Abstract
The gastropod infraclass Euthyneura comprises at least 30,000 species of snails and slugs, including nudibranch sea slugs, sea hares and garden snails, that flourish in various environments on earth. A unique morphological feature of Euthyneura is the presence of two pairs of sensory head tentacles with different shapes and functions: the anterior labial tentacles and the posterior rhinophores or eyestalks. Here we combine molecular phylogenetic and microanatomical evidence that suggests the two pairs of head tentacles have originated by splitting of the original single tentacle pair (with two parallel nerve cords in each tentacle) as seen in many other gastropods. Minute deep-sea snails of Tjaernoeia and Parvaplustrum, which in our phylogeny belonged to the euthyneurans’ sister group (new infraclass Mesoneura), have tentacles that are split along much of their lengths but associated nerves and epidermal sense organs are not as specialized as in Euthyneura. We suggest that further elaboration of cephalic sense organs in Euthyneura closely coincided with their ecological radiation and drastic modification of body plans. The monotypic family Parvaplustridae nov., superfamily Tjaernoeioidea nov. (Tjaernoeiidae + Parvaplustridae), and new major clade Tetratentaculata nov. (Mesoneura nov. + Euthyneura) are also proposed based on their phylogenetic relationships and shared morphological traits.
Similar content being viewed by others
Introduction
A distinct part of molluscan biodiversity—about 40% of the estimated 73,000 described species1—is comprised by the long-recognized infraclass Euthyneura in the gastropod subclass Heterobranchia2. Euthyneurans play important and diverse ecological roles as predators, prey, or pests in various marine habitats (both benthic and pelagic), in freshwater realms, and on land3,4. Nudibranch sea slugs, sea hares, and garden snails with stalked eyes (Fig. 1c–e) are well-known euthyneurans that exemplify a fraction of derived morphologies found in this clade. Research has profited from their complex hermaphroditism (e.g.5,6) and sophisticated acquisition of defensive chemicals or ‘stolen’ cnidocytes and chloroplasts from their food (7 for review). Moreover, their so-called ‘giant’ neurons have enabled us to study individual nerve cells, neuronal circuits, and learning8,9.
Comparative head-foot morphology of living Caenogastropoda and Heterobranchia. Macrophotographs of European species taken from above or right side. (a, b) Caenogastropoda and ‘lower’ Heterobranchia have one pair of smooth head tentacles and a short snout bearing the mouth at its tip. (c–e) Euthyneuran heterobranchs, in contrast, have one pair of anterior tentacles that form an upper lip above the mouth (labial/oral tentacles) and another separate pair of posterior tentacles (rhinophores or ommatophores). (a) Freshwater caenogastropod Sadleriana bavarica (2.2 mm in shell diameter; family Hydrobiidae). (b) Freshwater ‘lower’ heterobranch Valvata cristata (3 mm; Valvatidae); unpaired tentacle on right side is secondary and non-cephalic. (c) Marine nudibranch Favorinus branchialis (6 mm in body length; Favorinidae) with modified rhinophores; anterior foot extended to form tentacle-like projections. (d) Juvenile of marine euopisthobranch Aplysia punctata (10 mm; Aplysiidae) with two pairs of enrolled tentacles. (e) Terrestrial panpulmonate Cepaea hortensis (20 mm; Helicidae) with stalked eyes on posterior tentacles. Abbreviations: CE, cerata (dorsal appendages of mantle); EY, eye (highlighted by orange arrowheads); FT, foot; MT, unpaired tentacle extending from mantle margin; PP, parapodia (lateral extensions of foot covering shell); SH, shell; SN, snout; T, head tentacles; TA, anterior head tentacles; TP, posterior head tentacles; UL, upper lip or velum above mouth (highlighted by dotted lines).
The clade Euthyneura was named after its nervous system that lacks much of the asymmetry caused by gastropod torsion (twisting of the posterior body); ‘euthyneury’ or secondary detorsion was contrasted to ‘streptoneury’ with crossing posterior nerve fibres (10, Ponder et al. 2019: p. 340–346, 378–38011). Euthyneurans can more easily be distinguished from other gastropods by their head morphology in having two distinct pairs of head tentacles (with diverse shapes or reductions) formed by independent sets of anterior and posterior sensory areas (Fig. 1c–e;12,13,14). The lower, anterior tentacles (termed oral or labial tentacles) are specialized for contact chemoreception or ‘tasting,’ and fused in the middle to form the upper lip above the mouth. The upper, posterior tentacles (called rhinophores or eyestalks) are variable in shape but primarily responsible in distance chemoreception or ‘smelling’ (olfaction) and also the detection of water or air currents3,12,14,15,16,17.
In contrast, the other Gastropoda, namely the paraphyletic ‘prosobranchs’ (including the grade of the so-called ‘lower’ Heterobranchia; Fig. 1a,b), generally are streptoneurous. They usually have no horizontal upper lip but have a short, tube-shaped snout and only one pair of head tentacles which is, curiously, often innervated by a deeply forked nerve that sends two parallel branches into each tentacle (e.g.18, but see19,20 for discussion). Individual function of these parallel nerve branches has not been explored, but the latest phylogenetic analyses of Gastropoda21,22 confirm that this forked nerve is ancestral for Heterobranchia and its sister-group Caenogastropoda (collectively referred to as Apogastropoda19,20,23) (Figs. 1b, 2a–e). Some previous authors have suggested homology of the prosobranch (and thus lower heterobranch) head tentacle to only the labial tentacle of Euthyneura (see e.g.24,25). More recently, Staubach (e.g. fig. 32)13, based on detailed neuro-anatomical data for many taxa, hypothesized homology of the single tentacle pair of prosobranchs to both of the specialized euthyneuran tentacles (see also14,20).
Molecular phylogeny of Heterobranchia and its new infraclass Mesoneura as sister to Euthyneura. Tree reconstruction was performed in MrBayes and based on combined nucleotide sequences of nuclear 18S and 28S rRNA and mitochondrial 16S rRNA and COI genes. Numerals on branches denote bootstrap proportions from RAxML-HPC analysis (BP in %, left) and Bayesian posterior probabilities (PP, right); asterisks denote full support (BP: 100%, PP: 1). Three branches of Murchisonellidae were shortened by 50% for graphical purpose. Lower heterobranch Architectonicidae and Ammonicera not included due to very long branches (see Supplementary Fig. S2a). Orange asterisks indicate four novel clades retrieved in this analysis; from left to right: Tetratentaculata (no rank), new infraclass Mesoneura, new superfamily Tjaernoeioidea, and new family Parvaplustridae. (a–y) Exemplar species from major subgroups of Heterobranchia: (a) Tomura yashima (representing Cornirostridae), (b) Valvata cristata, (c) Boschitestella cf. eloiseae (Orbitestellidae), (d) Cima sp., (e) Larochella sp., (f) Tjaernoeia exquisita (redrawn and modified from Jensen, 1999: fig. 2A26), (g) Tj. exquisita (redrawn and modified from Warén, 1991: fig. 2627), (h) Parvaplustrum tenerum (redrawn and modified from Powell, 1951: fig. 9728), (i) Murchisonella cf. anabathron (photograph courtesy of Angela Dinapoli), (j) Rhodope veranii, (k) Rissoella diaphana, (l) Acteon tornatilis, (m) Ringicula doliaris, (n) Felimida krohni, (o) Berthella ocellata (Pleurobranchidae), (p) Trinchesia morrowae (Nudibranchia: Trinchesiidae), (q) Haminoea hydatis, (r) Aplysia punctata, (s) Thuridilla hopei, (t) Discus rotundatus, (u) Onchidella celtica, (v) Carychium pessimum, (w) Pseudunela marteli, (x) Turbonilla rufa, and (y) Physella acuta.
