Abstract
The cardiorenal syndrome (CRS) is defined as the confluence of heart-kidney dysfunction. This study investigates the molecular differences at the level of the urinary peptidome between CRS patients and controls and their association to disease pathophysiology. The urinary peptidome of CRS patients (n = 353) was matched for age and sex with controls (n = 356) at a 1:1 ratio. Changes in the CRS peptidome versus controls were identified after applying the Mann–Whitney test, followed by correction for multiple testing. Proteasix tool was applied to investigate predicted proteases involved in CRS-associated peptide generation. Overall, 559 differentially excreted urinary peptides were associated with CRS patients. Of these, 193 peptides were specifically found in CRS when comparing with heart failure and chronic kidney disease urinary peptide profiles. Proteasix predicted 18 proteases involved in > 1% of proteolytic cleavage events including multiple forms of MMPs, proprotein convertases, cathepsins and kallikrein 4. Forty-four percent of the cleavage events were produced by 3 proteases including MMP13, MMP9 and MMP2. Pathway enrichment analysis supported that ECM-related pathways, fibrosis and inflammation were represented. Collectively, our study describes the changes in urinary peptides of CRS patients and potential proteases involved in their generation, laying the basis for further validation.
Similar content being viewed by others
Introduction
The cardiorenal syndrome (CRS) is a complex pathological disorder, which reflects the interplay between heart and chronic kidney diseases1. Epidemiologic data reveal that about 45–63% of chronic heart failure (HF) patients developed chronic kidney disease (CKD)2. CRS is classified into five subtypes, referring to primary organ dysfunction, each with different underlying pathological mechanisms3. The term "CRS" has been applied to the relation of these two diseases, but the definition and classification have not been clearly established on a molecular pathophysiologic level until now4. However, multi-factorial mechanisms including hemodynamic changes, fibrosis, vascular calcification, neurohormonal activity, immunologic imbalance, inflammation, apoptosis, endothelial injury, thrombosis, and oxidative stress have been proposed to explain the general pathophysiology of CRS5.
In parallel to the efforts towards understanding disease pathophysiology, a number of biomarkers for heart and kidney disease have been studied in the context of CRS. These include changes in the levels of heart-specific biomarkers such as cardiac troponin T (cTnT) and B-type natriuretic peptide (BNP)6,7, but also kidney-specific biomarkers including albumin (ALB), kidney injury molecule-1 (KIM-1)8, liver-type fatty acid binding protein (L-FABP)9, neutrophil gelatinase-associated lipocalin (NGAL)10, β-2 microglobulin (B2M)11, n-acetyl-beta-d-glucosaminidase (NAG)12 and cystatin C13, (reviewed in14).
Urinary peptidomics focuses on the analysis of naturally occurring peptides and small proteins in urine. Application of the approach enabled identification of biomarkers associated with kidney and heart diseases including, among others, CKD15, kidney fibrosis16, diabetic kidney disease (DKD)17, acute kidney injury (AKI)18 as well as HF19, asymptomatic LV diastolic dysfunction (LVDD)20, and ischemic and dilated cardiomyopathy21; with a biomarker panel for CKD also having reached the level of clinical implementation for patient stratification in large clinical trials22.
Due to the lack of specific CRS biomarkers on a molecular level, we designed the current study to identify molecular differences at the level of the urinary peptidome between CRS and matched controls, prompted by the availability of high resolution-large sized datasets. In addition, we targeted to investigate if the observed urinary changes reflect molecular processes associated with CRS and its underlying pathologies.
Results
Cohort characteristics
The urine peptidomics datasets were screened for the availability of clinical information on both kidney and heart failure. Based on these criteria, 3463 urinary peptidomics datasets were retained for investigation. Matching resulted in the inclusion of a total of 709 datasets (CRS; n = 353, controls; n = 356) in the study, which comprised the main groups under comparison (CRS versus the respective matched controls). The CRS patients had an estimated glomerular filtration rate (eGFR) mean value of 43.1 ml/min/1.73 m2 whereas controls had 79.2 ml/min/1.73 m2. A number of the clinicopathological characteristics including the NT-proBNP levels, the serum creatinine levels, the systolic blood pressure, the diastolic blood pressure were statistically significant different between the two groups. The characteristics of the matched cases-controls are presented in Table 1.
Urine peptidome analysis
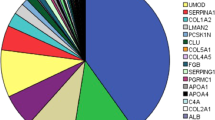
A total of 3184 peptides were detected and considered when comparing CRS patients with controls. After applying the Benjamini-Hochberg (BH) adjustment, 1480 differentially excreted peptides (BH, P < 0.05) were identified when comparing cases with controls. To shortlist the more representative features, a 30% frequency threshold, frequently used in such analyses23,24 was applied, which resulted in a final list of 559 differentially excreted peptides (see Fig. 1a and Supplementary Table S1). These peptides derived from 110 unique protein precursors, which based on functional annotation, reflected largely extracellular matrix (ECM) changes, cell-ECM interactions, collagen formation/degradation, metabolic and inflammatory processes. The top 15 most frequently observed protein precursors, associated with at least 400 differentially excreted peptides are presented in Fig. 1b. Of these 559 differentially excreted peptides, 313 (55.9%) originated from fibril-forming collagens and were fragments of collagen type I alpha 1 chain (COL1A1), collagen type I alpha 2 chain (COL1A2), and collagen type III alpha 1 chain (COL3A1). Together the collagens accounted for 72% of the quantified peptidome. The 20 most significant differentially excreted peptides included fragments of additional collagen types (COL4A1, COL4A3, COL9A3, COL5A1, COL5A2, COL5A3 and COL19A1) and other peptides originating from plasma proteins (such as apolipoprotein A1 (APOA1) and B2M) (see Table 2). Among the peptides showing the most prominent increase in abundance in patients versus controls were fragments of B2M, albumin, and alpha-1-antitrypsin (A1AT), whereas fragments of collagen types (IV, V and VI), clusterin (CLU) and ubiquitin-associated protein 1-like (UBAP1L) were the peptides with the most decreased abundance levels in CRS (see Fig. 1c).
