Abstract
Pulmonary fibrosis is a devastating disease, and the pathogenesis of this disease is not completely clear. Here, the medical records of 85 Covid-19 cases were collected, among which fibrosis and progression of fibrosis were analyzed in detail. Next, data independent acquisition (DIA) quantification proteomics and untargeted metabolomics were used to screen disease-related signaling pathways through clustering and enrichment analysis of the differential expression of proteins and metabolites. The main imaging features were lesions located in the bilateral lower lobes and involvement in five lobes. The closed association pathways were FcγR-mediated phagocytosis, PPAR signaling, TRP-inflammatory pathways, and the urea cycle. Our results provide evidence for the detection of serum biomarkers and targeted therapy in patients with Covid-19.
Similar content being viewed by others
Introduction
In December 2019, pneumonia with an unknown cause became prevalent in Wuhan, China. When RNA was extracted and sequenced from these patients in samples of bronchoalveolar lavage fluid, the causative agent was found to be a novel betacoronavirus, SARS-CoV-21. Subsequently, the outbreak was declared a public health emergency, and the novel coronavirus disease was named Covid-19 by the World Health Organization (WHO)2. As of November, 2020, over 42 million Covid-19 cases and 1 million deaths have been confirmed globally. However, the epidemic continues to spread, especially in the Americas and southeast Asia3. Currently, there are no effective therapies to treat Covid-19, although countries continue to develop new vaccines4,5. Therefore, developing new ways to control the progression of disease is urgent.
Pulmonary fibrosis is a phenomenon in patients with Covid-19, as evidenced by CT imaging and autopsy6,7. It is a progressive lung disease, and lung transplantation may be the only treatment to elevate life expectancy8. Recently, the Arg-Gly-Asp integrin-binging domain in SARS-CoV-2 spike protein was recognized by some researchers as one of the target pathways9,10, and the inflammatory factor IL-1 was implicated as possibly mediating its binding integrins11. TGF-β orchestrates multifunctional aspects of fibrogenesis, such as proliferation, deposition of extracellular matrix and fiber formation12,13. The TGF-β pathway is the main antifibrotic therapeutic pathway14,15. However, more pathogenetic profibrotic pathways need to be explored.
Proteomics and metabonomics are useful methods to screen markers of disease occurrence through differential expression of proteins and metabolic compounds16,17. In this study, we combined proteomic and metabonomic data with chest CT imaging to investigate the mechanism of pulmonary fibrosis. For the first time, to study the related pathways of fibrosis progression in Covid-19, patients with pulmonary fibrosis were divided into a nonprogressive group and a progressive group according to the degree of fibrosis progression. Our research may provide useful clues and strategies for controlling the epidemic of Covid-19 in other countries worldwide, particularly the prognosis and treatment of pulmonary fibrosis.
Results
Patient demographics and laboratory findings
By 17 Mar 2020, the clinical data of 85 laboratory-confirmed SARS-CoV-2-infected cases were collected in Guangxi, China. The median age for all patients was 41 years, with an interquartile range of 29–50.5 years (Tables 1 and S1). Sixty percent of the patients were female.
On admission, white blood cells (WBCs) were below the normal range in 9 (10.6%) patients and above the normal range in 6 (7.0%) patients. Seven (8.2%) patients showed neutropenia (neutrophil count < 1.7 × 109), and 5 (5.9%) showed lymphopenia (lymphocyte count < 0.7 × 109). The erythrocyte sedimentation rate (ESR) was higher in pulmonary fibrosis patients (median level 41.5 mm/h [interquartile 28.0–65.0]) than in nonpulmonary fibrosis patients (median level 24 mm/h [interquartile 9.8–45.3]). Levels of C-reactive protein, hypersensitive C-reactive protein, procalcitonin, D-dimer, and oxygenation index were increased in 13 (29.5%), 13 (29.5%), 18 (40.9%), 28 (63.6%) and 37 (84.1%) pulmonary fibrosis patients, respectively, while they were increased in 7 (17.1%), 8 (19.5%), 6 (14.6%), 22 (53.7%) and 36 (87.8%) nonpulmonary fibrosis patients.