Combined morphological and phylogenetic research has continuously refined the understanding of heterobranch and euthyneuran evolution29,30,31,32,33,34,35. Phylogenetic methods today have the potential to robustly reconstruct backbone trees of biological relationships even for ancient evolutionary events (see36,37), which in turn help to trace pathways of evolutionary changes on both large and fine scales. The inclusion of minute and difficult-to-collect taxa have further enhanced resolution and interpretative power of such approaches (e.g.33,38,39). Within the larger framework of reconstructing the early evolution of heterobranch gastropods, we here conducted a phylogenetic analysis using multi-locus molecular markers that are well-established for the study of the group33,35,40. We here added for the first time members of the enigmatic, primarily deep-water genera Tjaernoeia Warén & Bouchet, 1988 and Parvaplustrum Powell, 1951 (Fig. 2).
Tjaernoeia species are among the smallest living gastropods (< 1 mm). They have been classified in their own family Tjaernoeiidae Warén, 1991, and tentatively among the Euthyneura2. Five described species from the Atlantic and Antarctica have coiled shells with a characteristic dimpled surface26,27. The three described species of Parvaplustrum have been classified as dubious members of the euthyneuran clade Acteonoidea based on their fragile, oval ‘bubble’ shells and presence of an external copulatory organ28,41,42. Interestingly, although not commented upon by previous authors26,27,42,43, Tjaernoeia and Parvaplustrum principally resemble lower heterobranchs in the outline of head-foot morphology but also share a unique condition of long, basally forked head tentacles. We found that inclusion of these taxa in a phylogenetic analysis greatly improves resolution of relationships within Heterobranchia and provides a novel scenario on the evolution of the head tentacles and nervous system of Euthyneura.
Results and discussion
Mesoneura, a new clade sister to euthyneuran Heterobranchia
Our molecular phylogenetic reconstruction (Fig. 2, Supplementary Fig. S1; Table 1) recovered Tjaernoeia sp. from off Japan (Fig. 3a), Parvaplustrum tenerum Powell, 1951 from the South Atlantic and P. cadieni Valdés, Gosliner & Warén, 2017 from off California as a maximally supported clade in both Maximum-Likelihood (ML) and Bayesian analyses (bootstrap proportion, BP: 100%, transfer bootstrap expectation, TBE: 100%, Bayesian posterior probability, PP: 1). With this topology, we reclassify the genus Parvaplustrum into a new monotypic family of bubble snails, Parvaplustridae nov., which is sister to Tjaernoeiidae. A new superfamily Tjaernoeioidea is also erected to contain the families Tjaernoeiidae and Parvaplustridae nov. Surprisingly, this new superfamily formed a strongly supported sister (BP: 96%, TBE: 93%, PP: 1; Fig. 2f–h) to the recently recognized Allomorpha, which contains morphologically divergent rhodopid slugs and murchisonellid snails39. Allomorpha was confirmed in the present study as a monophyletic group with strong support (96%, 98%, 1; Fig. 2i, j). The clade Tjaernoeioidea nov. + Allomorpha, here named as a new infraclass Mesoneura, was identified as a robustly supported sister to the over 30,000 species of the infraclass Euthyneura (97%, 96%, 1; Fig. 2k–y). We here propose the name Tetratentaculata nov. for the clade of Mesoneura nov. and Euthyneura. Mesoneura presents a novel taxon in a phylogenetic position between the species-rich clade Euthyneura and a less-diverse grade of ‘lower’ heterobranchs (Fig. 2a–e).
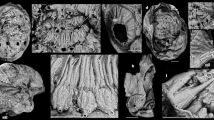
Micromorphology of superfamily Tjaernoeioidea nov. with focus on head tentacles and central nervous system. (a–c) Species of genus Tjaernoeia Warén & Bouchet, 1988. (a) Undescribed Tjaernoeia sp. from off Aomori, Japan (sequenced specimen, AORI YK#2783). Apertural view of shell, showing a dimpled surface and a slightly sinistral apex. (b) European Tjaernoeia exquisita, 3D reconstruction of preserved animal (ZSM Mol-20200024), right anterodorsal view. Shell removed, outer skin of mantle shown in transparency, organs of mantle roof omitted for clarity. (c) Same as b, anteroventral view, showing detail of head-foot with bifid tentacles (red). (d–f) South Atlantic Parvaplustrum tenerum Powell, 1951. (d) Apertural view of shell and preserved soft body inside (voucher specimen ZSM Mol-20020851); shell ‘bubble-shaped’ with a sunken apex at top. (e) 3D-reconstruction of central nervous system, right view (ZSM Mol-20021303). Cerebral nerves of left side (NTA, NTP, NS) omitted for clarity. (f) Schematic drawing of head and central nervous system showing innervation of bifid tentacles, dorsal view, anterior side up. Orange circles indicate positions of histologically distinct neurons, which are 3–4 times larger in diameter than other neurons and potential homologue of ‘giant’ neurons in Aplysia8. Abbreviations: DG, digestive gland; ES, oesophagus; FT, foot; GA, cluster of accessory ganglia near copulatory organ; GB, buccal ganglion; GCP, cerebropleural ganglion; GO, gonad; GP, pedal ganglion; GR, accessory (rhinophoral?) ganglion; G1, posteroventral visceral ganglion; G2, anterodorsal visceral ganglion ; HF, head-foot; I, intestine; M, mouth; MS, mantle skirt; NG, nidamental gland; NM, nerves to mantle; NS, nerves to snout; NTA, nerve of anterior tentacle; NTP, nerve of posterior tentacle; NP, nerves to foot (4 pairs); NV, nerve to visceral sac (two only on left side); O, sensory osphradium with ganglion; OC, mature oocytes; OT, oral tube; P, penis; RM, retractor muscle; SC, statocyst with single statolith; SD, seminal duct; TA, anterior head tentacle; TS, lateral projection on snout; TP, posterior head tentacle; VL, slightly streptoneurous visceral loop.
The present tree topology otherwise conformed to previous studies using either comparable marker and taxon sets33,35,40, mitogenomics44,45, or phylogenomics21,46 in retrieving the paraphyletic ‘lower’ heterobranchs on the one hand and the monophyletic Acteonacea (or Acteonimorpha), Ringipleura, Euopisthobranchia and Panpulmonata as constituting Euthyneura on the other (some previous authors classified Acteonacea as lower heterobranchs, e.g.33). Internal support values of derived clades (i.e., euopisthobranchs and panpulmonates) were lower than in comparable studies with more expansive taxon sampling of these groups33,47. Interestingly, careful BLAST searches for the present and previous Genbank sequences, followed by the removal of ambiguous or misidentified data (see Supplementary Table S2), resolved the topology of the lower heterobranchs differently from previous studies (Fig. 2a–e;39,40): Orbitestellidae with discoidal shells were recovered as the sister (77%, 87%, 1) to high-spired Cimidae (Cima and Larochella; full support), instead of having these taxa separate in a large grade40 or in a polytomy39.
This topology was stable to sensitivity analyses where taxa were selectively added or removed (Table 1, Supplementary Fig. S2). Inclusion of the only remaining lower heterobranch clades Architectonicoidea (presumed here to encompass Mathildidae based on morphology) and Ammonicera (currently classified as ‘Omalogyroidea’) into separate ML analyses found these to form a monophyletic taxon (BP: 100%, TBE: 100%) as sister to Valvatoidea but with low support (45%, 72%) and an extremely long branch (Supplementary Fig. S2). Inclusion of this long-branched clade resulted in the masking of more numerous alignment-ambiguous sites and thereby lowered nodal support over the tree, yet without significantly affecting the topology. Exclusion of either Rhodopidae or Murchisonellidae also resulted in the same topology with the monophyletic Mesoneura (BP: 78–83%, TBE: 64–93%) and Tetratentaculata (63–89%, 81–92%) (Table 1; Supplementary Fig. S2).