Urinary peptides in CRS. (a) Volcano plot showing the fold change (log2 CRS/Control) plotted against the BH P-value (− log10) of the 559 differentially excreted peptides between CRS and controls. The top five most representative protein precursors including COL1A1, COL3A1, COL1A2, FGA and UMOD are highlighted in different colors. (b) Pie chart of the top 15 most frequently observed protein precursors, associated with at least 400 differentially excreted CRS-associated peptides. The percentage of the differentially excreted peptides per protein precursor is presented. 55.9% of CRS-associated peptides originated from fibril-forming collagens and were fragments of COL1A1, COL1A2, and COL3A1. (c) Heatmap of the log2 transformed fold change of the top 10 most increased and decreased peptides in the CRS cohort. In each case, the peptide codes are provided in Supplementary Table S1.
Urinary peptide differences in CRS, CKD and HF
We further investigated if common peptides were identified between patients with HF and CRS as well as between CKD and CRS. We compared the 559 peptides of our study with the 218 and 577 differentially excreted peptides which were associated with CKD and HF, respectively, and described in previous studies15,19. Of these 218 CKD-associated peptides, 31 (14.2%) were commonly identified in our study as well. These common 31 peptides were sequenced fragments of albumin, B2M, A1AT, APOA1, alpha-1B-glycoprotein (A1BG), alpha-2-HS-glycoprotein (AHSG), sodium/potassium–transporting ATPase subunit γ (FXYD2), osteopontin (SPP1), collagen types I and III and other proteins (see Supplementary Table S2). In addition, 341 out of 577 (59%) of the HF-specific peptides were commonly identified in CRS patients (see Supplementary Table S3). These shared peptides showed the same directionality of difference and originated mostly from collagen types (I, II, III, IV, V, IX, XI), B2M, A1AT and uromodulin (UMOD).
Overall, 193 of 559 (34.5%) CRS-associated peptides were not among those identified in the HF and CKD studies (see Supplementary Table S4). Most of these 193 peptides, which were found in the CRS cohort only, were collagen fragments (n = 134, 69.4%), with collagen types I, II, III, IV and V represented by the largest number of collagen fragments (n = 117, 60.6%). The top most represented protein precursors were collagen types I (40.9%) and III (9.3%), UMOD (4.1%) fibrinogen alpha chain (FGA, 2.6%), COL5A2 (2.1%), CD99 (1.6%), CLU (1.6%), FXYD2 (1.6%) and polymeric immunoglobulin receptor (PIGR, 1.6%).
On a protein level, we further investigated if any of the protein precursors represented by 193 CRS-associated peptides was not identified in the CKD and HF urinary profiles. Totally, 30 protein precursors were detected only in CRS cohort but not in HF and CKD. These protein precursors included several collagen types (COL4A2, COL4A4, COL6A5, COL7A1, COL8A1 and COL13A1), PIGR, secreted and transmembrane protein 1 (SECTM1), ankyrin repeat domain-containing protein 17 (ANR17), ubiquitin-like protein ATG12 (ATG12), zinc-alpha-2-glycoprotein (AZGP1) and monocyte differentiation antigen CD14 (CD14) (and other proteins listed in Supplementary Table S5).
Protease prediction analysis
Proteases possibly implicated in the endogenous cleavage of the differentially excreted peptides were predicted by Proteasix tool (http://www.proteasix.org). The in silico analysis revealed that 117 of the 559 differentially excreted peptides were known substrates of 27 reported in the literature proteases. Of these 27, 18 shortlisted proteases had a percentage of cleavage events above 1% and are presented in Fig. 2a. Among the shortlisted proteases were multiple metalloproteinases (see Fig. 2a-in red), 4 proprotein convertases (see Fig. 2a-in blue), 2 cathepsins (see Fig. 2a-in green) and kallikrein 4 (KLK4). The 44% of the cleavage events were produced by 3 metalloproteinases (i.e. MMP13, MMP9 and MMP2). However, the most abundant diffrentially excreted peptides were putatively cleaved by the four proprotein convertases, KLK4 and MMP26, as shown in the heatmap (see Fig. 2b). For comparison, when analyzing the CRS-specific peptides (n = 193), similarly, 11 metalloproteinases and 4 cathepsins were predicted (see Fig. 2a marked with an asterisk), all included in the predictions from the 559 differentially excreted peptides.
In silico predicted proteases. (a) The 27 predicted proteases are presented. Of these, 18 were linked to ≥ 1% of cleavage events and are highlighted in red, blue and green whereas 9 had < 1% of cleavage events (highlighted in grey). The 15 predicted proteases which were involved in the generation of the 193 CRS-specific peptides are marked with an asterisk. The catalytic mechanism of the shortlisted proteases is also depicted. (b) Heat map of the log2 transformed abundance of the differentially excreted peptides predicted as substrates of 18 shortlisted proteases by the Proteasix. The predicted proteases which were associated with the 193 CRS-specific peptides are marked with an asterisk.