Chest CT findings
Of the 85 patients, 11 (12.9%) had no abnormality at admission on chest CT, including 10 (24.4%) of 41 nonpulmonary fibrosis patients and 1 (2.3%) of 44 pulmonary fibrosis patients (Table 2 and Fig. 1). Most of the patients (68 [80%]) had the single form of ground-glass opacities in the process of disease, while a few patients (10 [11.8%]) had the mixed form of ground-glass opacities and consolidation. Thirty-eight (44.7%) patients had fibrosis on admission, and 2 had fibrosis before discharge. In the nonpulmonary fibrosis group, the numbers of involved lobes were usually 1, 2, and 5. In contrast, the number of involved lobes in the pulmonary fibrosis group was mainly 5. The total lung severity score of the pulmonary fibrosis group was higher than that of the nonpulmonary fibrosis group at both stages of admission and discharge. Lesions were mainly peripheral and central distribution in the pulmonary fibrosis group (20 [50.0%]) at admission and then turned to peripheral distribution (27 [61.4%]) before discharge, whereas lesions were mainly peripherally distributed in the nonpulmonary fibrosis group. Importantly, almost half of the pulmonary fibrosis patients exhibited linear consolidation, bronchiectasis, and pleural incrassation in both stages of admission and discharge.
Representative chest CT images. (a) Ground-glass opacities in the dorsal segment of the left lower lobe of lung were seen in a 44-year-old female on admission, and partial absorption of lung lesions before discharge (b), Multiple fibrotic stripes shadows in dorsal segment of the lower lobe of the right lung were seen in a 62-year-old male after symptom onset (c), however, no obvious changes were observed before discharge (d, e), multiple fibrotic stripes shadows and patchy dense shadow (exudative lesion) were shown in a 65-year-old severe male on admission. (f) The exudative lesions were less and the fibrosis lesions were more than before in 20 days after hospitalization.
Then, we further evaluated the distribution of fibrosis in Covid-19 patients complicated with pulmonary fibrosis, including nonprogressive pulmonary fibrosis patients and progressive pulmonary fibrosis patients (Table 3, Fig. 1c–f.). In the progressive pulmonary fibrosis group, on admission, 6 (20%) patients had no fibrosis, and 14 (46.7%) patients had fibrosis in 5 lobes. However, 23 (76.7%) patients had fibrosis involving 5 lobes before discharge. Pulmonary fibrosis was widely distributed in both groups, especially in the left lower lobe and the right lower lobe.
Proteomic and metabolomic analysis of Covid-19 patients with pulmonary fibrosis
The Q-Exactive HF (Thermo Fisher Scientific, San Jose, CA) instrument collected mass spectrometry data from 27 samples (randomly sampled from each group) in DIA mode (Table S2). A total of 8055 peptide fragments and 1079 proteins were quantified (Table S3). A heat map of the sample correlation analysis is shown in Fig. S1a. After calculating the fold difference between Covid-19 patients with pulmonary fibrosis (n = 21) and Covid-19 patients without fibrosis (n = 6) by MSstats software, 20 differentially expressed proteins were found, including 13 downregulated and 7 upregulated proteins (Fig. 2a,b). GO and KEGG18 enrichment analysis indicated that DEPs were related to the immune system, biological adhesion, and glycosaminoglycan degradation (Fig. S1b–d and Table S4).
Differentially expressed proteins (DEPs) were identified and clustered in Covid-19 patients with pulmonary fibrosis. (a) Volcano plots of total protein levels. Red dots, significantly upregulated proteins; green dots, significantly downregulated proteins. (b) Cluster chart of DEPs. Red rows, significantly upregulated proteins; blue rows present significantly downregulated proteins.
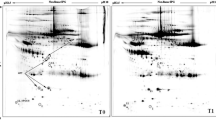
For metabolomic analysis, the data of positive and negative ions were collected from 28 samples (randomly sampled from each group) to improve the metabolite coverage. The coefficient of variation (CV) plot shows that there was a high degree of repeatability for samples (Fig. 3a,b). PLS-DA (Fig. 3c,d) was used to establish the relationship model between metabolite expression and sample types to predict sample categories and to determine the differential metabolism among groups. A total of 3020 positive compounds with identification information (Table S5) and 978 negative compounds were identified (Table S6), among which 102 positive compounds were upregulated and 21 were downregulated (Fig. 4a), while 38 negative compounds were upregulated and 3 were downregulated (Fig. S2a) between Covid-19 patients with pulmonary fibrosis (n = 22) and Covid-19 patients without fibrosis (n = 6). The cluster analysis of different metabolites is shown in Fig. 4b and Fig. S2b. Based on the KEGG database, pathways including the peroxisome proliferator-activated receptor (PPAR) signaling pathway, D-arginine and D-ornithine metabolism, inflammatory mediator regulation of TRP channels and alpha-linolenic acid metabolism were significantly enriched (Fig. 4c, S2c and Table S7).