Comparative microanatomy of Tjaernoeioidea
Past morphological studies on Tjaernoeia and Parvaplustrum were confounded by their extremely small body sizes, which resulted in the examination of only the shell, external anatomy and chitinous internal structures (26,27,28,41,42,43). Species of Tjaernoeia bear a broad, somewhat flattened, coiled shell, similar to those of some other heterobranchs, in particular lower heterobranch taxa such as the Valvatoidea27. Parvaplustrum species on the other hand have a fragile, oval and inflated ‘bubble’ shell with a sunken spire and without an umbilicus, resembling those of many euthyneuran groups (e.g. Hydatina and Haminoea, see Fig. 2q)28,41,42,43. Based on our study of aligned serial histological sections and three-dimensional reconstructions of the body shape and internal organs, we here show aspects of microanatomy for the type species of each genus, namely the North Atlantic Tjaernoeia exquisita (Jeffreys, 1883)(Fig. 3b,c) and South Atlantic P. tenerum (Fig. 3d–f). Particular focus is here placed on the head morphology and configuration of the central nervous system (CNS) in comparison with other gastropods including newly studied Ebala (Mesoneura: Allomorpha: Murchisonellidae) and Rissoella (Euthyneura: Rissoellidae) (see Supplementary Fig. S3). The CNS is of particular interest due to its conservativeness during gastropod evolution, with neuronal connections inside its ganglia conserved for hundreds of millions of years13,14.
The herein reconstructed, preserved animal of T. exquisita was reproductively mature, yet only 550-µm long (Fig. 3b) and much smaller than P. tenerum (ca. 2 mm; Fig. 3d). Although with different sizes and shell shapes, the two species are very similar to each other in head-foot morphology. They both resemble lower heterobranchs including the Cimidae and many of Valvatoidea (see e.g. Fig. 2a,b,d) in having a slender and anteriorly bifurcated foot and a short snout27. This snout bears a short, finger-shaped tentacle on either side of the tip (TS), as seen in some species of Valvatoidea18. More posteriorly, the head of T. exquisita and P. tenerum has two conspicuous pairs of long, deeply bifurcated and slightly flattened head tentacles of which the anterior branch is about 30–40% shorter than the posterior one (TA and TP in Fig. 3b,c,f). The surface of these tentacles is uniformly smooth, with interspersed gland cells and with ciliation mainly on the inner side. Eyes are lacking in both species, and the body is colourless (see also27). The posterior side of the foot lacks an operculum in both Tjaernoeia and Parvaplustrum, as in the Rhodopidae and the majority of Euthyneura, whereas the Murchisonellidae and lower heterobranchs are all operculate48,49.
The general similarity of their head-foot is also reflected in the nervous system of Tjaernoeia and Parvaplustrum. The CNS, shown here in the larger-bodied P. tenerum (Fig. 3e,f), contains a cerebral nerve ring with four ganglia (paired cerebropleural and pedal ganglia: GCP and GP) and two buccal ganglia (GB) below the pharynx. The asymmetric, twisted visceral nerve loop (VL) encircles the oesophagus and bears dorsal (G2) and posteroventral ganglia (G1). Parvaplustrum tenerum differs from T. exquisita in having small additional ganglia of unknown function joined to the sides of the cerebropleural ganglia (GR). Moreover, only P. tenerum has a cluster of accessory ganglia (GA) at the base of the external copulatory organ (P) on the right side of the head behind the bifid tentacles; this copulatory organ in P. tenerum is larger than that of T. exquisita and bears a chitinous stylet (not shown). In both species, several paired nerves emanate from the cerebropleural ganglia. The snout is innervated by two pairs (in P. tenerum) or a single pair (in T. exquisita) of nerves. The bifurcated head tentacles are each innervated by two independent nerves that emerge directly adjacent to each other (NTA, NTP); these nerves do not bear obvious lateral ramifications except two small branches near the nerve tip in Parvaplustrum.
In addition, certain ganglia of P. tenerum (GP, GB, G1, G2 and left GCP) have several large neurons (orange circles in Fig. 3f) that are histologically and topologically identifiable with the ‘giant’ neurons of the neurobiological model organism Aplysia (Figs. 1d, 2f) and many other euthyneurans (Gillette, 1991: p. 2358). These cells could not be identified in T. exquisita with its smaller body size, as was previously the case in Rhodopidae48 and Murchisonellidae49. The two species resemble lower heterobranchs in lacking specialized epidermal sensory areas innervated by smaller branches of the tentacle nerves, such as the so-called Hancock’s organs (which are present at the base of posterior head tentacles of most aquatic euthyneurans;14; see below). The osphradium, another chemosensory organ in the molluscan pallial cavity, is present in both T. exquisita and P. tenerum as a small ciliated patch of epidermis on the anterior roof of the mantle (O in Fig. 3e,f).
The different shell shapes of T. exquisita and P. tenerum are also reflected in the different organization of their mantle. Specifically, P. tenerum has its plicate gill, glands and kidney all located on the posterior right of the mantle (a condition called ‘detorted’ in Euthyneura). On the other hand, in T. exquisita the gill is a more medially lying, microscopic leaf without folds, glands are spread along the anterior margin of the mantle, and the kidney lies centrally, reflecting a more ancestral condition typical of non-euthyneuran heterobranchs (e.g.10,18). Opposing ciliary strips in the mantle cavity, regarded as an apomorphy of Heterobranchia10,29, could not be reliably identified in Tjaernoeia and Parvaplustrum and among the Mesoneura such structures are so far only confirmed for some Murchisonellidae50. Tjaernoeia and Parvaplustrum are similar in their complex hermaphroditic reproductive system (Fig. 3b) and in the simple digestive tract with a very narrow cuticular radula that was shown by other authors to bear only leaf-shaped lateral teeth27,41,43.
Morphological diagnoses of the newly proposed taxa can be summarized as follows: Tjaernoeioidea, a new superfamily for Tjaernoeia (Tjaernoeiidae) and Parvaplustrum (Parvaplustridae nov.): Small to minute heterobranch snails with a single pair of deeply bifid head tentacles (or, depending on perspective, two pairs of basally joined tentacles) and a slender snout with a lateral projection on either side; eyes lacking and skin unpigmented; external copulatory organ on the right side of the head-foot; shell fragile, globose to oval, smooth or with sculpture of small dimples, protoconch hyperstrophic; foot without an operculum. Parvaplustridae, a new monotypic family for Parvaplustrum [ZooBank registration (LSID): urn:lsid:zoobank.org:act:A7416D49-113A-4E60-86A7-6AE46818B20A]: Shell inflated, oval, with a large aperture and an involute spire; shell surface smooth or with minute, irregularly scattered pits; mantle detorted with the gill, kidney and large glands all located on the posterior right; copulatory organ with a tubular chitinous stylet. We regard these differences in external morphology to warrant separate family status. Mesoneura, a new infraclass for Tjaernoeioidea + Allomorpha: Named after its nervous system showing a mix of plesiomorphic and apomorphic conditions in their nervous system, with tentacle nerves that innervate independent areas of the head but otherwise largely lack ramifications; Hancock’s organ and median lip absent; visceral loop with torsion yet long; radula (if present) narrow without a rachidian tooth and with only one slender lateral tooth on either side of a transverse row. Tetratentaculata, a new clade name for Mesoneura + Euthyneura: Head with four individual sensory areas corresponding to four tentacles or two deeply bifurcated tentacles, although tentacles per se may be reduced secondarily; giant neurons present in the pedal, buccal, dorsal, posteroventral, and left cerebropleural ganglia (see below); visceral nerve cord at least slightly detorted or completely detorted. Accordingly, Euthyneura can be newly diagnosed as tetratentaculate heterobranchs distinguished from Mesoneura by having (1) labial tentacles that are medially fused to form an upper lip or velum and (2) tentacle nerves bearing many small ramifications that innervate sensory cells including those of the Hancock’s organ (see discussion below).