Gene Ontology (Go) enrichment analysis of the 18 shortlisted proteases showed the over-representation of metalloproteinases (i.e. MMP3, MMP12, MMP14, MMP20, MMP26, MMP1, MMP2, MMP13, MMP9, MMP20, MEP1B, MMP8) in zinc ion binding and transition metal ion binding molecular functions. Together, metalloproteinases (i.e. MMP12, MMP13, MMP9) along with 2 cathepsins (i.e. CTSK, CTSS) displayed collagen-binding activity.
Pathway enrichment analysis of the shortlisted proteases along with the protein precursors of the differentially excreted peptides was performed by Metascape. The analysis revealed that ECM-related pathways (i.e. ECM organization and degradation) were significantly affected. Together, cathepsins K and S along with various metalloproteinases (i.e. MMP1, MMP2, MMP3, MMP8, MMP9, MMP12, MMP13, MMP14, MMP20 and MMP26) and KLK4 were mostly involved in the structure, organization and degradation of ECM. Additionally, pathway analysis highlighted that the predicted proteases along with the protein precursors were also inolved in collagen formation, protein processing, regulation of inflammatory response and neutrophil mediated immunity (see Fig. 3).
Pathway enrichment analysis. Pathway enrichment analysis of the 18 shortlisted proteases along with the 110 protein precursors of the differentially excreted peptides. The six most significant clusters are presented in different colours and the most significant term of each cluster was selected as label. For each term the corrected P-value along with the associated gene names are presented. The associated proteases are highlited in red colour.
Discussion
In the present case–control matching study, a large-scale urine peptidome analysis of patients with CRS and matched controls was performed, targeting to identify the disease-specific urine peptidome profile and its potential links to pathophysiology. Importantly, our study identifies a high number of differentially excreted urine peptides between CRS patients and matched controls.
The results of the current study suggest that the urine peptidome of CRS patients integrates changes of 559 peptides originated from various protein precursors. These peptides originate to a large extent from proteins involved in ECM, collagen formation/degradation, inflammation, metabolism but also transcriptional regulation reflecting the underlying associated mechanisms25. A large number of overlapping urine peptides between CRS and HF as well as CRS and CKD patients were expected, as the examined patients of our study have combined both pathologies. These overlapping peptides derived, among others, from plasma proteins which are well-known renal and heart failure biomarkers including albumin, B2M, SPP1, AHSG, but also uromodulin and collagens26. In addition, to the above overlapping protein precursors, two proteins including, APOA1 (a protein which is associated with renal dysfunction in HF patients27) and FXYD2 (a protein which mediates the function of the Na, K-ATPase in mammalian kidney epithelial cells28) are commonly identified in CRS and CKD, similarly reflecting the common underlying molecular profiles of these diseases.
The highly abundant COL1A1, COL1A2, and COL3A1 proteins were represented by multiple urinary fragments, reflecting most likely fibrosis and changes in the ECM in both organs. In a healthy heart, the ECM is more than 95% composed of the fibrillar profibrotic collagens, including collagen types I and III29, important for maintaining the tensile strength, elasticity and extensibility of the myocardium29. Alterations of collagen type I and III expressions have been associated with myocardial fibrosis in HF patients30 and vascular calcification in the heart31 in animal studies. Similarly, collagen types I and III are the most abundant collagen types in the renal ECM32 linked to ECM dysregulation and CKD progression33. In addition, the activity of proteases may also be linked with the high number of the detected collagen-related peptides as it is well reported that metalloproteinases regulate cardiac and renal remodeling as well as fibrosis by facilitating ECM turnover and inflammatory cells34,35.
Further expected changes based on current knowledge were consistently detected. Among the most pronounced was the selective enrichment of increased albumin peptides in CRS, as expected for renal dysfunction and consequently albuminuria. Similarly, we observed an enrichment of peptides derived from COL9A3 and COL5A3, which are selectively highly expressed at the protein level in the heart and not detected in the kidneys based on proteomics databases (ProteomicsDB). Along the same lines, the abundance of a matrix GLA peptide (MGP) was strongly increased in CRS patients. MGP was already described as a calcification inhibitor protein with a strong association to HF indices and mortality36.
Similarly, in line with the literature, among protein precursors represented by decreased fragments in CRS patients were COL4A1 and COL4A3, collagens which are associated with nephropathy37. Furthermore, circulating proteins portrayed by increased fragments in CRS included AHSG (a protein which is associated with vascular calcification, cardiovascular mortality and kidney dysfunction38), APOA1 (a protein which is associated with HF and kidney dysfunction27), B2M (a protein which is associated with cardio-renal remodeling11 and inflammation39), COL18A1 (endostatin, generated from COL18A1, is associated with the development of cardiovascular events in CKD patients40), MGP and thymosin beta-4 (TMSB4X, a protein which is associated with renal fibrosis41).
Protein precursors detected only in CRS cohort but not in HF and CKD were also investigated. These included several collagen types, including COL4A2, COL4A4, COL6A5. Although few studies have aimed to investigate the role of collagen type IV and VI in cardio and renal failure, recently, it was suggested that COL4A2 is associated with cardiac fibrosis phenotype42, as well as with glomerular basement membrane alterations43, suggesting that COL4A2 may play an important role in CRS. In addition, COL4A4 and COL6A5 are associated with CKD44,45 but no evidence was found to link COL4A4 and COL6A5 with HF. Interestingly, two additional proteins, ROBO1 and HUWE1 are linked with cardiac and renal fibrosis46,47 whereas ANR17 protein may play a key role in the formation and maintenance of the blood vessels48. Moreover, several proteins associated with the immune system were uniquely found in the CRS cohort. These proteins were CD14 (a protein which is associated with heart and renal dysfunction49), CD99 and IRF6, suggesting the importance of the immune response in CRS. In addition, urinary PIGR peptides have been previously associated with cardiorenal dysfunction50, nevertheless, the exact role of the protein in the disease pathology remains unknown.
