Abstract
Coagulase-negative staphylococci (CoNS) are the most frequent contaminating bacteria; therefore, we aimed to investigate an indicator of CoNS to predict the increase in blood culture contamination rate (ConR). We performed a retrospective study of selected patients, who underwent blood culture testing. Contamination was defined as the presence of either one of two or more sets of skin-resident bacteria, except for cases with a low likelihood of contamination based on clinical aspects. We calculated the monthly ConR [(total number of contaminated cases per month)/(total number of blood culture sets collected per month) × 100] and analysed the ConR prediction ability using the following four indicators: the number of CoNS-positive sets of blood cultures, cases with at least one CoNS-positive blood culture set, cases with only one CoNS-positive blood culture set, and cases of contamination by CoNS. Cases with CoNS-positive blood cultures correlated with ConR (r = 0.85). Although the area under the receiver operating characteristic curve for the number of cases with ConR ≥ 2.5 differed significantly from that of the number of cases contaminated by CoNS, the negative predictive value was high, reaching up to 95.5% (95% confidential interval 87.3–99.1). The number of CoNS-positive cases could help predict an increase in ConR ≥ 2.5.
Similar content being viewed by others
Introduction
Relevant culture tests are important for the appropriate use of antimicrobial agents to combat antimicrobial-resistant bacteria. However, these tests can result in increased contamination, leading to an excessive use of antimicrobial agents, contributing to longer hospital stays and higher costs1,2. The blood culture contamination rate (ConR) is calculated retrospectively based on certain criteria3; however, its calculation is time-consuming and requires additional labour. Coagulase-negative staphylococci (CoNS) are the most frequently detected bacteria in blood culture contamination2. Therefore, we intended to investigate a simple real-time indicator of CoNS for predicting the increase in ConR. The values of the four indicators were aggregated as follows: the number of CoNS-positive sets of blood cultures, cases with at least one CoNS-positive blood culture set, cases with only one CoNS-positive blood culture set, and cases of contamination by CoNS.
Results
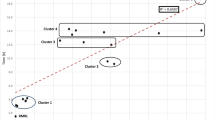
A total of 103,339 sets of blood cultures were collected during the study period. Of these, 12,670 sets were culture-positive. Of these, 594 cases (778 sets) were excluded, because of pending determination in 190 cases and no determination in 404 cases. Eligible blood cultures were collected from a total of 56,843 cases. Of these, 520 (0.9%) had three or more sets of specimens collected on the same day. CoNS was positive in 3126 sets, of which Staphylococcus epidermidis was included in 1771 sets (56.7%). A total of 2142 cases were determined as contaminated; of these, 1689 (78.9%) cases were contaminated with CoNS. The ConR was 2.5% or higher for 29 months of the total study period (26.8%) (Supplementary Fig. 1). The correlation coefficients with ConR for indicators A–D were 0.71, 0.85, 0.91, and 0.92, respectively (Fig. 1). The ROC curve is shown in Fig. 2, with the area under the curves (AUCs) (95% confidence interval) of 0.84 (0.76–0.92), 0.92 (0.86–0.97), 0.95 (0.91–0.99), and 0.96 (0.92–0.99), respectively. When AUCs for each indicator were compared, we found that A vs. B, A vs. C, A vs. D, B vs. C, and B vs. D were statistically significant with Holm correction (Supplementary Table 1). The sensitivity was 86.2%, 89.7%, 89.7%, and 89.7% and the specificities were 67.1%, 79.7%, 87.3%, and 87.3%, with cut-off values of 29, 23, 19, and 19 cases for indicators A–D, respectively; all with a high negative predictive value (95% confidential interval) of 93.0% (83.0–98.1), 95.5% (87.3–99.1), 95.8% (88.3–99.1), and 95.8% (88.3–99.1), respectively. We compared the predictive ability of monthly contamination rate of 2.5 or higher depending on whether the detected organism was S. epidermidis or other CoNS for each sample, but there was no significant difference (AUC 0.784 vs. 0.801, p = 0.76, Supplementary Figure).
Discussion
These results suggest that the indicators A and B, which can be counted simply, correlated with ConR to predict a ConR of ≥ 2.5, or higher, but their prediction ability was inferior to that of indicators C and D in multiple comparisons of ROC curves. High negative predictive rates were observed for all indicators, indicating that if the number of cases with CoNS detection or CoNS positivity remained low, it was unlikely that the ConR would increase. The sensitivity and specificity for predicting ConR ≥ 2.5 were higher when indicators C or D were used, the former requires time until another blood culture set collected at the same time is determined to be negative, and the latter requires human resources and time to determine contamination. In contrast, indicators A and B do not require much human resources and can be displayed in real time. Most hospitals calculate ConR once a month; however, approximately 30% of the facilities calculate it over a longer time span4. Indicators A and B could be good predictors for ConR in such institutes.
