Abstract
Inter-individual differences of drug responses could be attributed to genetic variants of pharmacogenes such as cytochrome P450 (CYP), phase 2 enzymes, and transporters. In contrast to extensive studies on the genetic polymorphisms of CYP gene, genetic mutation spectrum of other pharmacogenes was under-representative in the pharmacogenetics investigations. Here we studied the genetic variations of 125 pharmacogenes including drug transporters, non-CYP phase 1 enzymes, phase 2 enzymes, nuclear receptors and others in Chinese from the Chinese Millionome Database (CMDB), of which 38,188 variants were identified. Computational analyses of the 2554 exonic variants found 617 deleterious missense variants, 91.1% of which were rare, and of the 54 loss-of-function (splice acceptor, splice donor, start lost, and stop gained) variants, 53 (98.1%) were rare. These results suggested an enrichment of rare variants in functional ones for pharmacogenes. Certain common functional variants including NUDT15 13:48611934 G/A (rs186364861), UGT1A1 2:234676872 C/T (rs34946978), and ALDH2 12:112241766 G/A (rs671) were population-specific for CMDB Chinese because they were absent (with a zero of variant allele frequency) or very rare in other gnomAD populations. These findings might be useful for the further pharmacogenomics research and clinical application in Chinese.
Similar content being viewed by others
Introduction
Increasing whole genome sequences data provide a goldmine for genetic studies on the disease etiology and drug therapeutics. Genetic variants of cytochrome P450 (CYP) genes performed an important role in the inter-individual differences of drug response1, and clinical implementation of several CYP variation-drug pairs such as CYP2C19*2–3 and clopidogrel, CYP2C9*2–3 and warfarin, and CYP3A5*3 and tacrolimus has been applied substantially worldwide2,3. Recently, pharmacogenes other than CYP such as ABCC2 (ATP-binding cassette subfamily C member 2) transporters, UGT2B7 (uridine-5'-diphosphate (UDP) glucuronosyltransferase family 2 member B7) and F2R (coagulation factor II (thrombin) receptor) were found to be of importance in the drug efficacy and adverse drug reactions4,5,6. However, information on the genetic variations of these pharmacogenes including non-CYP phase 1 enzymes, phase 2 enzymes, nuclear receptors and drug transporters in mainland Chinese was limited, to our knowledge. In addition, limited number of subjects in these studies was a common shortcoming, which weakened their power of application.
As the price of whole genome sequencing reduced to about $1000 dollars per genome, more genome sequencing projects across nations have been performed including the 1000 Genomes Project, the UK100K Genomes Project and the Genome Aggregation Database (gnomAD)7,8. Analysis of more than 130,000 whole genome sequences identified genetic variants with important clinical relevance in the ABC transporters, solute carrier (SLC) superfamily of transporters and organic anion transporting polypeptides (OATP) genes9,10,11. Mining of the genome sequence database could identify highly population-specific variants. For instance, SLC22A1*2-*5 involved in drug response of imatinib, metformin and opioids are absent in East Asians while own allele frequencies up to 21.9% in other world populations. Whereas, SLC22A1 L160F is common in East Asians with a minor allele frequency of 0.142 but lowest in African population (minor allele frequency = 0.038)10. In addition, the ratios of population-specific variations differed among ABC genes from 70% in ABCA7 to 92% in ABCE1, while only 0.3% of variations were shared among all seven world groups11. These results demonstrated that the need to better understand drug efficacy in participants of diverse ancestral backgrounds. However, these whole genome sequences database mainly comprised European populations while Chinese population were underrepresented.
A large-scale Chinese population sequencing project named the Chinese Millionome Database (CMDB) recently investigated 141,431 whole genome sequences from Chinese women who took a non-invasive prenatal testing12. Our previous study of 57 CYP genes and POR (cytochrome P450 oxidoreductase) in CMDB provided a comprehensive data set of P450 genes covering the whole mainland China provinces and 37 ethnicities13. In the present study we investigated the genetic variations of 125 pharmacogenes including non-CYP phase 1 enzymes, phase 2 enzymes, drug transporters, nuclear receptors and others in Chinese from the CMDB.