Our protease prediction analysis suggested the activity of 18 proteases responsible for more than 1% of the cleavage events. These predictions can be mostly supported by the existing bibliography on cardiovascular and renal pathologies. Notably, 9 out of the 11 predicted metalloproteases (MMP13, MMP9, MMP2, MMP20, MMP3, MMP12, MMP14, MMP8 and MMP1) were previously positively correlated with HF and were increased after myocardial infarction34,51. Interestingly, the transcriptional activation of MMP13 induces the vascular smooth muscle cell (VSMC) apoptosis and ECM breakdown via the FOXO3a activation52. These observations can be reflected by the contribution of these metalloproteases to vascular and kidney damage. Specifically, MMP2, MMP3, MMP8, MMP9 and MMP12 are involved in the ECM deposition in the glomeruli; MMP2, MMP3, MMP9, MMP13 and MMP14 induce epithelial-to-mesenchymal transition that leads to kidney fibrosis; and MMP2, MMP3 and MMP9 are correlated with vascular calcification, arterial stiffening and atherogenesis53. As reflected also by the cleavage events in our results, MMP2 and MMP9 have a prominent role in these processes and their levels have been associated with cardiovascular events and HF in various studies54,55. The role of MEP1B and MMP26 predicted by our analysis to mediate a large number of cleavage events, yet not previously linked to renal or heart dysfunction, apparently merits further investigation in the context of CRS.
From the 4 predicted proprotein convertases, three PCSK6 PSCK5 and PCSK7, have been previously associated with heart failure, to our knowledge. PCSK6 converts procorin to corin which in turn can activate natriuretic peptides, regulating cardiovascular and renal function56. Interestingly, PCSK6 is a key regulator of smooth muscle cell function (SMCs) in vascular remodeling and a novel player in cardiac remodeling after myocardial infraction57,58. Additionally, the inactivation of PCSK6 along with PCSK5 in endothelial cells leads to decreased collagen deposition and cardiovascular hypotrophy via IGF-1/Akt/mTOR signaling59. Moreover, PCSK7 is associated with both cardiovascular disease (CVD)60 and end-stage kidney disease61. However, the role of PCSK4 and KLK4 has not yet been linked to HF or CKD and would merit further investigation in the context of CRS based on our results.
Regarding cathepsins, elevated levels of CTSK were correlated with the presence of chronic HF62 and with major adverse cardiac and cerebrovascular events in CKD patients63, while circulating CTSS levels increase with CKD progression and GFR decline64. Levels of the mRNA, protein and activity of CTSS were found increased in the left ventricular myocardium of humans and rats with HF compared with controls, suggesting its participation in cardiac remodeling65. CTSS was also shown to affect epithelial-to-mesenchymal transition and ECM deposition in mouse models of mild and severe hydronephrosis, indicating its role in the regulation of renal fibrosis66.
Collectively, the pathology of CRS is complex and a number of pathways including ECM-related, fibrosis and inflammation are involved. Urinary peptidomics analysis reflects such CRS-associated changes, occasionally overlapping, as expected, with changes earlier observed in HF and CKD; but also alterations such as collagen type IV (COL4A2 and COL4A4), type VI (COL6A5), HUWE1, CD14, ANR17, PIGR and ROBO1 as well as a number of predicted proteases including MEP1B, MMP26, PCSK4 and KLK4, meriting further investigation in the context of CRS.
Among the strengths of our study was the large sample size allowing for meaningful patient matching. Based on this large sample size as well as on the case–control matching, we are confident in data validity and reliability. Additionally, all urinary peptides were analyzed using the same analytical platform and protocols (CE-MS). In contrast to any MS/MS approach, (CE-)MS does not provide sequence information. However, also as a result of the excellent reproducibility, the dataspace can be well defined, which results in the identification of over 4000 urine peptides and assignment of sequence based on mass and migration time with very high confidence67, which would not be possible based on LC–MS/MS. A detailed comparison of LC–MS/MS and CE-MS/MS is presented in Klein et al.68. In consequence, the approach combining highly reproducible CE-MS analysis with a large database on CE- and LC–MS/MS analyses, results in a much higher number of peptide identifications and in higher confidence in the results in comparison to LC–MS/MS only.
The study has some limitations: these include that it is retrospective, based on already available published data. However, the sample size, the multicenter design and the very high consistency and significance of the observed changes reduce the risk of this bias. Further, we limited our study to a subgroup of CRS: to subjects with CKD and HF. This is owed to the fact that we aimed towards depicting molecular changes. Such an approach requires homogeneity in molecular pathophysiology. In addition, a number of known kidney-specific and heart-specific protein biomarkers (such as KIM-1, L-FABP, NGAL, and NAG)14 could not be detected in our study; this is in fact expected as the applied technique (CE-MS) resolves the peptidome (< 10 kDa molecular mass peptides). Finally, the analysis is descriptive which, however, still opens multiple research avenues towards understanding the functional impact from the generation of the presented significant fragments.
