ampicillin (1 mg/mL) and gentamicin (1 mg/mL) for 15 min to eliminate Acinetobacter on the body surface. Nematodes were further transferred to a new NGM plate containing ampicillin (1 mg/mL) and gentamicin (1 mg/mL) for 30 min to remove any residual Acinetobacter. After that, the nematodes were lysed with a motorized pestle. The lysates were serially diluted in M9 solution and plated on Luria–Bertani plates. After overnight incubation at 37 °C, the colonies were counted to determine the CFU per nematode. The experiments are repeated for three times.
Lifespan assay
After exposure of L4-larvae nematodes to different Acinetobacter strains for 24-h, the survival of worms was counted every day at 20 °C52. If no response was observed after prodding using platinum wire, the worms were considered as dead. The animals were transferred daily during the first 7-day. For the lifespan assay, 60 animals were examined for each treatment. Three replicates were carried out. We used log-rank test to analyze the lifespan curve data. Survival curves were considered to have significant difference if P values were ≤ 0.01.
Locomotion behavior
Locomotion behavior reflects the functional state of motor neurons53. Body bend and head thrash were selected as the endpoints54. After exposure, the worms were first washed with M9 buffer. After that, assuming that animals traveled along x axis, a body bend is defined as a change of bending direction at the mid-body. A head thrash is defined as a change of posterior bulb direction along y axis. For each treatment, 40 animals were analyzed.
Activation of oxidative stress
Production of reactive oxygen species (ROS) reflects the activation of oxidative stress55. The method was performed as described56. After the exposure to different Acinetobacter strains, the animals were labeled for 3 h using CM-H2DCFDA (1 µM). After that, the animals were observed at 488 nm (excitation wavelength)/510 nm (emission filter) under a laser scanning confocal microscope. Using Image J software, we semi-quantified intestinal fluorescence intensity in comparison to intestinal autofluorescence. For each treatment, 50 animals were examined.
In nematodes, sod-3 encodes mitochondrial Mn-SOD9. Using Image J software, fluorescence intensity of SOD-3::GFP signals in the intestine was semi-quantified. For each treatment, 50 animals were examined.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The total RNAs of control and exposed nematodes were extracted using Trizol. Using a spectrophotometer, concentration and purity of the obtained RNAs were determined. We performed the reverse transcriptase reaction with Mastercycler gradient PCR system for cDNA synthesis. With the aid of SYBR Green qRT-PCR master mix, transcriptional expression of spp-1, lys-8, lys-7, lys-1, spp-12, dod-6, dod-22, and F55G11.4 were determined in real-time PCR system. The reference gene was tba-1. Three biological replicates were carried out. Primer information is provided in Table S3.
Polymerase chain reaction (PCR) of virulence genes
Primers were designed according to the published sequences of virulence genes of Acinetobacter on GenBank. Primer information was provided in Table S4. All experimental strains were inoculated onto beef extract agar and cultured at 37 ℃ for 24 h. A single colony was selected and inoculated in beef extract broth at 37 ℃ for 12 h, and the bacterial culture was used as the template. PCR was undertaken in a final volume of 25 μL using the PCR kit (Sangong Bioengineering (Shanghai) Co., Ltd.) with 1μL of each primer and 1 μL of the template. The thermal cycling parameters were 30 s at 98 °C for, followed 35 cycles of 5 s at 98 °C, 5 s at 59 °C and 60 s at 72 °C. 5μL PCR products were analyzed on agarose 1.4% (w/v) gels.
Statistical analysis
Statistical analysis was carried out using SPSS Statistics 19.0 Software (SPSS Inc., USA.). Probability level of 0.01 was considered statistically significant. Using one-way analysis of variance (ANOVA), the differences between groups were tested.
References
Li, Z. et al. Soil-air greenhouse gas fluxes influenced by farming practices in reservoir drawdown area: A case at the Three Gorges Reservoir in China. J. Environ. Manag. 181, 64–73 (2016).