Origin of euthyneuran head tentacles
The herein recovered phylogenetic position of the Tjaernoeioidea corroborates a recent morphology-based hypothesis13,20 that the two pairs of specialized euthyneuran head tentacles might have originated through bifurcation of an ancestral single pair of tentacles, where each tentacle was already innervated by two nerve cords, as now seen in the Caenogastropoda and lower Heterobranchia3. It was previously assumed that only the anterior pair (labial tentacles) of the Euthyneura was homologous to the plesiomorphic head tentacles of the Gastropoda (see13,24,51). The posterior pair (rhinophores or ommatophores or eyestalks) was regarded as secondarily acquired or even repeatedly acquired (e.g.25). However, convincing evidence for the homology of rhinophores across the Euthyneura came from the use of axonal backfilling techniques12,14,15 that reliably identified and correlated individual tentacle nerves across a broad set of taxa. Furthermore, Staubach13 identified highly conserved neuron clusters associated with the tentacle nerves in the cerebral ganglia of the periwinkle (Caenogastropoda: Littorina) and giant African snail (Heterobranchia: Stylommatophora: Achatina). This conservatism in details of neuronal architecture of tentacle innervation further supports homology of tentacles across the Apogastropoda (see Fig. 1, Supplementary Fig. S3).
In combination with these results, our new findings lead to an evolutionary scenario of two steps. First, at the origin of Tetratentaculata, the single ancestral tentacle with double nerves split into two tentacles, each with one of the two ancestral nerve cords. Second, at the origin of Euthyneura, the two tentacles became specialized into the anterior and posterior tentacles, with different shapes and with more elaborate sensory areas such as the rhinophores and Hancock’s organs. The first of these evolutionary steps might still be visible in the extant Tjaernoeioidea, which have a pair of deeply bifurcated yet basally joined tentacles. With only few exceptions, the two pairs of ramified tentacle nerves—acquired in the second step—persist across the Euthyneura including groups with atypical tentacles (Fig. 4, Supplementary Fig. S3, see3,11,13,14,24,35).
Time-calibrated phylogeny of heterobranch gastropods and distributions of different types of head and shell morphologies. Tree reconstruction was based on combined four-gene sequences and calibration priors placed on three nodes and performed in BEAST. Numerals on branches are Bayesian posterior probabilities; asterisks denote full support (PP: 1). Numbered circles at nodes indicate major clades, including (1) Heterobranchia, (2) Tetratentaculata nov., (3) Mesoneura nov., (4) Allomorpha, (5) Tjaernoeioidea nov., (6) Parvaplustridae nov., (7) Euthyneura, (8) Acteonimorpha, (9) Ringipleura, (10) Nudipleura, (11) Tectipleura, (12) Euopisthobranchia, and (13) Panpulmonata. Pictograms of head morphology suggest two pairs of tentacles existed at node 2 (hence new name Tetratentaculata: ‘bearing four tentacles’). Colour-coded circles at terminals show phenotypic plasticity of shells in Tetratentaculata, particularly in Euthyneura. Abbreviations: CG, cerebral/cerebropleural ganglion; FT, foot; S + N, snout with snout nerves; T + 2N, single tentacle with two nerve cords (green area); TA + N, anterior/labial tentacle with one or two nerve pairs (blue area); TP + N, posterior tentacle or rhinophore/ommatophore with single nerve (yellow area). Head schemes of Aplysia, Archidoris and Achatina after13, anterior side is up. See Supplementary Fig. S3 for further comparison.
The Allomorpha, although being sister to the Tjaernoeioidea, do not share the long and bifid head tentacles and thus might question the evolutionary scenario proposed above. However, allomorph snails and slugs may have modified or lost the bifid tentacles in relation to their sediment-dwelling or even infaunal lifestyles39,48,49 as have many euthyneurans12,13,29,35. Murchisonellids bear a pair of posterior, oftentimes broad, tentacles that are innervated with two pairs of essentially unbranched nerve cords (see49, and Supplementary Fig. S3 for reexamination of Ebala). Rhodopids have entirely lost the head tentacles per se but there remain the two pairs of the tentacle nerves in the head39,48. After a confused taxonomic history (see39), the Allomorpha are now found to resemble their previously unrecognized sistergroup Tjaernoeioidea more than other heterobranchs in having four separate tentacle nerves, but lacking unequivocal Hancock’s organs, distal ramifications of tentacle nerves, and a typical medially-fused upper lip49. Absence of the Hancock’s organ and upper lip also distinguishes the Tjaernoeioidea from the shallow-water, herbivorous euthyneuran snails of the superfamily Rissoelloidea (Supplementary Fig. S3), regardless of the short, bifid head tentacles of Rissoella that externally might recall those of tjaernoeioids (Fig. 2k;52,53).
Shell shapes and radiation of Tetratentaculata over geologic time
Our BEAST analysis with the ages of three fossil heterobranchs as calibration points (Fig. 4) estimated that Tetratentaculata (node 2) originated sometime in the Devonian–Carboniferous period with a posterior mean age of 359 million years ago (Mya) and 95% highest posterior density (HPD) intervals of 406–307. The split between Tjaernoeioidea and Allomorpha (node 3; mean: 325 Mya, 95% HPD: 377–270) potentially predated any other split within the Tetratentaculata, whereas Tjaernoeia and Parvaplustrum appear to have diverged much more recently (node 5; 103 Mya, 169–47). The rhodopid stemline may extend into the late Palaeozoic (node 4; 281 Mya, 339–217), further back in time than other extant lineages of slugs (Fig. 4: black circles at branch terminals), save potentially the Nudipleura (node 9; 254 Mya, 303–203) or the Nudibranchia (node 10; 201 Mya, 253–144).
The speciose radiation of Euthyneura (node 7), leading also to diverse shell-shapes and instances of shell loss (Fig. 4: circles at branch terminals), was estimated to have started in the Carboniferous–Permian time (296 Mya, 345–248). This predates the oldest known euthyneuran fossils, the diverse Cylindrobullinoidea occurring since the Early Triassic (245 Mya) (see54,55 for discussion). Cylindrobullinoids have been suspected to contain paraphyletic or polyphyletic members of already diverged euthyneuran lineages (see55 for review), some of which may lead to the extant Acteonimorpha, Ringipleura, Euopisthobranchia and Panpulmonata. The early Euthyneura are suggested to have a characteristic bubble shell with a large body whorl—a morphology found in several lineages of the extant Euthyneura (Fig. 4: blue circles at terminals)—as well as a hypertrophied foot and headshield for an oftentimes infaunal mode of life35. The morphological variability found in Mesoneura does not allow unequivocal reconstruction, but they display bubble-shell and slug morphotypes (Parvaplustridae and Rhodopidae) in parallel with the Euthyneura. Fossils of putatively ancestral murchisonellids (as Donaldinidae and Streptacididae, see56) come from the strata of 350–260 Mya, which is much older than the first occurrences of the fossil Cylindrobullinoidea and closely fit the herein proposed age of divergence (node 3 in Fig. 4, see above). Some of those Palaeozoic taxa57,58,59are indeed fairly similar to the modern Murchisonella60,61 in teleoconch and protoconch morphology. On the other hand, certain early-Triassic fossils have a murchisonellid-like protoconch and a cylindrobullinoid-like teleoconch and were therefore interpreted as a phylogenetic link between the Streptacididae and Cylindrobullinoidea56.