In conclusion, this study reports the detection of a high number of urinary CRS-associated peptides when compared with controls. The underlying molecular mechanisms for the CRS pathology, as reflected by these peptides, represent fibrosis, ECM-related pathways, collagens formation/degradation and inflammation, in line with the existent knowledge. However, a number of peptides/protein precursors, not highlighted previously in association to HF or CKD, such as peptides of PIGR, CD14, ANR17, COL4A2 COL4A4, COL6A5, ROBO1 and HUWE1 may be important players in the mechanisms of CRS. As the present work is the first attempt to explore the urine peptidome profile of CRS patients, the findings of our study require further validation.
Methods
Study design: patient data and selection
Urinary peptidomics datasets from patients with CRS (in this study, CRS patients simultaneously combine both pathologies; HF and CKD), as well as individuals with no signs of diseases (controls) at urine sampling were used. These datasets corresponded to urine samples from cohorts described in several published studies investigating mainly renal failure (including Syskid15, FSGS-Aachen24), or heart and cardiovascular failure (including NTCVD-Urin69, FROG-ICU70, PCHF-Urin71 and HOMAGE72). The data were investigated for the availability of information on both kidney (i.e. CKD) and heart (i.e. HF) disease. The kidney function was assessed via the eGFR assessed based on ´Chronic Kidney Disease Epidemiology Collaboration´ (CKD-EPI). The ´European Society of Cardiology´ guidelines were used for the HF diagnosis and subtyping73. Clinical, pathophysiological, and molecular variables such as ejection fraction, systolic blood pressure, diastolic blood pressure, serum creatinine, hypertension, NT-proBNP levels, left ventricular ejection fraction (LVEF) as well as information on the age and sex, were retrieved (see Table 1). These datasets (corresponding to 3463 individuals) were then separated into the CRS cohort consisting of patients with both pathologies; HF and CKD with an eGFR < 60 ml/min/1.73 m2. The control group with no signs of heart disease and preserved kidney function (eGFR > 60 ml/min/1.73 m2) was selected (see Table 1). The study had received ethics approval (ΕΚ163/19 Ethik-Commission of the medical faculty of the RWTH Aachen), fulfilling all the requirements on the protection of the individuals participating in medical research and in accordance with the principles of the Declaration of Helsinki74. All data sets received were anonymized. All experiments were performed in accordance with relevant named guidelines and regulations. Each patient has written informed consent to use part of the tissue for scientific research.
Case–control matching
CRS patients were matched on age and sex with controls at a 1:1 ratio. Following the case–control matching procedure, the final groups consisted of 353 patients with CRS compared with 356 individuals with no signs of either disease as listed in Table 1.
CE-MS
The urine samples have been prepared and measured by CE-MS as stated before75. The P/ACE MDQ capillary electrophoresis system (Beckman Coulter, USA) connected to a micro-TOF–MS (Bruker Daltonic, Germany) was used for the CE-MS analysis. The probabilistic clustering algorithm along with isotopic distribution and conjugated masses for charge have been used for RAW MS data evaluation as described previously67. Totally, twenty-nine fragments of collagens that were not affected by disease were used for the normalization of the CE-MS data.
Peptide sequencing
Urinary peptides were fragmented using Orbitrap MS coupled to CE (CE-MS/MS) or liquid chromatography (LC–MS/MS)68. The fragmentation spectra were matched to the protein sequences from up-to-date databases (International Protein Index76, Reference sequence database at NCBI77 and UniProt Knowledgebase78) using Proteome Discoverer 1.4 (Thermo Scientific, Bremen, Germany) with an integrated Sequest search engine79). The following search parameters were applied: (1) precursor mass tolerance: 5 ppm, (2) fragment mass tolerance: 50 mDa and (3) variable post-translational modifications: hydroxylation of lysine and proline, and oxidation of methionine, and (4) Enzyme name: No enzyme (Unspecific). Identified biomarkers were also assigned in silico to sequenced peptides from the Human Urinary Proteome database.
Bioinformatic analysis
Information on the function and expression of the parental proteins was extracted from Gene Cards (http://www.genecards.org) and linked databases ProteomicsDB (http://www.proteomicsdb.org), Uniprot (http://www.uniprot.org), and Human Protein Atlas (http://www.proteinatlas.org). Pathway enrichment analysis and visualization of data were performed by the Metascape80. Gene Ontology (GO) annotation of molecular functions was performed by the Gene Ontology resource (GO; http://geneontology.org).
Proteases analysis
The Proteasix (http://www.proteasix.org), an open-source tool was used for the protease prediction analysis81. As such, the potentially involved proteases were linked with the generation of the identified CRS-associated peptides. In brief, Proteasix uses information about the protease/cleavage site associations from a number of protease databases including the MEROPS, the CutDB, the UniProt Knowledgebase and the literature. The generated list of proteases is divided into two types; (a) “predicted” and (b) “observed”. For the “observed” proteases the protease/cleavage site association is collected from the literature, whereas for the “predicted” proteases the predicted proteolysis is determined by the MEROPS database. To improve the reliability of the proteolytic data, we decided to focus only on the “observed” proteases.
Statistical analysis
The Kolmogorov–Smirnov normality test was used to determine the distribution of the urine peptidome data. Statistical analysis of the abundance of urinary peptides was performed using the non-parametric Mann–Whitney U test, followed by correction for multiple testing using the Benjamini-Hochberg (BH) method. Statistical analysis was performed using SPSS software version 20.0 (SPSS, Inc., Chicago, Illinois). A BH-adjusted P-value < 0.05 was considered to be statistically significant. The abundance of urinary peptides was analyzed and plotted using GraphPad Prism 7 (GraphPad Software, La Jolla, California, USA). Data are presented as mean ± standard deviation (s.d.) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
References
Ronco, C. et al. Cardio-renal syndromes: Report from the consensus conference of the acute dialysis quality initiative. Eur. Heart J. 31, 703–711. https://doi.org/10.1093/eurheartj/ehp507 (2010).