Quality control of samples in Covid-19 patients with pulmonary fibrosis. (a, b) Coefficient of variation (CV) plots for positive and negative ion modes. (c,d) PLS-DA model plots for positive and negative ion modes. A, Covid-19 patients without pulmonary fibrosis (n = 6). B, Covid-19 patients with pulmonary fibrosis (n = 22).
Positive differentially expressed metabolites were identified, clustered and enriched in Covid-19 patients with pulmonary fibrosis. (a) Volcano plots of positive compounds. Red dots, significantly upregulated metabolites; green dots, significantly downregulated metabolites. (b) Heatmap of different positive metabolites (“pheatmap” package in R software v3.5.0, https://www.r-project.org/). Each row denotes a different metabolite, and each column denotes a sample. A, Covid-19 patients without pulmonary fibrosis (n = 6). B, Covid-19 patients with pulmonary fibrosis (n = 22). (c) Bubble plot of KEGG enrichment of positive metabolic pathways. The size of the circle dot denotes the number of different metabolites. RichFactor is defined as the number of differential metabolites annotated to the pathway divided by all identified metabolites annotated to the pathway.
Proteomic and metabolomic analyses of Covid-19 patients with progressive pulmonary fibrosis
To further search for factors related to the progression of pulmonary fibrosis, we divided the patients with pulmonary fibrosis into a progressive group and a nonprogressive group and continued to conduct proteomic and metabolomic tests on their serum. It was found that 34 proteins were differentially expressed between the progressive pulmonary fibrosis Covid-19 patients (n = 16) and the nonprogressive pulmonary fibrosis Covid-19 patients (n = 5) (Fig. 5a). After clustering (Fig. 5b) and enrichment analysis of DEPs (Fig. 5c–e, and Table S4), it was found that DEPs were mainly distributed in the immune system, including primary immunodeficiency, intestinal immune network for IgA production, FcγR-mediated phagocytosis, natural killer cell-mediated cytotoxicity, the B cell receptor signaling pathway, and the FcεRI signaling pathway. Moreover, the NF-kappa B, calcium, hematopoietic cell lineage, and phospholipase D signaling pathways were also involved.
Differentially expressed proteins (DEPs) were identified and analyzed using proteomics in Covid-19 patients with progressive pulmonary fibrosis. (a) Volcano plots of total protein level. (b) Cluster chart of DEPs. (c) Enrichment analysis of DEPs by GO function. (d) Bubble plot of the top 20 KEGG enrichment pathways of DEPs. RichFactor is defined as the number of differential metabolites annotated to the pathways divided by all identified metabolites annotated to the pathway. (e) Pathway network of DEPs (“igraph” package in R software v3.5.0, https://www.r-project.org/). The red and blue dots represent up-regulated and down-regulated differential proteins, respectively. The purple balls represent the top 10 pathways of enrichment. The area is directly proportional to the enrichment degree.
In terms of the metabolomic analysis, we found that the PLS-DA model had a good prediction effect (Figs. 6a and S3a). A total of 511 differential positive metabolites and 160 negative metabolites were screened (Figs. 6b and S3b) between the progressive pulmonary fibrosis Covid-19 patients (n = 16) and the nonprogressive pulmonary fibrosis Covid-19 patients (n = 6) and clustered (Figs. 6c and S3c). Using the KEGG database, metabolic pathway enrichment analysis revealed the following pathways with differential metabolite enrichments: PPAR signaling pathway, mTOR signaling pathway, inflammatory mediator regulation of TRP channels, FcγR-mediated phagocytosis, arginine metabolism, and alpha-Linolenic acid metabolism (Figs. 6d, S3d and Table S7).
Positive differentially expressed metabolites were identified and analyzed using proteomics in Covid-19 patients with progressive pulmonary fibrosis. (a) PLS-DA model plot for positive ion mode. (b) Volcano plots of positive compounds. (c) Heatmap of different positive metabolites (“pheatmap” package in R software v3.5.0, https://www.r-project.org/). (d) Bubble plot of the top 20 KEGG enrichment of positive metabolic pathways. C, Nonprogressive pulmonary fibrosis of Covid-19 patients. D, Progressive pulmonary fibrosis of Covid-19 patients. RichFactor is defined as the number of differential metabolites annotated to the pathway divided by all identified metabolites annotated to the pathway.