Materials and methods
Data sources
One-hundred and twenty-five pharmacogenes, including 16 ABC transporters, 50 SLC transporters, 17 phase 1 enzymes, 26 phase 2 enzymes, 9 nuclear receptors and 7 others (including ACE, CDA, POR, NUDT15, CACNA1S, G6PD, IFNL3), were obtained through the pipeline in Fig. 1a. Briefly, the automated annotation text-mining system in PharmGKB (https://www.pharmgkb.org) was performed to scan sentences in literatures from PubMed. When the sentences included information linking a chemical and a variation, the text-mining system would annotate the sentence as pharmacogenomics information. And 1055 automated annotation pharmacogenes were identified (accessed 1 March, 2020), which were compared to pharmacogenes studied in reference (Ingelman- Sundberg et al.)14. Whole-genome sequencing data from 141,431 Chinese including 37 ethnicities living in 31 provinces of mainland China in CMDB were used to collect genetic variants information across the above 125 pharmacogenes (accessed 21 March, 2020). In brief, raw variants were gained using a P value less than 10–6 by the maximum likelihood model in the accessible genomic regions. Then, a Bayesian Gaussian mixture model was applied to assign every variant candidate a Phred-scaled probabilistic score (VQSR score) demonstrating the probability that this variant was a genuine polymorphic variant. The higher VQSR score indicated the higher probability that the variant candidate was a true polymorphic variation. High transition versus transversion (Ti/Tv) ratio is found for the raw call set (maximum 8.9 for novel variants and 3.4 for known variants) but it would decrease when the filtration threshold for VQSR score increases. A 35 of filtration threshold of VQSR score was applied indicating a Ti/Tv ratio of 2.4 for novel variants and of 2.2 for known variants12. Novel variant (without one rs number) was defined relative to dbSNP release 135. Variants with a variation allele frequency (VAF) less than 1% were defined as rare and variants with a VAF more than 1% were defined as common. The VAF in different gnomAD populations were collected from gnomAD browser (http://www.gnomad-sg.org/) in version 3.1.
The genetic landscape of 125 pharmacogenes. (a) Flowchart of screen of the studied pharmacogenes. (b) Pie chart showing the distribution of the identified 38,188 variants among ABC transporters, SLC transporters, non-CYP phase 1 enzymes, phase 2 enzymes, nuclear receptors, and others. (c) 1038 (2.7%) of these 38,188 variants we identified were novel as compared to dbSNP release 135. (d) Number of variants with different variant allele frequencies. (e) Number of variants with different nucleotide mutation type.
Variant effect prediction
The functional consequences of missense variants were predicted through a panel of online computational algorithms including SIFT (http://sift-dna.org), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), and PROVEAN (http://provean.jcvi.org/index.php). Missense variation was categorized as deleterious when the ≥ 50% of the above algorithms predicted a damaging effect on the variation. In addition, splice acceptor and splice donor (in total of 8) were assessed by Combined Annotation Dependent Depletion (CADD) (https://cadd.gs.washington.edu/) to be deleterious, and start lost, and stop gained variants were regarded as putatively loss-of-function variants.
Statistical analysis
Kruskal–Wallis test was utilized to compare the differences between variants number across pharmacogene subfamilies (GraphPad Prism version 8) (www.graphpad.com). Linear regression was used to analyze the relationship between missense, deleterious and total variants numbers and the corresponding gene length in different pharmacogenes groups. The chi-square test or Fisher’s exact test when needed was used for comparative analysis of the variant allele frequencies for the ABCC4, SLCO1B1, ALDH2, TPMT, UGT1A1, VDR and NUDT15 polymorphisms between different populations in gnomAD and CMDB Chinese using R version 4.1.0 (https://www.r-project.org/). P < 0.05 was recognized as statistically significant, while the Bonferroni Correction was used to adjust the significance threshold (P < 0.002 (0.05/18)) for the multiple testing of the linear regression analysis between the number of total variants, missense variants and deleterious variants and the corresponding gene length among the 6 pharmacogenes groups, and to adjust the significance threshold (P < 0.0006 (0.05/72)) for the multiple testing of the comparison for variants allele frequencies between CMDB Chinese and diverse gnomAD populations.