Hebert, K. et al. Epidemiology and survival of the five stages of chronic kidney disease in a systolic heart failure population. Eur. J. Heart Fail. 12, 861–865 (2010).
Ronco, C. Cardiorenal syndromes: Definition and classification. Contrib. Nephrol. 164, 33–38 (2010).
He, T. et al. Proteomic Biomarkers in the Cardiorenal Syndrome: Toward Deciphering Molecular Pathophysiology. Am. J. Hypertens. hpaa201, https://doi.org/10.1093/ajh/hpaa201. (2021).
Kumar, U., Wettersten, N. & Garimella, P. S. Cardiorenal syndrome: Pathophysiology. Cardiol. Clin. 37, 251–265. https://doi.org/10.1016/j.ccl.2019.04.001 (2019).
Sun, L. et al. High-sensitive cardiac troponin T: A biomarker of left-ventricular diastolic dysfunction in hemodialysis patients. J. Nephrol. 31, 967–973 (2018).
Testani, J. M. et al. A combined-biomarker approach to clinical phenotyping renal dysfunction in heart failure. J. Card. Fail. 20, 912–919 (2014).
Atici, A. et al. The role of kidney injury molecule-1 in predicting cardiorenal syndrome type 1 after diuretic treatment. Arch. Med. Sci. Atheroscler. Dis. 4, e208–e214 (2019).
Hishikari, K. et al. Urinary liver-type fatty acid-binding protein level as a predictive biomarker of acute kidney injury in patients with acute decompensated heart failure. CardioRenal Med. 7, 267–275 (2017).
Palazzuoli, A. et al. Admission plasma neutrophil gelatinase associated lipocalin (NGAL) predicts worsening renal function during hospitalization and post discharge outcome in patients with acute heart failure. Acute Card. Care 16, 93–101 (2014).
Vianello, A. et al. β2-Microglobulin and TIMP1 are linked together in cardiorenal remodeling and failure. CardioRenal Med. 5, 1–11 (2015).
Jungbauer, C. G. et al. Kidney injury molecule-1 and N-acetyl-ß-d-glucosaminidase in chronic heart failure: Possible biomarkers of cardiorenal syndrome. Eur. J. Heart Fail. 13, 1104–1110 (2011).
Rafouli-Stergiou, P. et al. Prognostic value of in-hospital change in cystatin C in patients with acutely decompensated heart failure and renal dysfunction. Int. J. Cardiol. 182, 74–76 (2015).
Petra, E., Zoidakis, J. & Vlahou, A. Protein biomarkers for cardiorenal syndrome. Expert Rev. Proteomics 16, 325–336. https://doi.org/10.1080/14789450.2019.1592682 (2019).
Schanstra, J. P. et al. Diagnosis and prediction of CKD progression by assessment of urinary peptides. J. Am. Soc. Nephrol. 26, 1999–2010 (2015).
Magalhães, P. et al. Association of kidney fibrosis with urinary peptides: A path towards non-invasive liquid biopsies?. Sci. Rep. 7, 16915 (2017).
de Almeida Brondani, L. et al. Urinary peptidomics and bioinformatics for the detection of diabetic kidney disease. Sci. Rep. 10, 1242 (2020).
Metzger, J. et al. Urinary excretion of twenty peptides forms an early and accurate diagnostic pattern of acute kidney injury. Kidney Int. 78, 1252–1262 (2010).
He, T. et al. Urinary peptides in heart failure: A link to molecular pathophysiology. Eur. J. Heart Fail. https://doi.org/10.1002/ejhf.2195 (2021).
Zhang, Z. Y. et al. Novel urinary peptidomic classifier predicts incident heart failure. J. Am. Heart Assoc. 6, e005432 (2017).
Farmakis, D. et al. Urine proteome analysis in heart failure with reduced ejection fraction complicated by chronic kidney disease: Feasibility, and clinical and pathogenetic correlates. Eur. J. Heart Fail. 18, 822–829 (2016).
Tofte, N. et al. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): A prospective observational study and embedded randomised placebo-controlled trial. Lancet. Diabetes Endocrinol. 8, 301–312 (2020).
Good, D. M. et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol. Cell. Proteomics 9, 2424–2437 (2010).
Siwy, J. et al. Noninvasive diagnosis of chronic kidney diseases using urinary proteome analysis. Nephrol. Dial. Transplant. Off. Publ. Eur. Dial. Transpl. Assoc. Eur. Ren. Assoc. 32, 2079–2089 (2017).
Takahama, H. & Kitakaze, M. Pathophysiology of cardiorenal syndrome in patients with heart failure: Potential therapeutic targets. Am. J. Physiol. Heart Circ. Physiol. https://doi.org/10.1152/ajpheart.00215.2017 (2017).
Lousa, I. et al. New potential biomarkers for chronic kidney disease management—A review of the literature. Int. J. Mol. Sci. 22, 43 (2020).
Zhang, H. et al. Association between the lipid profile and renal dysfunction in the heart failure patients. Kidney Blood Press. Res. 44, 52–61 (2019).
Gong, X.-M., Ding, Y., Yu, J., Yao, Y. & Marassi, F. M. Structure of the Na, K-ATPase regulatory protein FXYD2b in micelles: Implications for membrane-water interfacial arginines. Biochim. Biophys. Acta 1848, 299–306 (2015).