Wang, W., Ndungu, A. W. & Wang, J. Monitoring of endocrine-disrupting compounds in surface water and sediments of the Three Gorges Reservoir Region, China. Arch. Environ. Contam. Toxicol. 71, 509–517 (2016).
Wu, J. et al. The three gorges dam an ecological perspective. Front. Ecol. Environ. 2, 241–248 (2004).
Zhang, J. F. & Deng, W. Industrial structure change and its eco-environmental influence since the establishment of municipality in Chongqing, China. Proc. Environ. Sci. 2, 517–526 (2010).
Zhao, X., Li, T., Zhang, T., Luo, W. & Li, J. Distribution and health risk assessment of dissolved heavy metals in the Three Gorges Reservoir, China (section in the main urban area of Chongqing). Environ. Sci. Pollut. Res. Int. 24, 2697–2710 (2017).
Li, Z. et al. Responses of spatial-temporal dynamics of bacterioplankton community to large-scale reservoir operation: A case study in the Three Gorges Reservoir, China. Sci. Rep. 7, 42469 (2017).
Xiao, G. et al. Occurrence and infection risk of waterborne pathogens in Wanzhou watershed of the Three Gorges Reservoir, China. J. Environ. Sci. 25, 1913–1924 (2013).
Xiao, G. et al. Occurrence and potential health risk of Cryptosporidium and Giardia in the Three Gorges Reservoir, China. Water Res. 47, 2431–2445 (2013).
Wang, D.-Y. Molecular Toxicology in Caenorhabditis elegans (Springer Nature Singapore Pte Ltd, 2019).
Liu, H., Tian, L., Qu, M. & Wang, D. Acetylation regulation associated with the induction of protective response to polystyrene nanoparticles in Caenorhabditis elegans. J. Hazard. Mater. 411, 125035 (2021).
Yang, Y., Dong, W., Wu, Q. & Wang, D. Induction of protective response associated with expressional alterations in neuronal G protein-coupled receptors in polystyrene nanoparticle exposed Caenorhabditis elegans. Chem. Res. Toxicol. 34, 1308–1318 (2021).
Liu, H., Tian, L. & Wang, D. Notch receptor GLP-1 regulates toxicity of simulated microgravity stress by activating germline-intestine communication of insulin signaling in C. elegans. Biochem. Biophys. Res. Commun. 534, 248–253 (2021).
Kumar, A. et al. Caenorhabditis elegans: A model to understand host–microbe interactions. Cell. Mol. Life Sci. 77, 1229–1249 (2020).
Madende, M., Albertyn, J., Sebolai, O. & Pohl, C. H. Caenorhabditis elegans as a model animal for investigating fungal pathogenesis. Med. Microbiol. Immunol. 209, 1–13 (2020).
Liu, H., Tian, L., Wang, S. & Wang, D. Size-dependent transgenerational toxicity induced by nanoplastics in nematode Caenorhabditis elegans. Sci. Total Environ. 790, 148217 (2021).
Wang, D.-Y. Nanotoxicology in Caenorhabditis elegans (Springer Nature Singapore Pte Ltd, 2018).
Sun, L., Liao, K. & Wang, D. Comparison of transgenerational reproductive toxicity induced by pristine and amino modified nanoplastics in Caenorhabditis elegans. Sci. Total Environ. 768, 144362 (2021).
Liu, H., Kwak, J. I., Wang, D. & An, Y. Multigenerational effects of polyethylene terephthalate microfibers in Caenorhabditis elegans. Environ. Res. 193, 110569 (2021).
Xiao, G. et al. Biosafety assessment of water samples from Wanzhou watershed of Yangtze Three Gorges Reservior in the quiet season in Caenorhabditis elegans. Sci. Rep. 8, 14102 (2018).
Xiao, G. et al. Toxicity evaluation of Wanzhou watershed of Yangtze Three Gorges Reservior in the flood season in Caenorhabditis elegans. Sci. Rep. 8, 6734 (2018).
Doughari, H. J., Ndakidemi, P. A., Human, I. S. & Benade, S. The ecology, biology and pathogenesis of Acinetobacter spp.: An overview. Microbes Environ. 26, 101–112 (2011).