ConR of ≤ 3.0 is often used as a standard for the quality of blood culture tests5. In this study, we set the predicted ConR to be ≥ 2.5. When the cut-off was increased, the predictive power of each index increased, because the number of months covered decreased; however, there was no difference in the trend of AUC between 2.5% or higher and 3.0% or higher (Supplementary Table 2). ConR is related to the disinfection of the puncture site, collection method, hand hygiene, education, and feedback methods regarding collection6. Recently, the usefulness of a blood collection device was reported7. These relevant factors can be reviewed when CoNS-positive cases increase. In addition, feedback from monitoring the results alone can improve the ConR6,8. Therefore, establishing a system that provides real-time feedback on the number of cases of CoNS detection could be a countermeasure to reduce contamination without requiring additional labour. However, the situation regarding blood cultures varies from hospital to hospital4. For example, in facilities with many patients with central venous catheters, true infection by CoNS is more common. In addition, the target ConR varies; and therefore, the results should not be applied directly to facilities that have not set a ConR target of 2.5 or higher. The results of this study, especially the cut-off values, may not be directly applicable to other facilities. However, as CoNS is the most commonly detected organism in contamination, a similar result can be predicted if a cut-off is set and the correlation with ConR at least once at one’s own institution is determined before using it as a simple indicator.
Methods
Study design
We performed a retrospective study of patients, who underwent blood culture testing at the National Center for Global Health and Medicine between April 2012 and March 2021. The need for informed consent was waived, because of the retrospective nature of the study design. The study information was presented on the Web for the possibility of opting out of consent. This was substituted for the participants’ consent. The protocol of this study including the opt-out consent method was approved by the Certificate Review Board of National Center for Global Health and Medicine (NCGM-G-004168-00) and conformed to the amended Declaration of Helsinki. The data were compiled from the registry of blood culture surveillance, including data on contamination, and the microbiology laboratory.
Identification of bacterial species
All blood culture samples were collected into standard aerobic and anaerobic culture bottles (92F or 94F and 93F, 23F or 20F and 24F Becton Dickinson Microbiology Systems, Sparks, MD, USA) and processed using the BACTEC 9240, 9120, and FX systems (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). These samples were routinely monitored for at least 144 h. The bottles that tested positive were removed and subjected to Gram staining. The specimens were then inoculated into 5% sheep blood agar and BTB agar media (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan), and incubated at 35 °C (Depending on the situation, other media may be added, or the environment may be changed, such as anaerobic incubation). Conventional bacterial identification and susceptibilities to the predefined antimicrobials were determined in accordance with the Clinical and Laboratory Standard Institutions criteria (M100)9,10 using matrix assisted laser desorption/ionization-time of flight mass spectrometry system (MALDI Biotyper system; Bruker, Billerica, MA, USA) and automated broth micro dilution system (MicroScan WalkAway 96 SI system; Beckman Coulter, Brea, CA, USA). All Staphylococci species, except S. aureus and S. lugdunensis, were treated as CoNS.
Blood sampling
The standard procedure recommended that the puncture site be cleaned with an alcohol swab, disinfected twice with poppidone-iodine, and then puncturing a vein or artery to collect a blood specimen. It was recommended to collect blood on two or more different sites. The use of blood collection during catheterization and catheter regurgitation is not recommended, but was allowed. The recommended collection volume is 10 mL of blood per bottle for adult bottles (92F and 93F, 23F and 24F) and 1–3 mL per bottle for paediatric bottles (94F and 20F). Compliance with disinfection methods and sample collection volumes was not monitored.
Definition of contamination
Contamination was defined as the presence of either one of the two or more sets of skin-resident bacteria (CoNS, Bacillus spp. excluding B. anthracis, Corynebacterium spp., Cutibacterium spp., Streptococcus Viridans group, Aerococcus spp., Micrococcus spp.) listed in the CUMITECH3. However, even in cases where these bacteria were detected, if a central venous catheter or an intravascular device showed obvious signs of infection such as redness, fever, pus drainage, or pus accumulation, and the patient's condition clearly improved with the administration of a susceptible antimicrobial agent, these cases were not be treated as contamination. In case of enterococci and Clostridium spp., if only one set was positive out of two or more sets collected, and there was no obvious foci of infection and the symptoms improved without administration of a susceptible antimicrobial agent, it was considered a contamination. These decisions were made retrospectively, based on the medical records, by infectious disease specialists who perform blood culture surveillance.