Li, L., Zhao, Q. & Kong, W. Extracellular matrix remodeling and cardiac fibrosis. Matrix Biol. 68, 490–506. https://doi.org/10.1016/j.matbio.2018.01.013 (2018).
Querejeta, R. et al. Increased collagen type I synthesis in patients with heart failure of hypertensive origin: Relation to myocardial fibrosis. Circulation 110, 1263–1268 (2004).
Alesutan, I. et al. Inhibition of osteo/chondrogenic transformation of vascular smooth muscle cells by MgCl2 via calcium-sensing receptor. J. Hypertens. 35, 523–532 (2017).
Genovese, F., Manresa, A. A., Leeming, D. J., Karsdal, M. A. & Boor, P. The extracellular matrix in the kidney: A source of novel non-invasive biomarkers of kidney fibrosis?. Fibrogenesis Tissue Repair 7, 1–14. https://doi.org/10.1186/1755-1536-7-4 (2014).
Djudjaj, S. & Boor, P. Cellular and molecular mechanisms of kidney fibrosis. Mol. Aspects Med. 65, 16–36. https://doi.org/10.1016/j.mam.2018.06.002 (2019).
DeLeon-Pennell, K. Y., Meschiari, C. A., Jung, M. & Lindsey, M. L. Matrix metalloproteinases in myocardial infarction and heart failure. Prog. Mol. Biol. Transl. Sci. 147, 75–100 (2017).
Zakiyanov, O., Kalousová, M., Zima, T. & Tesař, V. Matrix metalloproteinases in renal diseases: A critical appraisal. Kidney Blood Press. Res. 44, 298–330 (2019).
Ueland, T. et al. Undercarboxylated matrix Gla protein is associated with indices of heart failure and mortality in symptomatic aortic stenosis. J. Intern. Med. 268, 483–492 (2010).
Groopman, E. E. et al. Diagnostic utility of exome sequencing for kidney disease. N. Engl. J. Med. 380, 142–151 (2019).
Westenfeld, R., Jahnen-Dechent, W. & Ketteler, M. Vascular calcification and fetuin-A deficiency in chronic kidney disease. Trends iCardiovasc. Med. 17, 124–128. https://doi.org/10.1016/j.tcm.2007.02.005 (2007).
Topçiu-Shufta, V., Miftari, R., Haxhibeqiri, V. & Haxhibeqiri, S. Association of beta-2 microglobulin with inflammation and dislipidemia in high-flux membrane hemodialysis patients. Med. Arch. 70, 348–350 (2016).
Kanbay, M. et al. Endostatin in chronic kidney disease: Associations with inflammation, vascular abnormalities, cardiovascular events and survival. Eur. J. Intern. Med. 33, 81–87 (2016).
Zuo, Y. et al. Thymosin β4 and its degradation product, Ac-SDKP, are novel reparative factors in renal fibrosis. Kidney Int. 84, 1166–1175 (2013).
Maron, B. A. et al. Individualized interactomes for network-based precision medicine in hypertrophic cardiomyopathy with implications for other clinical pathophenotypes. Nat. Commun. 12, 873 (2021).
Masuda, Y. et al. Glomerular basement membrane injuries in IgA nephropathy evaluated by double immunostaining for α5(IV) and α2(IV) chains of type IV collagen and low-vacuum scanning electron microscopy. Clin. Exp. Nephrol. 19, 427–435 (2015).
Cameron-Christie, S. et al. Exome-based rare-variant analyses in CKD. J. Am. Soc. Nephrol. 30, 1109–1122 (2019).
Yamada, Y. et al. Identification of 13 novel susceptibility loci for early-onset myocardial infarction, hypertension, or chronic kidney disease. Int. J. Mol. Med. 42, 2415–2436 (2018).
Liu, Y. et al. Crosstalk between the activated Slit2-Robo1 pathway and TGF-β1 signalling promotes cardiac fibrosis. ESC Heart Fail. 8, 447–460 (2021).
Zhu, Q. et al. HUWE1 promotes EGFR ubiquitination and degradation to protect against renal tubulointerstitial fibrosis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 34, 4591–4601 (2020).
Hou, S.-C. et al. Ankrd17, an ubiquitously expressed ankyrin factor, is essential for the vascular integrity during embryogenesis. FEBS Lett. 583, 2765–2771 (2009).
Verbree-Willemsen, L. et al. Extracellular vesicle Cystatin C and CD14 are associated with both renal dysfunction and heart failure. ESC Heart Fail. 7, 2240–2249 (2020).
He, T. et al. Associations of urinary polymeric immunoglobulin receptor peptides in the context of cardio-renal syndrome. Sci. Rep. 10, 8291 (2020).
Toba, H. & Lindsey, M. L. Extracellular matrix roles in cardiorenal fibrosis: Potential therapeutic targets for CVD and CKD in the elderly. Pharmacol. Ther. 193, 99–120 (2019).
Yu, H. et al. FOXO3a (Forkhead Transcription Factor O Subfamily Member 3a) links vascular smooth muscle cell apoptosis, matrix breakdown, atherosclerosis, and vascular remodeling through a novel pathway involving MMP13 (Matrix Metalloproteinase 13). Arterioscler. Thromb. Vasc. Biol. 38, 555–565 (2018).
Provenzano, M. et al. The association of matrix metalloproteinases with chronic kidney disease and peripheral vascular disease: A light at the end of the tunnel?. Biomolecules 10, 154 (2020).