The Euthyneura comprise almost half of all molluscan species richness —why could they have radiated into so many ecological niches and diversified into so many species? Kano et al.35 hypothesized that early euthyneurans were freed from the strict connection of the shell and mantle margin, thereby releasing the mantle from morphological constraints and allowing the creation of evolutionary novelty. We here add that the modification of the head, although not evident in the fossil record, seems to present another overlooked key event in their evolutionary history. The diverse nature of head tentacles is considered important for the taxonomy of euthyneuran subgroups, particularly those of sea slugs14,25,34. However, understanding of their role in euthyneuran evolution as a whole has been overshadowed by a focus on shell loss and coinciding chemical defence. The diverse rhinophores, eyestalks, and Hancock’s organs of Euthyneura play crucial roles in directional chemosensing11,13,14,25,62 and are much more specialized than head tentacles in other gastropods (shown in Fig. 4 at right; Supplementary Fig. S3). For the other (aquatic) gastropods, the osphradium in the mantle cavity is the primary chemosensory organ3, but it is generally simplified in euthyneuran and non-euthyneuran heterobranchs63,64. This suggests that, at least in Euthyneura, sensory capacity of the head has not only become more pronounced relative to other Gastropoda but it has also functionally replaced much of mantle-based chemosensing, as implied by previous authors (see Morton, 1972: p. 33765; Gosliner, 1994: p. 34625). We here suggest that the acquisition of the more elaborate head sensors has determined the observed shift of euthyneuran ecology towards more motile and predatory lifestyles, often with specialized prey items and habitats66,67. Furthermore, reduced reliance on the osphradium for chemosensing may have removed constraints to reorganization of the mantle, allowing additional evolutionary plasticity and innovations in the morphology of the posterior body and shell. The acquisition of the enhanced head sensors may therefore be linked to the shell loss and also to the explosive radiation and speciation of Euthyneura.
This study highlights that inclusion of rare, microscopic taxa in an integrated analysis has the potential to greatly improve the resolution of phylogenetic relationships and provides a novel scenario on a large-scale evolutionary process, in the present case within heterobranch and euthyneuran gastropods.
Methods
Sampling and preparation of specimens
Living snails of the following seven heterobranch species were collected from coastal to bathyal waters using various methods (see below; Supplementary Tables S1, S2 and Supplementary Fig. S3 for additional data). Specimens were preserved either (1) directly in 95–99% ethanol, or (2) fixed in a 10% formalin-seawater solution after anesthetization in isotonic magnesium chloride solution and then transferred to 80% ethanol. All sectioned specimens and DNA extracts are deposited at the Mollusca section of the Bavarian State Collection of Zoology, Munich, Germany (ZSM) or at the Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa, Japan (AORI).
Parvaplustrum tenerum Powell, 1951 [museum voucher numbers ZSM Mol-20021303/2 (DNA aliquot B103), ZSM Mol-20020851 (embedded specimen block 1W7), ZSM Mol-20021303/1 (block 2W5, 3D-reconstructed)]: Collected in 2002 at Burdwood Bank, SW of Falkland Islands, South Atlantic (272 m, 54°02’S, 62°02’W, Agassiz Trawl) during LAMPOS expedition, R/V Polarstern cruise ANTXIX-5, station PS61/145–1. Vouchers preserved in 96% ethanol; DNA extracted and used for phylogenetic analysis (Figs. 2, 4) or embedded in Epon epoxy resin and serially sectioned, used for 3D reconstruction (Fig. 3d–f).
Parvaplustrum cadieni Valdés, Gosliner & Warén, 2017 [Swedish Museum of Natural History SMNH-111919 (tissue clip AORI YK#2783)]: Collected in 2010 at Hydrate Ridge, off Oregon, USA (795 m, 44° 34′N, 125° 09′W) during R/V Atlantis (AGOR-25) cruise AT15-68, DSV ALVIN dive 4635, by Anders Warén. Voucher preserved in pure ethanol, clipped tissue used for phylogenetic analysis (Figs. 2, 4).
Tjaernoeia exquisita (Jeffreys, 1883) [ZSM Mol-20200024 (block 33B)]: Collection date and details unknown, presumably before 2000 in Skagerrak, Sweden by subtidal dredging, collected and prepared by the late Cristoffer Schander. Voucher fixed and preserved in Formalin (?), embedded in Araldite epoxy resin and serially sectioned, 3D-reconstructed (Fig. 3b,c). [Note that Appolloni et al.68 consider Tj. imperspicua (Monterosato, 1875) (non Chaster, 1895) to be the oldest valid name for this species, although it is generally regarded a nomen nudum27].
Tjaernoeia sp. (AORI YK#2783): Collected in 2012 from off Hachinohe, Aomori Prefecture, Honshu, Japan (459–498 m, 40°58’N, 141°46’E) during R/V Tansei-maru cruise KT-12-18, by Yasunori Kano. Voucher preserved in 99% ethanol, clipped tissue used for phylogenetic analysis (Figs. 2, 4), shell imaged (Fig. 3a).
Helminthope cf. psammobionta Salvini-Plawen, 1991 (Smithsonian Institution No. SI-CBC2010KJ01_B05, a DNA aliquot): Collected 2010 at Carrie Bow Cay, Belize during meiofauna workshop of Smithsonian Institution (Station 4, ridge of outer reef slope, 15 m), from bulk sample of coarse subtidal sand, by Katharina M. Jörger, Jon L. Norenburg, Katrine M. Worsaae. Voucher preserved in 96% ethanol, tissue clipping sequenced (Figs. 2, 4).
Ebala sp. [ZSM Mol-20200025 (block 2b*7)]: Collected in 2017 at Yura, Sumoto, Awaji Island, Hyogo Prefecture, Japan (34°16’N, 134°57’E), from intertidal seagrass bed, by Sho Kashio. Voucher preserved in 80% ethanol after Formalin fixation, serially sectioned and used for 3D reconstruction of CNS (Fig. S3).
Rissoella sp. [ZSM Mol-20200026 (block 4b*7)]: Collected in 2017 at Kinchaku-jima Island, Uchiumi, Miyazaki Prefecture, Kyushu, Japan (31°44’N, 131°29’E), from upper subtidal coralline algae, by Yasunori Kano and Bastian Brenzinger. Voucher preserved in 80% ethanol after Formalin fixation, serially sectioned and used for 3D reconstruction of CNS (Fig. S3).
DNA extraction, PCR amplification and sequencing
Full genomic DNA was extracted from clipped foot or mantle tissue using DNeasy Blood and Tissue Kit (Qiagen) or Macherey–Nagel Blood and Tissue Set and following the manufacturers’ instructions. Partial sequences of nuclear (18S and 28S rRNA) and mitochondrial (16 s rRNA and COI) markers were amplified using primers shown in Supplementary Table S1; see69 and35 for amplification conditions and other details. Amplicons were purified with ExoSAP-IT (Affymetrix) and then sequenced with Big Dye Terminator Cycle Sequence Kit 3.1 (Applied Biosystems) and amplification and sequencing primers (Supplementary Table S1). The reaction mixtures were analyzed on an ABI PRISM 3130xl sequencer (at AORI) or an ABI 3730 sequencer (at the Department of Biology Genomic Service Unit of the Ludwig-Maximilians-University Munich) after purification with Big Dye XTerminator Purification Kit (ABI). New DNA sequences have been deposited in the DDBJ⁄EMBL⁄GenBank with accession numbers LC631476–LC631487 (Supplementary Table S2).