Results
Genetic variability overview in 125 human pharmacogenes
We analyzed the genetic variations in 125 genes with importance for drug response utilizing whole genome sequencing data from 141,431 unrelated Chinese subjects. Totally, we identified 38,188 variants distributed across transporter genes (26,769 variants in 66 genes), genes encoding phase 1 (2476 variants in 17 genes) and phase 2 enzymes (5201 variants in 26 genes), nuclear receptors (2554 variants in 9 genes), and other pharmacogenes with diverse functions (1188 variants in 7 genes; Fig. 1b). In addition, 1038 (2.7%) of these 38,188 variants identified were novel as compared to dbSNP release 135 (Fig. 1c). Notably, 14,294 (37.4%) variants were rare with a VAF less than 1% (Fig. 1d). Of the twelve nucleotide transversions and transitions, C > T (25.3%) and G > A (24.8%) constituted the most common types of mutations, representing over half of the mutated nucleotides in 125 pharmacogenes (Fig. 1e).
Of these 38,188 variants, 35,634 variants were non-coding ones (intronic, upstream and downstream regions) while 2554 (6.7%) were exonic variants (Fig. 2a). The majority of 2554 exonic variants were missense (n = 1063; 41.6%), followed by untranslated region (UTR) (n = 799; 31.3%) and synonymous (n = 638; 25.0%; Fig. 2a). The most variants in ABC transporters, SLC transporters, non-CYP phase 1 enzymes, phase 2 enzymes, nuclear receptors, and others were observed in ABCC4 (n = 993), SLC14A2 (n = 1582), AOX1 (n = 384), UGT1A1 (n = 495), RXRA (n = 696), and POR (n = 343), respectively (Fig. 2b). The least variants in ABC transporters, SLC transporters, non-CYP phase 1 enzymes, phase 2 enzymes, nuclear receptors, and others were observed in ABCC2 (n = 126), SLC30A4 (n = 71), ADH5 (n = 32), GSTM1 (n = 22), NR1I3 (n = 54), and IFNL3 (n = 34), respectively. Intronic variants were the most type across the 125 pharmacogenes (Fig. 2b).
Gene region pattern of 125 pharmacogenes. (a) Of these 38,188 variants, 35,634 variants were non-coding ones (intronic, upstream and downstream regions) while 2554 (6.7%) were exonic variants. Pie chart showing the distribution of the identified 2554 exonic variants. (b) Number of variants with different gene region distributions in each of ABC transporters, SLC transporters, non-CYP phase 1 enzymes, phase 2 enzymes, nuclear receptors, and others pharmacogenes.
The majority of variants with putative clinical relevance were rare
Neutral missense variants were comprised of 446 while deleterious missense and loss-of-function variants were comprised of 671 (see the “Methods” section for details; Fig. 3a). Of the 617 deleterious missense variants, 562 (91.1%) were rare while only 55 (8.9%) were common (Fig. 3b), which suggested an enrichment of rare missense variants in functional ones for pharmacogenes. The number and percentage of rare deleterious missense variants in ABC transporters, SLC transporters, non-CYP phase 1 enzymes, phase 2 enzymes, nuclear receptors, and others were 157 (96.3%), 211 (93.0%), 54 (81.8%), 73 (82.0%), 31 (96.9%), and 36 (90.0%), respectively. Of the 54 loss-of-function (splice acceptor, splice donor, start lost, and stop gained) variants, 53 (98.1%) were rare, except one common splice donor variant in SULT1A1 (16:28,631,383 C/G; rs79527462; c.-266 + 1 N > C) with a VAF of 8.71%. A significant different distribution of loss-of-function variants among ABC transporters (n = 18), SLC transporters (n = 18), non-CYP phase 1 enzymes (n = 3), phase 2 enzymes (n = 7), nuclear receptors (n = 6), and others (n = 2) were observed (Fig. 3c; P = 0.0047). Totally, the average distribution of functional variants (deleterious missense plus loss-of-function variants) in ABC transporters (mean ± SD: 11.2 ± 9.03) was significantly more than those in SLC transporters (4.2 ± 3.22), non-CYP phase 1 enzymes (4.1 ± 5.09), phase 2 enzymes (3.7 ± 3.15), nuclear receptors (4.1 ± 2.86), and others (5.9 ± 5.60) (Fig. 3d; P = 0.0048). However, we re-test the relationship between pharmacogenes family and loss-of-function burden and functional variants adjusting for gene size and no significant difference was found among the pharmacogenes families (P = 0.0916 for LoF variants and P = 0.1212 for functional variants). The significantly larger average distribution of functional variants in ABC transporters than those in other groups could be attributed to the gene size and the large number of exons across the ABC superfamily relative to the other genes/gene families tested. To evaluate the functional importance of rare variants in individual pharmacogene, we calculated the percentage of rare and common functional variants (deleterious missense plus loss-of-function variants) and aggregated frequencies of splice acceptor, splice donor, start lost, stop gained and putatively deleterious missense variants in each of the 125 studied pharmacogenes (Fig. 4). In