Correlation analysis between proteomics and metabolomics in Covid-19 patients complicated with progressive pulmonary fibrosis
To further explore the relationship between biological network regulation, we conducted proteome and metabolomic association analyses between the nonprogressive pulmonary fibrosis group and the progressive pulmonary fibrosis group. By performing regularized canonical correlation analysis, correlation clustering of DEPs and differential metabolites can be observed in the heatmap (Fig. 7a). After multivariate dimensionality reduction and relationship visualization of omics (Fig. 7b), it was found that the DEPs and differential metabolites in pulmonary fibrosis group (n = 21) were correlated with each other.
Association analysis of proteomics and metabolomics in Covid-19 patients with progressive pulmonary fibrosis. (a) Cluster heat map of the correlation between proteomics and metabolomics (“pheatmap” package in R software v3.5.0, https://www.r-project.org/). Each row denotes a differentially expressed metabolite, and each column denote a DEP. Blue, negative correlation; Red, positive correlation. (b) Concentric circles of correlation between differential proteins and differential metabolites. If the angle between a differential protein and differential metabolite is an acute angle (less than 90 degrees), the correlation is positive. If the angle between the differential protein and the differential metabolite is an oblique angle (greater than 90 degrees and less than 180 degrees), the correlation is negative. Starting from the center of the circle, the lines are connected to the differential metabolites and differential proteins. The longer the connection length, the stronger the relationship.
Discussion
To our knowledge, this is the first paper describing an integrated multiomics study in Covid-19 complicated with pulmonary fibrosis patients at the molecular and ionic levels. First, after reviewing the medical records of 85 Covid-19 patients hospitalized in the People's Hospital of Guangxi Zhuang Autonomous Region Yongwu branch, we found that as many as 44.7% of the patients had fibrotic stripe shadows in the early onset, and the incidence rate was close to that of SARS outbreaks in 200319. The most common manifestations of chest CT in Covid-19 fibrosis in our 85 cases were summarized as follows: lesions located in bilateral lower lobes, involved in five lobes, and complicated with linear consolidation, bronchiectasis, and pleural incrassation, which partially agreed with a previous report6. Second, we described the detailed profile of CT images in Covid-19 complicated with pulmonary fibrosis patients, especially focusing on progressive pulmonary fibrosis patients. Third, using proteomics and metabonomics methods, we explored the mechanism of the information pathway related to the occurrence and development of progressive fibrosis, which is expected to predict disease progression and to provide evidence for targeted therapy.
In recent years, proteomics has become an accurate method to screen disease biomarkers20 and has also been used in the pathogenesis of SARS-CoV-2 infection16,17. In this study, we found significant differential expression levels of immune system and biological adhesion markers and glycosaminoglycan degradation in pulmonary fibrosis patients. Previous experimental and clinical data demonstrated that the accumulation and deposition of glycosaminoglycan in the extracellular matrix contributed to progressive pulmonary fibrosis21,22. Moreover, glycosaminoglycan hyaluronan (HA) on AEC2s (stem cells in the adult lung) is critical for restoring renewal capacity and limiting the scope of fibrosis23. The general properties of cell adhesion molecules (CAMs) involve cell recruitment; they participate in cell–cell interactions and cell-extracellular matrix interactions24 and then promote the formation of fibrosis by intervention in the microenvironment25. For example, platelet/endothelial cell adhesion molecule-1 (PECAM-1) is expressed at intercellular junctions on platelets and leukocytes, including NK cells, T cells, monocytes and neutrophils26, which are involved in chemokine-mediated migration to the inflammatory site27. Moreover, PECAM-1 participates in the maintenance and restoration of vascular integrity following alveolar-capillary barrier disruption in ARDS28. In addition, CAMs modulate signal transduction by interacting with extracellular matrix proteins29. However, the results of this omics analysis are limited. The specific adhesion molecule biomarkers and the occurrence of diseases need further accurate detection.