Towner, K. J. Acinetobacter: An old friend, but a new enemy. J. Hosp. Infect. 73, 355–363 (2009).
Jung, J. & Park, W. Acinetobacter species as model microorganisms in environmental microbiology: Current state and perspectives. Appl. Microbiol. Biotechnol. 99, 2533–2548 (2015).
van der Kolk, J. H., Endimiani, A., Graubner, C., Gerber, V. & Perreten, V. Acinetobacter in veterinary medicine, with an emphasis on Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 16, 59–71 (2019).
Wang, D.-Y. Target Organ Toxicology in Caenorhabditis elegans (Springer Nature Singapore Pte Ltd, 2019).
Yang, Y., Dong, W., Wu, Q. & Wang, D. Response of G protein-coupled receptor CED-1 in germline to polystyrene nanoparticles in Caenorhabditis elegans. Nanoscale Adv. 3, 1997–2006 (2021).
Sun, L., Li, D., Yuan, Y. & Wang, D. Intestinal long non-coding RNAs in response to simulated microgravity stress in Caenorhabditis elegans. Sci. Rep. 11, 1997 (2021).
Alper, S., McBride, S. J., Lackford, B., Freedman, J. H. & Schwartz, D. A. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol. Cell. Biol. 27, 5544–5553 (2007).
Evans, E. A., Kawli, T. & Tan, M. W. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 4, e1000175 (2008).
Hoeckendorf, A., Stanisak, M. & Leippe, M. The saposin-like protein SPP-12 is an antimicrobial polypeptide in the pharyngeal neurons of Caenorhabditis elegans and participates in defence against a natural bacterial pathogen. Biochem. J. 445, 205–212 (2012).
Liu, J., Hafting, J., Critchley, A. T., Banskota, A. H. & Prithiviraj, B. Components of the cultivated red seaweed Chondrus crispus enhance the immune response of Caenorhabditis elegans to Pseudomonas aeruginosa through the pmk-1, daf-2/daf-16, and skn-1 pathways. Appl. Environ. Microbiol. 79, 7343–7350 (2013).
Mallo, G. V. et al. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12, 1209–1214 (2002).
Pinkston-Gosse, J. & Kenyon, C. DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat. Genet. 39, 1403–1409 (2007).
Ren, M., Zhao, L., Lv, X. & Wang, D. Antimicrobial proteins in the response to graphene oxide in Caenorhabditis elegans. Nanotoxicology 11, 578–590 (2017).
Antunes, L. C., Visca, P. & Towner, K. J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 71, 292–301 (2014).
Harding, C. M., Hennon, S. W. & Feldman, M. F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102 (2018).
Liu, H., Qiu, Y. & Wang, D. Alteration in expressions of ion channels in Caenorhabditis elegans exposed to polystyrene nanoparticles. Chemosphere 273, 129686 (2021).
Bai, L. et al. Comparative genomics analysis of Acinetobacter haemolyticus isolates from sputum samples of respiratory patients. Genomics 112, 2784–2793 (2020).
Tsubouchi, T. et al. Complete genome sequence of Acinetobacter baumannii ATCC 19606(T), a model strain of pathogenic bacteria causing nosocomial infection. Microbiol. Resour. Announc. 9, e0028920 (2020).
Qiu, Y., Liu, Y., Li, Y., Li, G. & Wang, D. Effect of chronic exposure to nanopolystyrene on nematode Caenorhabditis elegans. Chemosphere 256, 127172 (2020).
Chen, D. Guidelines for the Investigation of Aquatic Organisms in Rivers (China Science Publishing & Media Ltd, 2014).
Liu, L. et al. Phytohalomonas tamaricis gen. nov., sp. nov., an endophytic bacterium isolated from Tamarix ramosissima roots growing in Kumtag desert. Arch. Microbiol. 202, 143–151 (2020).
Grujovic, M. Z., Mladenovic, K. G., Nikodijevic, D. D. & Comic, L. R. Autochthonous lactic acid bacteria-presentation of potential probiotics application. Biotechnol. Lett. 41, 1319–1331 (2019).