total, we could observe a substantially different distribution and pattern of genetic diversity among the 125 pharmacogenes studied. No functional variants with a zero of aggregated functional variants frequency were identified in pharmacogenes including SLC10A2, SLC26A11, SLC30A4, SLC47A2, SLCO1B3, ADH1B, ADH1C, ADH4, ADH5, ADH7, ALDH1A3, FMO5, GSTM1, GSTM5, UGT2B7, UGT2B28, and PPARG. Several common variants constituted the majority (common variants fraction ≥ 50%) of genetic variability with functional importance for such pharmacogenes as SLC2A9, SLC6A4, SLC28A3, EPHX1, FMO2, GSTA5, GSTO1, NAT2, UGT2B4, and NUDT15, with an aggregated functional variants frequency of 66.56%, 2.93%, 18.42%, 41.67%, 58.59%, 88.17%, 15.53%, 3.40%, 21.90%, 1.61%, respectively. Whereas, the remaining 98 (78.4%) pharmacogenes including ABCB1, SLC10A1, SLCO1B1, ALDH2, GSTP1, TPMT, SULT1A1, UGT1A1, AHR, G6PD, and IFNL3, the majority of the functionality of which was governed by rare genetic variations (rare variants fraction ≥ 50%; Fig. 4). The most highly polymorphic pharmacogenes were GSTA5 (aggregated functional variants frequency 88.17%), SLC22A2 (87.33%), ABCG8 (86.09%), SLC15A2 (69.86%), SLC2A9 (66.56%), FMO2 (58.59%), and VDR (52.47%), whereas the least numbers of functional variants were identified for SLC16A7 (0.14%), SLC22A8 (0.13%), SLC12A1 (0.13%), NAT1 (0.10%), and IFNL3 (0.09%). The fold between the highest frequency of genetic variants predicted to impact the function of the gene product (GSTA5 88.17%) and lowest (IFNL3, 0.09%) was approximately 979.7. Accordingly, overall genetic variation as well as the fraction of functional variants that was allotted to rare variants differs significantly among the 125 studied pharmacogenes.
Rare variants were the majority of putatively functional pharmacogenes variants. (a) Number of neutral missense variants were 446 while deleterious missense and loss-of-function variants were 671 for the studied 125 pharmacogenes. (b) Number and fraction of deleterious missense variants with different variant allele frequencies in diverse pharmacogenes families. (c) Number of loss-of-function variants in different pharmacogenes families. (d) Number of functional variants (deleterious missense plus loss-of-function variants) in different pharmacogenes families.
Frequency distribution of functional variants in individual pharmacogene. The aggregated putatively functional variants frequency of each of ABC transporters, SLC transporters, non-CYP phase 1 enzymes, phase 2 enzymes, nuclear receptors, and others pharmacogenes is plotted in log scale and indicated as dots connected by the black line (right y-axis). The fraction of the functional variation that is allotted to common (blue) or rare (red) variants is indicated on the left y-axis.
Different correlation patterns between number of variants and gene length among pharmacogene groups
Through linear regression analysis, we investigated the relationship between the number of total variants, missense variants and deleterious variants and the corresponding gene length among the 6 pharmacogenes groups. After the Bonferroni Correction for the multiple test, no significant correlation between the number of total variants (P = 0.0083), missense variants (P = 0.0234), and deleterious variants (P = 0.0081) and the corresponding gene length was found in phase 2 enzymes group (Fig. 5). Moreover, no correlation between the number of total variants (P = 0.9141), missense variants (P = 0.8818), and deleterious variants (P = 0.7784) and the corresponding gene length was found in nuclear receptors group. The trend upwards between the nuclear receptor total variant category and gene length was damaged by a single outlier (AHR, aryl hydrocarbon receptor). AHR, a member of the basic helix-loop-helix-period-aryl hydrocarbon receptor nuclear translocator-single-minded (bHLH-PAS) family of transcription factors, has a gene length of 429.79 kb but only 114 variants in total, which might be attributed to its conservative role as the biological sensor in initiating gene expression procedures in responses to exogenous and endogenous signals. In addition, the relationship between the number of total variants and gene length was found to be significant in SLC transporters (P < 0.0001), and non-CYP phase 1 enzymes (P = 0.0007) respectively, but not significant in ABC transporters (P = 0.0038). Furthermore, no significant correlation between the number of missense and deleterious variants and gene length was found in the three pharmacogenes groups (all P > 0.05). The others pharmacogenes group (totally 7 genes) had the significant correlation between the number of total variants (P < 0.0001) and gene length, while the relationship between the number of deleterious variants (P = 0.0559) and missense variants (P = 0.0299) and the corresponding gene length in this group were not significant.