The registry data of blood culture surveillance
For every case, two or more infectious disease physicians of the National Center for Global Health and Medicine determined whether the case was contaminated from a clinical point of view, by reviewing clinical records and laboratory data in accordance with the aforementioned criteria. Undetermined cases and those with pending determination were excluded from the study.
Indicators
We calculated the monthly ConR [(total number of contaminated cases per month)/(total number of blood culture sets collected per month) × 100]3. The values of the four indicators were aggregated as follows: the number of CoNS-positive sets of blood culture (Indicator A), cases with at least one CoNS-positive blood culture set (Indicator B), cases with only one CoNS-positive blood culture set (Indicator C), and cases of contamination by CoNS (Indicator D).
Statistical analysis
Correlation coefficients were calculated using Pearson’s correlation test. Receiver operating characteristic (ROC) curves were prepared for all indicators with a ConR of ≥ 2.5, as the objective variable, and cut-off values were calculated using Youden's index. The AUCs were compared using the Delong method with the Holm correlation. Statistical analysis was performed using EZR for Windows version 1.5411. Figures were created using IBM SPSS Statistics software for Windows (version 26.0; IBM Corp., Armonk, NY, USA). The probability of significance was calculated to be 5%.
Ethics approval
The need for informed consent was waived due to the retrospective nature of the study design. The study information was presented on the Web for the possibility of opting out of consent. The protocol was approved by the institutional review board of the National Center for Global Health and Medicine (NCGM-G-004168-00).
Consent to participate
The study information was presented on the Web for the possibility of opting out of consent.
References
Souvenir, D. J. et al. Blood cultures positive for coagulase-negative staphylococci: Antisepsis, pseudobacteremia, and therapy of patients. J. Clin. Microbiol. 36, 1923–1926. https://doi.org/10.1128/JCM.36.7.1923-1926.1998 (1998).
Alahmadi, Y. M. et al. Clinical and economic impact of contaminated blood cultures within the hospital setting. J. Hosp. Infect. 77, 233–236. https://doi.org/10.1016/j.jhin.2010.09.033 (2011).
Baron, E. J. et al. CUMITECH 1C, Blood Cultures IV (ASM Press, 2005).
Garcia, R. A., Spitzer, E. D., Kranz, B. & Barnes, S. A national survey of interventions and practices in the prevention of blood culture contamination and associated adverse health care events. Am J. Infect Control. 46, 571–576. https://doi.org/10.1016/j.ajic.2017.11.009 (2018).
Schifman, R. B., Strand, C. L., Meier, F. A. & Howanitz, P. J. Blood culture contamination: A College of American Pathologists Q-Probes study involving 640 institutions and 497134 specimens from adult patients. Arch. Pathol. Lab. Med. 122, 216–221 (1998) (PMID: 9823858).
Dargère, S., Cormier, H. & Verdon, R. Contaminants in blood cultures: Importance, implications, interpretation and prevention. Clin. Microbiol. Infect. 24, 964–969. https://doi.org/10.1016/j.cmi.2018.03.030 (2018).
Rupp, M. E., Cavalieri, R. J., Marolf, C. & Lyden, E. Reduction in blood culture contamination through use of initial specimen diversion device. Clin. Infect. Dis. 65, 201–205. https://doi.org/10.1093/cid/cix304 (2017).
Gibb, A. P., Hill, B., Chorel, B. & Brant, R. Reduction in blood culture contamination rate by feedback to phlebotomists. Arch. Pathol. Lab. Med. 121, 503–507 (1997) (PMID: 9167605).
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement M100-S22 (Clinical and Laboratory Standards Institute, 2012).
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Sixth Informational Supplement M100-S26 (Clinical and Laboratory Standards Institute, 2015).
Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 48, 452–458. https://doi.org/10.1038/bmt.2012.244 (2013).
Author information
Authors and Affiliations
Contributions
K.Y.: conception and design of the study, and analysed and interpretated of data; K.Y. and K.M.: acquired data; K.Y.: wrote the draft of the article; and K.M. and N.O.: revised it critically for important intellectual content. All authors reviewed the manuscript and approved the final version to be submitted.
Corresponding author
Ethics declarations
Competing interests
K.Y. has received research grants from Fujirebio, Inc., Mizuho Medy, Co., Ltd., and VisGene Inc., outside the submitted work. N.O. has received grants from Sanofi Pasteur and Eiken Chemical Co., Ltd., outside the submitted work. K.M. declares no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yamamoto, K., Mezaki, K. & Ohmagari, N. Simple indictor of increased blood culture contamination rate by detection of coagulase-negative staphylococci. Sci Rep 11, 17538 (2021). https://doi.org/10.1038/s41598-021-96997-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-96997-y
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.