Hsu, T.-W. et al. Progression of kidney disease in non-diabetic patients with coronary artery disease: Predictive role of circulating matrix metalloproteinase-2, -3, and -9. PLoS ONE 8, e70132 (2013).
Kelly, D. et al. Plasma matrix metalloproteinase-9 and left ventricular remodelling after acute myocardial infarction in man: A prospective cohort study. Eur. Heart J. 28, 711–718 (2007).
Li, H., Zhang, Y. & Wu, Q. Role of corin in the regulation of blood pressure. Curr. Opin. Nephrol. Hypertens. 26, 67–73 (2017).
Rykaczewska, U. et al. PCSK6 is a key protease in the control of smooth muscle cell function in vascular remodeling. Circ. Res. 126, 571–585 (2020).
Kuhn, T. C. et al. Secretome analysis of cardiomyocytes identifies PCSK6 (Proprotein Convertase Subtilisin/Kexin Type 6) as a novel player in cardiac remodeling after myocardial infarction. Circulation 141, 1628–1644 (2020).
Marchesi, C. et al. Inactivation of endothelial proprotein convertase 5/6 decreases collagen deposition in the cardiovascular system: Role of fibroblast autophagy. J. Mol. Med. 89, 1103–1111 (2011).
Yao, C. et al. Integromic analysis of genetic variation and gene expression identifies networks for cardiovascular disease phenotypes. Circulation 131, 536–549 (2015).
Zeng, X. et al. A network-based variable selection approach for identification of modules and biomarker genes associated with end-stage kidney disease. Nephrology 25, 775–784 (2020).
Zhao, G. et al. Increased circulating cathepsin K in patients with chronic heart failure. PLoS ONE 10, e0136093 (2015).
Izumi, Y. et al. Impact of circulating cathepsin K on the coronary calcification and the clinical outcome in chronic kidney disease patients. Heart Vessels 31, 6–14 (2016).
Sena, B. F., Figueiredo, J. L. & Aikawa, E. Cathepsin S as an inhibitor of cardiovascular inflammation and calcification in chronic kidney disease. Front. Cardiovasc. Med. 4, 88 (2017).
Cheng, X. W. et al. Elastolytic cathepsin induction/activation system exists in myocardium and is upregulated in hypertensive heart failure. Hypertension 48, 979–987 (2006).
Yao, X. et al. Cathepsin S regulates renal fibrosis in mouse models of mild and severe hydronephrosis. Mol. Med. Rep. 20, 141–150 (2019).
Latosinska, A., Siwy, J., Mischak, H. & Frantzi, M. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: The past, the present, and the future. Electrophoresis 40, 2294–2308 (2019).
Klein, J., Papadopoulos, T., Mischak, H. & Mullen, W. Comparison of CE-MS/MS and LC-MS/MS sequencing demonstrates significant complementarity in natural peptide identification in human urine. Electrophoresis 35, 1060–1064 (2014).
Seidel, U. K. et al. The prevalence, severity, and association with HbA1c and fibrinogen of cognitive impairment in chronic kidney disease. Kidney Int. 85, 693–702 (2014).
Mebazaa, A. et al. Post-ICU discharge and outcome: Rationale and methods of the The French and euRopean Outcome reGistry in Intensive Care Units (FROG-ICU) observational study. BMC Anesthesiol. 15, 1–9 (2015).
Campbell, R. T. et al. Palliative care needs in patients hospitalized with heart failure (PCHF) study: Rationale and design. ESC Heart Fail. 2, 25–36 (2015).
Jacobs, L. et al. Heart ‘omics’ in AGEing (HOMAGE): Design, research objectives and characteristics of the common database. J. Biomed. Res. 28, 349–359 (2014).
McMurray, J. J. V. et al. ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure 2012: The Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure 2012 of the European Society of Cardiology. Developed in collaboration with the Heart. Eur. Heart J. 33, 1787–1847 (2012).
World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 310, 2191–2194 (2013).
Mischak, H., Vlahou, A. & Ioannidis, J. P. A. Technical aspects and inter-laboratory variability in native peptide profiling: The CE-MS experience. Clin. Biochem. 46, 432–443 (2013).
Kersey, P. J. et al. The International Protein Index: An integrated database for proteomics experiments. Proteomics 4, 1985–1988 (2004).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Klein, J. et al. Proteasix: A tool for automated and large-scale prediction of proteases involved in naturally occurring peptide generation. Proteomics 13, 1077–1082 (2013).
Acknowledgements
The project is funded by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 764474” as well as by the ´Deutsche Forschungsgemein-schaft´ (DFG, German Research Foundation) by the Transregional Collaborative Research Centre (SFB/TRR 219; Project-ID 322900939; S-03; C-04).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
E.P., H.M., A.V., and J.J. designed the study; E.P., wrote the manuscript text, performed data analysis and interpretation; T.H., A.L., and H.M. compiled peptidomics datasets; V.L. contributed to the pathway enrichment analysis; all authors revised the manuscript; all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
H.M. is the co-founder and co-owner of Mosaiques Diagnostics GmbH. T.H. and A.L. are employed by Mosaiques Diagnostics GmbH. E.P, V.L, A.V, J.J: The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Petra, E., He, T., Lygirou, V. et al. Urine peptidome analysis in cardiorenal syndrome reflects molecular processes. Sci Rep 11, 16219 (2021). https://doi.org/10.1038/s41598-021-95695-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-95695-z
This article is cited by
-
Salivary peptidome analysis and protease prediction during orthodontic treatment with fixed appliances
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.