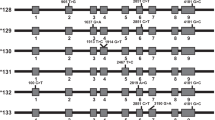
Different VAF with clinical relevance among CMDB and gnomAD populations
To demonstrate the VAF in CMDB Chinese and other populations, we chose the common functional variants in ABCC4, SLCO1B1, ALDH2, TPMT, UGT1A1, VDR and NUDT15 from the 6 pharmacogenes groups. Diverse variant allele frequencies of the chosen 8 variants in different gnomAD populations (African, Ashkenazi Jewish, Amish, Middle Eastern, Latino, European (non-Finnish), European (Finnish), East Asian, and South Asian) and CMDB Chinese were found as shown in Table 1. The P values are shown in Supplementary Table S1. Especially some common functional variants were population-specific for CMDB Chinese because they were absent (with a zero of VAF) or very rare in other gnomAD populations, including NUDT15 13:48611934 G/A (rs186364861), UGT1A1 2:234676872 C/T (rs34946978), and ALDH2 12:112241766 G/A (rs671). These genetic polymorphisms in the pharmacogenes in different populations might impact the drug response in the kinds of ethnicities.
Discussion
The present study provided a comprehensive and systematic genetic overview of 125 pharmacogenes including 16 ABC transporters, 50 SLC transporters, 17 non-CYP phase 1 enzymes, 26 phase 2 enzymes, 9 nuclear receptors and 7 others in Chinese from 141,431 whole genome sequences preserved in CMDB. To the best of our knowledge, this is the first study on the genetic variation of 125 pharmacogenes in such a large-scale population in mainland Chinese. Europeans were over-representative in genomes sequencing projects such as gnomAD worldwide, whereas other populations including Chinese were under-representative in these projects. Therefore, it was not appropriate to use the results from these projects directly on other populations. Moreover, drug development trials neglected to study participants of diverse ancestries would result in poor generalizability of marketed drug efficacy information. Additionally, the withdrawal of marketed drugs due to adverse drug reactions might be attributed to the patients with specific genetic variation, while in fact they would be efficacious for other patients without such certain genetic variant. For example, functional alleles SLCO1A2*2 and SLCO1A2*3 involved in drug uptake of opioid receptor agonist deltorphin II and methotrexate were prevalent in Ashkenazim (VAF = 0.145 and 0.034, respectively) and Europeans (VAF = 0.136 and 0.062, respectively) while absent in East Asians (both VAF < 0.001)9. And our present study found some common functional pharmacogene variants such as NUDT15 rs186364861, UGT1A1 rs34946978 and ALDH2 rs671 in CMDB Chinese were population-specific but absent in other populations (Table 1). These results were of key importance in designing the pharmacogenomics testing panel in Chinese. FDA (Food and Drug Administration) had provided 457 drug-gene pairs in the table of pharmacogenomics biomarkers in drug labeling (https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling), and before the whole genome sequencing becomes the routine testing in clinics, a cost-effective pharmacogenomics testing panel covering an individualized pharmacogenes variants testing items would be needed15.
The limitation of our present study is that lacking of haplotype information across pharmacogenes which is a main source of actionable variation for translating these data to clinically meaningful conclusions. According to a review by the Human Genetic Resources Administration of China (HGRAC), the individual genetic data from CMDB is unavailable12. We could obtain detailed summaries of the data such as allele frequencies and GWAS summary statistics. Unfortunately, ethnicity-specific allele frequencies across pharmacogenes are unavailable. However, the minorities in mainland China such as Mongolian, Tibetan and Uyghur populations owned different alleles and genotypes frequencies of pharmacogenes from those in Han Chinese16. Therefore, more detailed pharmacogenomic studies in these minorities would be needed to perform in future.