Phylogenetic reconstruction
For molecular phylogenetic analyses we selected 49 heterobranch species including one Tjaernoeia and two Parvaplustrum (Supplementary Table S2). The selection was made on the basis of covering the phylogenetic diversity of Heterobranchia and consistent evolutionary rates of all four targeted genes. The lower heterobranch genera Architectonica and Ammonicera were excluded from the main analyses due to their extremely long branches in a preliminarily tree. Dubious sequences, identified by BLAST searches and by careful comparison in the context of larger alignments, were excluded from succeeding analyses (see Supplementary Table S2 for notes on excluded sequences). Three species of Caenogastropoda were included in the dataset for outgroup comparison, resulting in a total of 52 taxa. The sequences of the four genes were aligned individually with MAFFT 7.182 using the L-INS-i strategy70; COI sequences were aligned as amino acids. Each aligned dataset was masked to remove alignment ambiguous sites on Gblocks Server 0.91b with all three options for a less stringent selection71.
Phylogenetic trees were reconstructed from single-gene and concatenated multi-gene datasets using the Maximum-Likelihood (ML) method implemented in raxmlGUI 2.0 (RAxML-HPC and RAxML-NG;72,73,74). Each gene and codon position was allowed to have different parameters, resulting in six partitions for the four-gene dataset. The RAxML-HPC analyses were performed using following commands: a rapid bootstrap analysis with 1000 replicates and search for the best-scoring ML tree in a single program run under the default GTR + G model, following the software manual. The RAxML-NG runs were carried out with the same setting to calculate transfer bootstrap expectation (TBE)75 values with 1000 replicates. The concatenated four-gene dataset was also analysed under Bayesian inference using MrBayes 3.1.276. Substitution models used (estimated with jModeltest 2.1.10;77) were GTR + G for the 3rd codon of COI and GTR + I + G for all other partitions. Two parallel runs were made for 10 M generations with a sample frequency of 1000, using the default value of four Markov chains. The first 5000 trees for each run were discarded to make sure the four chains reached stationarity by referring to the average standard deviation of split frequencies76. The consensus tree and posterior probabilities (PP) were computed from the remaining 10,000 trees (5000 trees, two runs). Bootstrap proportion (BP) and TBE of ≥ 80% and PP of ≥ 0.99 were considered significant support.
The stability of clades was further tested in sensitivity analyses where taxa were selectively added or removed. The sequences of individual genes were aligned and masked by adding the long-branched clade of Architectonica and Ammonicera (55 taxa), or excluding rhodopid slugs or murchisonellid snails (50 taxa each), to generate three additional sets of four-gene matrices. These datasets were analysed in raxmlGUI2 with 1000 replicates to obtain BP and TBE values.
Divergence times were calculated from the four-gene dataset used in the main analyses (52 taxa) with the relaxed molecular clock model implemented in BEAST 1.5.478. The tree was calibrated by setting ages for three nodes with reliable fossil records: (1) the split of Heterobranchia and Caenogastropoda by Early Devonian time (Gamma distribution, Shape: 1, Offset: 400, Scale: 13.34), (2) the first split in Euopisthobranchia by Early Jurassic (Offset: 190, Scale: 6.33), and (3) first split in Ellobioidea by Late Jurassic (Offset: 152, Scale: 5.07). See35 for the details of the calibration points and other settings for BEAST analysis.
Microanatomical sampling and reconstruction
Ethanol-preserved specimens of Tjaernoeia exquisita, Parvaplustrum tenerum (Figs. 3, 4, Supplementary Fig. S3), and for comparison of the nervous systems, Ebala sp. and Rissoella sp. (Supplementary Fig. S3) were washed in 0.1 M phosphate buffer, decalcified using 3.5% ascorbic acid, stained in a solution of 3.5% Safranin in ethanol, then dehydrated in an ascending acetone series (70–100%), and finally embedded in Epon epoxy resin (except T. exquisita in Araldite resin). From the resin blocks, ribbons of serial semithin sections with a thickness of 1.0–1.5 µm were cut using a Diatome HistoJumbo diamond knife (Biel, Switzerland) with a Zeiss Microm rotation microtome (Jena, Germany)79. Sections were stained using Richardson’s stain80, sealed with Araldite resin and coverslips, and individually photographed using dotSlide software in combination with a semi-automated Olympus BX16VS microscope (both Olympus Soft Imaging Solutions, Tokyo). Photographs were resized, adjusted and changed to greyscale using scripts in Adobe Photoshop and imported into AMIRA 5.3 software (Visage Imaging, Berlin) for both manual and semi-automated realignment, segmentation, 3D reconstruction, and rendering of models. Presented images are surface renderings of histologically distinct structures or schematic drawings derived thereof.
References
Rosenberg, G. A new critical estimate of named species-level diversity of the recent Mollusca. Am. Malac. Bull. 32(2), 308–322 (2014).
Bouchet, P. et al. Revised classification, nomenclator and typification of gastropod and monoplacophoran families. Malacologia 61, 1–526 (2017).
Ponder, W.F., Lindberg, D.R. & Ponder J.M. Gastropoda III: The Heterobranchia. In Biology and Evolution of the Mollusca. Volume Two. 419–525 (CRC Press Inc, Boca Raton, 2019).
Barker, G. M. The Biology of Terrestrial Molluscs (CABI Publishing, New York, 2001).
Beeman, R. D. Gastropoda: Opisthobranchia. In: Reproduction of Marine Invertebrates, Vol. 4 (eds Giese, A. C. & Pearse, J. S.) 115–179 (1977).
Koene, J. M. & Schulenburg, H. Shooting darts: Co-evolution and counter-adaptation in hermaphroditic snails. BMC Evol. Biol. 5(1), 25. https://doi.org/10.1186/1471-2148-5-25 (2005).
Wägele, H. & Klussmann-Kolb, A. Opisthobranchia (Mollusca, Gastropoda)–more than just slimy slugs. Shell reduction and its implications on defence and foraging. Front. Zool. 2, 3 (2005).
Gillette, R. On the significance of neuronal giantism in gastropods. Biol. Bull. 180(2), 234–240 (1991).
Voronezhskaya, E. E. & Croll, R. P. Mollusca: Gastropoda. In Structure and Function of Invertebrate Nervous Systems (eds Schmidt-Rhaesa, A. et al.) 196–221 (Oxford University Press, Oxford, 2016).
Haszprunar, G. On the origin and evolution of major gastropod groups, with special reference to the Streptoneura. J. Molluscan Stud. 54, 367–441 (1988).
Ponder, W. F., Lindberg, D. R. & Ponder J. M. Chapter 7: Nervous system, sense organs, learning and behaviour. In Biology and Evolution of the Mollusca. Volume One. 380–420 (CRC Press Inc, Boca Raton, 2019).
Staubach, S. & Klussmann-Kolb, A. The cephalic sensory organs of Acteon tornatilis (Linnaeus, 1758) (Gastropoda Opisthobranchia)—cellular innervation patterns as a tool for homologisation. Bonner Zool. Beitr. 55, 311–318 (2007).
Staubach, S. The Evolution of the Cephalic Sensory Organs within the Opisthobranchia. 1–155 (Dissertation, Johann Wolfgang Goethe-Universität, Frankfurt am Main, 2008).