Chen, L. et al. Dyadobacter luteus sp. nov., isolated from rose rhizosphere soil. Arch. Microbiol. 202, 191–196 (2020).
Ghajavand, H., Esfahani, B. N., Havaei, S. A., Moghim, S. & Fazeli, H. Molecular identification of Acinetobacter baumannii isolated from intensive care units and their antimicrobial resistance patterns. Adv. Biomed. Res. 4, 110 (2015).
Rossett, S. et al. Isolation and identification of an Eikelboom type 1863 strain as Acinetobacter johnsonii. Water Res. 31, 657–660 (1997).
Rojas, R., Miranda, C. D., Romero, J., Barja, J. L. & Dubert, J. Isolation and pathogenic characterization of Vibrio bivalvicida associated with a massive larval mortality event in a commercial hatchery of scallop Argopecten purpuratus in Chile. Front. Microbiol. 10, 855 (2019).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Wang, S., Zhang, R. & Wang, D. Induction of protective response to polystyrene nanoparticles associated with methylation regulation in Caenorhabditis elegans. Chemosphere 271, 129589 (2021).
Zhi, L., Yu, Y., Li, X., Wang, D. & Wang, D. Molecular control of innate immune response to Pseudomonas aeruginosa infection by intestinal let-7 in Caenorhabditis elegans. PLoS Pathog. 13, e1006152 (2017).
Yu, Y., Zhi, L., Wu, Q., Jing, L. & Wang, D. NPR-9 regulates innate immune response in Caenorhabditis elegans by antagonizing activity of AIB interneurons. Cell. Mol. Immunol. 15, 27–37 (2018).
Wang, D., Cao, M., Dinh, J. & Dong, Y. Methods for creating mutations in C. elegans that extend lifespan. Methods Mol. Biol. 1048, 65–75 (2013).
Yang, Y., Wu, Q. & Wang, D. Dysregulation of G protein-coupled receptors in the intestine by nanoplastic exposure in Caenorhabditis elegans. Environ. Sci. Nano 8, 1019–1028 (2021).
Liu, H., Zhao, Y., Bi, K., Rui, Q. & Wang, D. Dysregulated mir-76 mediated a protective response to nanopolystyrene by modulating heme homeostasis related molecular signaling in nematode Caenorhabditis elegans. Ecotoxicol. Environ. Saf. 212, 112018 (2021).
Zhao, Y. et al. Induction of protective response to polystyrene nanoparticles associated with dysregulation of intestinal long non-coding RNAs in Caenorhabditis elegans. Ecotoxicol. Environ. Saf. 212, 111976 (2021).
Wang, S., Liu, H., Qu, M. & Wang, D. Response of tyramine and glutamate related signals to nanoplastic exposure in Caenorhabditis elegans. Ecotoxicol. Environ. Saf. 217, 112239 (2021).
Acknowledgements
This study was funded by the grants from Natural Science Foundation of Chongqing (cstc2018jcyj-AX0639, cstc2019jcyj-msxmX0533, and cstc2020jcyj-msxmX0317) and Scientific and Technological Research Program of Chongqing Municipal Education Commission (KJQN201801225 and KJQN201901222).
Author information
Authors and Affiliations
Contributions
Y.D. and G.X. conceived and designed the research. Y.D., M.T., Q.W., Y.H., F.C., F.Z., and D. W. carried out the experiments. Y.D., H.D. and Q.H. analyzed the data. G.X., H.D. and D.W. wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Deng, Y., Du, H., Tang, M. et al. Biosafety assessment of Acinetobacter strains isolated from the Three Gorges Reservoir region in nematode Caenorhabditis elegans. Sci Rep 11, 19721 (2021). https://doi.org/10.1038/s41598-021-99274-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99274-0
This article is cited by
-
Seasonal pollution and surface characteristics of microplastics in surface water in the Wanzhou section of the Three Gorges Reservoir, China
Environmental Science and Pollution Research (2023)
-
Alternatives to animal models to study bacterial infections
Folia Microbiologica (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.