Genetic variability of the well-known pharmacogenes alleles could explain only part of the inter-individual differences in drug responses, and rare genetic variants in pharmacogenes such as SLC30A8 could modulate antidepressant (desipramine or fluoxetine) treatment17. The potential role on the disease development and drug efficacy of the rare variants in these genetically polymorphic pharmacogenes would be interesting and needed to be studied further18,19. The present study demonstrated that 91.1% of the 617 deleterious missense variants in the 125 pharmacogenes were rare (Fig. 3b), which suggested the necessity to focus on the rare pharmacogenes variants in studying inter-individual variability in drug response. Analysis of 25 clinically relevant pharmacogenes in 291 genomes of the Thai population identified 121 putatively functional variants, majority of which were rare and specific to the Thais but absent from gnomAD database20. As more and more rare pharmacogene variants were found, the method to interpret their clinical implication were needed to be developed urgently21,22. Therefore, the database comprised of both the pharmacogenes genotype and drug response phenotype information should be constructed across the world.
Most of the drugs are metabolized by several enzymes, and often a combination of CYPs and non-CYPs. Although the majority of drug biotransformation is performed by the CYP enzymes, other pharmacogenes such as drug transporters and nuclear receptors might participate in many clinically important drugs23,24. Constitutive androstane receptor (CAR) rs2502815 polymorphism and the carbamazepine response in epilepsy patients was potentially relevant. However, our present study found the rare variants in CAR (NR1I3) formed the 100% fraction of the functional variation and the aggregated putatively functional variants frequency of CAR was 0.00305. These results might explain the minor role played by the non-CYP pharmacogenes in the pharmacogenetics studies, where functional alleles in these pharmacogenes were not considered because of the very low VAF in the studied populations. Twin studies on the pharmacokinetics of metoprolol and torsemide suggested that up to 90% of the variation in their pharmacokinetic parameters could be allotted to the subjects’ genetic makeup, whereas the known genetic variations of CYP2D6, CYP2C9, and SLCO1B1 explained only 39%, 2%, and 39% of the pharmacokinetics variability, respectively25. These findings combined with our present results indicated that a substantial fraction of the heritable variation in the drug responses of clinically important medicines remained to be elucidated.
In conclusion, we comprehensively mapped the genetic landscape of 125 pharmacogenes in mainland Chinese from CMDB and identified 38,188 variants. Computational analyses of the 2554 exonic variants identified 617 deleterious missense variants, 91.1% of which were rare, and of the 54 loss-of-function (splice acceptor, splice donor, start lost, and stop gained) variants, 53 (98.1%) were rare. These results suggested an enrichment of rare variants in functional ones for pharmacogenes. Certain common functional variants including NUDT15 13:48611934 G/A (rs186364861), UGT1A1 2:234676872 C/T (rs34946978), and ALDH2 12:112241766 G/A (rs671) were population-specific for CMDB Chinese because they were absent (with a zero of VAF) or very rare in other gnomAD populations. These findings might be useful for the further pharmacogenomics research and clinical application in Chinese.
References
Zanger, U. M. & Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141 (2013).
Anderson, H. D., Crooks, K. R., Kao, D. P. & Aquilante, C. L. The landscape of pharmacogenetic testing in a US managed care population. Genet. Med. 22, 1247–1253 (2020).
Youssef, E., Kirkdale, C. L., Wright, D. J., Guchelaar, H. J. & Thornley, T. Estimating the potential impact of implementing pre-emptive pharmacogenetic testing in primary care across the UK. Br. J. Clin. Pharmacol. 2, 2 (2021).
Liu, N. et al. Association of ABCC2 polymorphism and gender with high-density lipoprotein cholesterol response to simvastatin. Pharmacogenomics 19, 1125–1132 (2018).
Du, Z. et al. Influence of UGT2B7 and UGT1A6 polymorphisms on plasma concentration to dose ratio of valproic acid in Chinese epileptic children. Xenobiotica 2, 1–6 (2021).
Pan, Y. et al. F2R polymorphisms and clopidogrel efficacy and safety in patients with minor stroke or TIA. Neurology 96, 1–9 (2021).
Abecasis, G. R. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Zhang, B. & Lauschke, V. M. Genetic variability and population diversity of the human SLCO (OATP) transporter family. Pharmacol. Res. 139, 550–559 (2019).