Klussmann-Kolb, A., Croll, R. P. & Staubach, S. Use of axonal projection patterns for the homologisation of cerebral nerves in Opisthobranchia, Mollusca and Gastropoda. Front. Zool. 10, 20 (2013).
Faller, S., Staubach, S. & Klussmann-Kolb, A. Comparative immunohistochemistry of the cephalic sensory organs in Opisthobranchia (Mollusca, Gastropoda). Zoomorphology 127, 227–239 (2009).
Matsuo, R., Kobayashi, S., Yamagishi, M. & Ito, E. Two pairs of tentacles and a pair of procerebra: Optimized functions and redundant structures in the sensory and central organs involved in olfactory learning of terrestrial pulmonates. J. Exp. Biol. 214, 879–886 (2011).
Wyeth, R. C. Olfactory navigation in aquatic gastropods. J. Exp. Biol. https://doi.org/10.1242/jeb.185843 (2019).
Haszprunar, G., Speimann, E., Hawe, A. & Heß, M. Interactive 3D anatomy and affinities of the Hyalogyrinidae, basal Heterobranchia (Gastropoda) with a rhipidoglossate radula. Org. Divers. Evol. 11(3), 201–236 (2011).
Ponder, W. F. & Lindberg, D. R. Towards a phylogeny of gastropod molluscs: An analysis using morphological characters. Zool. J. Linn. Soc. 119, 83–265 (1997).
Koller, K., Brenzinger, B. & Schrödl, M. A caenogastropod in 3D: Microanatomy of the Munich endemic springsnail Sadleriana bavarica Boeters, 1989. Spixiana 37, 1–19 (2013).
Zapata, F. et al. Phylogenomic analyses of deep gastropod relationships reject Orthogastropoda. Proc. R. Soc. B Biol. Sci. 281(1794), 20141739 (2014).
Cunha, T. J. & Giribet, G. A congruent topology for deep gastropod relationships. Proc. R. Soc. B Biol. Sci. 286(1898), 20182776 (2019).
Salvini-Plawen, L. V. & Haszprunar, G. The Vetigastropoda and the systematics of streptoneurous Gastropoda (Mollusca). J. Zool. 211, 747–770 (1987).
Huber, G. On the cerebral nervous system of marine Heterobranchia (Gastropoda). J. Molluscan Stud. 59, 381–420 (1993).
Gosliner, T.M. Chapter 5. Gastropoda: Opisthobranchia. In Microscopic Anatomy of Invertebrates, Volume 5: Mollusca I. 253–355 (Wiley-Liss, 1994).
Jensen, K. R. Sjælden mikroskopisk snegl fundet levende i nordlige Kattegat. Flora og Fauna 105, 41–44 (1999).
Warén, A. New and little known Mollusca from Iceland and Scandinavia. Part 1. Sarsia 76, 53–124 (1991).
Powell, A. W. B. Antarctic and subantarctic Mollusca: Pelecypoda and Gastropoda. Discovery Reports 26, 47–196 (1951).
Haszprunar, G. The Heterobranchia—a new concept of the phylogeny of the higher Gastropoda. J. Zool. Syst. Evol. Res. 23, 15–37 (1985).
Dayrat, B. & Tillier, S. Evolutionary relationships of euthyneuran gastropods (Mollusca): A cladistic re-evaluation of morphological characters. Zool. J. Linn. Soc. 135, 403–470 (2002).
Mordan, P. & Wade, C. Heterobranchia II. The Pulmonata. In Phylogeny and Evolution of the Mollusca (eds Ponder, W. F. & Lindberg, D. R.) 409–426 (University of California Press, Berkeley, 2008).
Wägele, H., Klussmann-Kolb, A., Vonnemann, V. & Medina, M. Heterobranchia I. The Opisthobranchia. In Phylogeny and Evolution of the Mollusca (eds Ponder, W. F. & Lindberg, D. R.) 383–406 (University of California Press, Berkeley, 2008).
Jörger, K. M. et al. On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BMC Evol. Biol. https://doi.org/10.1186/1471-2148-10-323 (2010).
Wägele, H., Klussmann-Kolb, A., Verbeek, E. & Schrödl, M. Flashback and foreshadowing - a review of the taxon Opisthobranchia. Org. Divers. Evol. 14, 133–149 (2014).
Kano, Y., Brenzinger, B., Nützel, A., Wilson, N. G. & Schrödl, M. Ringiculid bubble snails recovered as the sister group to sea slugs (Nudipleura). Sci. Rep. 6, 30908 (2016).
Laumer, C. E. et al. Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc. R. Soc. B 86, 20190831 (2019).
Giribet, G. & Edgecombe, G. D. The Invertebrate Tree of Life (Princeton University Press, Princeton, 2020).
Laumer, C. E. et al. Spiralian phylogeny informs the evolution of microscopic lineages. Curr. Biol. 25, 2000–2006 (2015).
Wilson, N. G., Jörger, K. M., Brenzinger, B. & Schrödl, M. Phylogenetic placement of the enigmatic worm-like Rhodopemorpha slugs as basal Heterobranchia. J. Molluscan Stud. 83, 399–408 (2017).
Dinapoli, A. & Klussmann-Kolb, A. The long way to diversity—phylogeny and evolution of the Heterobranchia (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 55, 60–76 (2010).
Marcus, E. & Marcus, E. Opisthobranchian and lamellarian gastropods collected by the “Vema”. American Museum Novitates 2368, 1–33 (1969).
Valdés, Á., Gosliner, T. M. & Warén, A. A new species of Parvaplustrum Powell, 1951 (Gastropoda: Heterobranchia: Aplustridae) from the northeastern Pacific. Nautilus 131, 97–100 (2017).
Chaban, E. M. & Chernyshev, A. V. New and little-known shell-bearing heterobranch mollusks (Heterobranchia: Aplustridae and Cephalaspidea) from the bathyal zone of the northwestern part of the Sea of Japan. Deep Sea Res. II(86), 156–163 (2013).
Kurabayashi, A. & Ueshima, R. Complete sequence of the mitochondrial DNA of the primitive opisthobranch gastropod Pupa strigosa: Systematic implication of the genome organization. Mol. Biol. Evol. 17, 266–277 (2000).
Varney, R. M. et al. Assessment of mitochondrial genomes for heterobranch gastropod phylogenetics. BMC Ecol. Evol. 21(6), 1–14 (2021).
Kocot, K. M., Halanych, K. M. & Krug, P. J. Phylogenomics supports Panpulmonata: Opisthobranch paraphyly and key evolutionary steps in a major radiation of gastropod molluscs. Mol. Phylog. Evol. 69, 764–771 (2013).
Dayrat, B. et al. Phylogenetic relationships and evolution of pulmonate gastropods (Mollusca): New insights from increased taxon sampling. Mol. Phylog. Evol. 59, 425–437 (2011).
Brenzinger, B., Haszprunar, G. & Schrödl, M. At the limits of a successful body plan—3D microanatomy, histology and evolution of Helminthope (Mollusca: Heterobranchia: Rhodopemorpha), the most worm-like gastropod. Front. Zool. 10, 37 (2013).
Brenzinger, B., Wilson, N. G. & Schrödl, M. Microanatomy of shelled Koloonella cf. minutissima (Laseron, 1951) (Gastropoda: ‘lower’ Heterobranchia: Murchisonellidae) does not contradict a sister-group relationship with enigmatic Rhodopemorpha slugs. J. Molluscan Stud. 80, 518–540 (2014).
Wise, J. B. Reassignment of Henrya morrisoni Bartsch, 1947 from the Family Aclididae to the Ebalidae (Gastropoda: Heterobranchia). Nautilus 113, 64–70 (1998).