Schaller, L. & Lauschke, V. M. The genetic landscape of the human solute carrier (SLC) transporter superfamily. Hum. Genet 138, 1359–1377 (2019).
Xiao, Q., Zhou, Y. & Lauschke, V. M. Ethnogeographic and inter-individual variability of human ABC transporters. Hum. Genet 139, 623–646 (2020).
Liu, S. et al. Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and Chinese population history. Cell 175, 347-359.e14 (2018).
Qi, G., Han, C., Sun, Y. & Zhou, Y. Genetic insight into cytochrome P450 in Chinese from the Chinese Millionome Database. Basic Clin. Pharmacol. Toxicol 126, 341–352 (2020).
Ingelman-Sundberg, M., Mkrtchian, S., Zhou, Y. & Lauschke, V. M. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum. Geno. 12, 26 (2018).
Zhu, Y. et al. A model-based cost-effectiveness analysis of pharmacogenomic panel testing in cardiovascular disease management: Preemptive, reactive, or none. Genet. Med. 23, 461–470 (2021).
Qi, G., Yin, S., Zhang, G. & Wang, X. Genetic and epigenetic polymorphisms of eNOS and CYP2D6 in mainland Chinese Tibetan, Mongolian, Uygur, and Han populations. Pharmacogenomics J 20, 114–125 (2020).
Wong, M. L. et al. Rare functional variants associated with antidepressant remission in Mexican-Americans. j. Affect. Disord. 279, 491–500 (2021).
Liu, X. Y. et al. Alflutinib (AST2818), primarily metabolized by CYP3A4, is a potent CYP3A4 inducer. Acta Pharmacol. Sin. 41, 1366–1376 (2020).
Yasar, U. & Babaoglu, M. O. Increased risk for cerebral ischemic stroke in diabetes: Genetically polymorphic CYP mediated production of neuroprotective EETs and sulfonylurea metabolism in relation with K(ATP) channels. Acta Pharmacol. Sin. 40, 569–570 (2019).
Mauleekoonphairoj, J. et al. Phenotype prediction and characterization of 25 pharmacogenes in Thais from whole genome sequencing for clinical implementation. Sci. Rep. 10, 18969 (2020).
Zhou, Y., Fujikura, K., Mkrtchian, S. & Lauschke, V. M. Computational methods for the pharmacogenetic interpretation of next generation sequencing data. Front. Pharmacol. 9, 1437 (2018).
Zhou, Y. & Lauschke, V. M. Computational tools to assess the functional consequences of rare and noncoding pharmacogenetic variability. Clin. Pharmacol. Ther. 2, 2 (2021).
Banerjee, A., Moreno, A., Pata, J., Falson, P. & Prasad, R. ABCG: A new fold of ABC exporters and a whole new bag of riddles. Adv. Protein Chem. Struct. Biol. 123, 163–191 (2021).
Kong, F. C., Ma, C. L., Lang, L. Q. & Zhong, M. K. Association of xenobiotic receptor polymorphisms with carbamazepine response in epilepsy patients. Gene 771, 145359 (2021).
Matthaei, J. et al. Heritability of metoprolol and torsemide pharmacokinetics. Clin. Pharmacol. Ther. 98, 611–621 (2015).
Acknowledgements
The study was supported by the grants from Young Investigators Foundation of The First Affiliated Hospital of Zhengzhou University (Grant No. YNQN2017125), Joint Projects of Medical Science and Technology Problem-Tackling Plan of Henan Province (Grant No. LHGJ20190270), Natural Science Foundation of Henan Province (Grant No. 212300410246) and Natural Science Foundation of China (Grant No. 81800546).
Author information
Authors and Affiliations
Contributions
G.Z.Q., J.M.Z., P.F.M. and C.H., conceived the project; G.Z.Q., supervised the project; G.Z.Q., J.M.Z., C.H., Y.B.Z. and D.L.L., performed the research; G.Z.Q., J.M.Z., and C.H., analyzed the data; and G.Z.Q. and C.H., wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Qi, G., Zhang, J., Han, C. et al. Genetic landscape of 125 pharmacogenes in Chinese from the Chinese Millionome Database. Sci Rep 11, 19222 (2021). https://doi.org/10.1038/s41598-021-98877-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-98877-x
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.