Schmekel, L. Aspects of evolution within the opisthobranchs. In The Mollusca. Evolution Vol. 10 (eds Trueman, E. R. & Clarke, M. R.) 221–267 (Academic Press, San Diego, 1985).
Fretter, V. The structure and life history of the some minute prosobranchs of rock pools: Skeneopsis planorbis (Fabricius), Omalogyra atomus (Alder) and Rissoella opalina (Jeffreys). J. Mar. Biol. Ass. U.K. 27, 597–632 (1948).
Ponder, W. F. & Yoo, E. K. A revision of the Australian species of the Rissoellidae (Mollusca: Gastropoda). Rec. Aust. Mus. 31, 133–185 (1977).
Nützel, A. Recovery of gastropods in the Early Triassic. C.R. Palevol. 4, 501–51 (2005).
Gründel, J. & Nützel, A. On the early evolution (Late Triassic to Late Jurassic) of the Architectibranchia (Gastropoda: Heterobranchia), with a provisional classification. Neues Jahrb Geol. Paläontol. Abh. 264, 31–59 (2012).
Pan, H.-Z., Erwin, D. H., Nützel, A. & Zhu, X.-S. Jiangxispira, a new gastropod genus from the Early Triassic of China with remarks on the phylogeny of the Heterostropha at the Permian/Triassic boundary. J. Paleontol. 77, 44–49 (2003).
Yoo, E. K. Early Carboniferous Gastropoda from the Tamworth Belt, New South Wales, Australia. Rec. Aust. Mus. 46(1), 63–120 (1994).
Bandel, K., Nützel, A. & Yancey, T. E. Larval shells and shell microstructures of exceptionally well-preserved Late Carboniferous gastropods from the Buckhorn Asphalt Deposit (Oklahoma, USA). Senckenbergiana Iethaea 82(2), 639–689 (2002).
Isaji, S. & Okura, M. Microgastropods from the late Carboniferous limestone in Fukuji, Gifu Prefecture, central Japan. Paleontol. Res. 24(3), 192–202 (2020).
Warén, A. Murchisonellidae: Who are they, where are they and what are they doing? (Gastropoda, lowermost Heterobranchia). Vita Malacologica 11, 1–14 (2013).
Bandel, K. Living fossils among tiny Allogastropoda with high and slender shell from the reef environment of the Gulf of Aqaba with remarks on fossil and recent relatives. Mitt. Geol. Paläont Inst. Univ. Hamburg 89, 1–24 (2005).
Croll, R. P. Gastropod chemoreception. Biol. Rev. 58, 293–319 (1983).
Haszprunar, G. The fine morphology of the osphradial sense organs of the Mollusca I. Gastropoda, Prosobranchia. Philos. Trans. R. Soc. Lond. B Biol. Sci. 307(1133), 457–496 (1985).
Haszprunar, G. The fine morphology of the osphradial sense organs of the Mollusca II. Allogastropoda (Architectonicidae, Pyramidellidae). Trans. R. Soc. Lond. B Biol. Sci. 307(1133), 497–505 (1985).
Morton, J. E. The form and function of the pallial organs in the opisthobranch Akera bullata, with a discussion on the nature of the gill in Notaspidea and other tectibranchs. Veliger 14, 337–349 (1972).
Wägele, H. Potential key characters in Opisthobranchia (Gastropoda, Mollusca) enhancing adaptive radiation. Org. Div. Evol. 4, 175–188 (2004).
Göbbeler, K. & Klussmann-Kolb, A. Molecular phylogeny of the Euthyneura (Mollusca, Gastropoda) with special focus in Opisthobranchia as a framework for reconstruction of evolution of diet. Thalassas 27, 121–154 (2011).
Appolloni, M. et al. Catalogue of the primary types of marine molluscan taxa described by Tommaso Allery di Maria, Marquis of Monterosato, deposited in the Museo Civico di Zoologia, Roma. Zootaxa 4477(1), 1–138 (2018).
Takano, T. & Kano, Y. Molecular phylogenetic investigations of the relationships of the echinoderm-parasite family Eulimidae within Hypsogastropoda (Mollusca). Mol. Phylogenet. Evol. 79, 258–269 (2014).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinform. 35(21), 4453–4455 (2019).
Edler, E., Klein, J., Antonelli, A. & Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 12, 373–377 (2021).
Lemoine, F. et al. Renewing Felsenstein’s phylogenetic bootstrap in the era of Big Data. Nature 556(7702), 452–456 (2018).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinform. 19, 1572–1574 (2003).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Ruthensteiner, B. Soft Part 3D visualization by serial sectioning and computer reconstruction. Zoosymposia 1, 63–100 (2008).
Richardson, K. C., Jarett, L. & Finke, E. H. Embedding in epoxy resins for ultrathin sectioning in electron microscopy. Stain Technol. 35, 313–323 (1963).
Acknowledgements
We thank Katrin Linse (Cambridge) for her assistance during LAMPOS expedition aboard R/V Polarstern (cruise leader Wolf Arntz) and Shigeaki Kojima and Hiroaki Fukumori (both AORI) during their help in R/V Tansei-maru cruise KT-12-18. We gratefully acknowledge Anders Warén (Stockholm), Sho Kashio (Osaka) and the late Christoffer Schander (Bergen) for providing the specimens of Parvaplustrum cadieni, Ebala sp. and Tjaernoeia exquisita, respectively. Bernhard Ruthensteiner, Timea P. Neusser and Sebastian Thal (ZSM and LMU Munich) provided assistance in anatomical analysis; Peter C. Kohnert, Katharina M. Jörger (both Munich), Tsuyoshi Takano and H. Fukumori (both AORI) assisted us in molecular lab work; T.T., H.F., Takuya Yahagi (AORI) and Franziska S. Bergmeier (LMU Munich) further assisted us in fieldwork. Kathe R. Jensen (Copenhagen) is thanked for providing useful literature on Tjaernoeia, Angela Dinapoli (Frankfurt) for sharing her photo of a live Murchisonella cf. anabathron. Sid Staubach and Annette Klussmann-Kolb (both Frankfurt) kindly explained to us details of euthyneuran cerebral nerves elaborated in S.S.’s thesis, and Gerhard Haszprunar (Munich) discussed with us the condition of sensory nerves in prosobranch gastropods. Invaluable feedback on the manuscript was made by Katharina M. Jörger and two referees. Financial support was provided by grants from JSPS (Postdoctoral Fellowship Programme Short Term: PE17703 to BB, JSPS KAKENHI: numbers 15H04412, 18H02494 and 19KK0385 to YK) and DFG (BR 5727/1-1 to BB and SCHR 667/4, 9, 13 and 15 to MS).
Author information
Authors and Affiliations
Contributions
All authors provided specimens and taxonomic expertise; B.B. and Y.K. designed the study; Y.K. performed molecular analyses and created supplementary files; B.B. performed morphological analyses, drafted the figures and manuscript; B.B. and Y.K. wrote the manuscript; all authors read, discussed and added to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brenzinger, B., Schrödl, M. & Kano, Y. Origin and significance of two pairs of head tentacles in the radiation of euthyneuran sea slugs and land snails. Sci Rep 11, 21016 (2021). https://doi.org/10.1038/s41598-021-99172-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99172-5
This article is cited by
-
A chromosome-level genome for the nudibranch gastropod Berghia stephanieae helps parse clade-specific gene expression in novel and conserved phenotypes
BMC Biology (2024)
-
One-health approach on the future application of snails: a focus on snail-transmitted parasitic diseases
Parasitology Research (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.