It has been well documented that the immune system plays an important role in the occurrence and development of fibrosis30,31 and Covid-1932. The association of FCGR3B (encodes the lgG receptor FcγRIIIb) copy number variations with susceptibility to idiopathic pulmonary fibrosis (IPF) has been previously reported33. FcγR binds to serum amyloid P (SAP), inhibits the differentiation of monocytes into fibrocytes30,31 and affects neutrophil adhesion30. Moreover, SAP binds to Ca2+-dependent ligands to induce FcγR-mediated phagocytosis in vivo34. Recently, it was reported that segregated-nucleus-containing atypical monocytes, which were derived from FcεRI+ granulocyte/macrophage progenitors, were involved in fibrosis35. NK cells lacking FcεRIγ are associated with lower levels of fibrosis36. Furthermore, samples from 18 patients with IPF showed that the proportion of Breg cells (i.e., Breg cells to total B cells) was decreased, even though this proportion was positively correlated with the diffusion capacity of the lungs37. Consistent with the results of the above references, our study supported the cross-regulation among pathways of the immune system, calcium signaling and cell adhesion.
As a member of the nuclear receptor and transcription factor family, PPAR is a classical receptor for diabetic therapy38. However, recent emerging literature suggests a correlation between PPAR and lung fibrosis39,40,41. Activation of the PPARγ pathway provided protection against organ fibrosis in the lung. PPAR agonists are beneficial in treating preclinical models such as bleomycin-induced pulmonary fibrosis42 and genetically engineered mice43. Moreover, inhibiting PPARγ promoted fibroproliferative ARDS, which was observed in clinical bronchoalveolar lavage fluid (BAL) samples44. Our results from both pulmonary fibrosis patients and progressive pulmonary fibrosis patients in metabolomic analysis confirmed the differential expression of the PPAR signaling pathway, which suggested that PPAR signaling is a core pathway in the formation and development of lung fibrosis. TRP channels respond to a series of heterogeneous stimuli and are essential for pathophysiological homeostasis45. It has been reported that TRP channels participate in lung disease mainly through calcium signaling, recruitment of proinflammatory cells, and neurogenic inflammatory pathways46. In our study, pathway enrichment revealed that inflammatory mediator regulation of the TRP channel was an important pathway to regulate the occurrence and development of pulmonary fibrosis. Both arginine and ornithine are essential components in the urea cycle. L-arginine is the precursor of nitric oxide47, which is involved in the vascular system48. Treatment with L-arginine protects against pulmonary fibrosis progression in a mouse model49. D-arginine is an inactive form of L-arginine. The results from our study indicated that lung fibrosis was associated with dysfunction of the urea cycle metabolic pathway. D-arginine and D-ornithine could be used as an early warning and a sign of progression of pulmonary fibrosis disease.
This study has several limitations. First, although the 85 Covid-19 cases were all confirmed patients in our hospital, their numbers are relatively few and cannot represent the whole of China or the international situation or even secondary infection. Second, the CV of proteomics was large because the samples were taken from patients' serum. Third, the immune system, PPAR, and TRP pathways screened by proteomics and metabonomics in this study need to be further verified in vivo and in vitro, while the pathways related to nervous system diseases need to be further explored, although it has been reported that Covid-19 patients have nervous system damage50. Fourth, long-term follow-up of the progression of fibrosis is required.
In conclusion, in this study, proteomics, metabonomics, and correlation analysis were used to screen the related pathways of fibrosis formation and progression in Covid-19 patients, and the results showed that pathways including the immune system (especially the action of FcγR-mediated phagocytosis), PPAR signaling, TRP-inflammatory pathways, and the urea cycle were closely related. Our data provide new ideas for the treatment of Covid-19 and fibrosis patients.
Methods
Patient involvement and data collection
For this retrospective, single-center study, we recruited patients from Jan 15 to Mar 17, 2020, at the branches of the People's Hospital of Guangxi Zhuang Autonomous Region (Yongwu Hospital), a designated hospital for patients infected with SARS-CoV-2 in Guangxi Province. According to the guidelines of the National Health Commission of China, a case of confirmed infection with SARS-CoV-2 was diagnosed51. Patients enrolled in this study were laboratory confirmed. The study was approved by the ethics committee of the People's Hospital of Guangxi Zhuang Autonomous Region, China (KY-LW-2020–22) and was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all patients.
Laboratory confirmation
Specimens including nasal and pharyngeal swabs were collected for SARS-CoV-2 detection by real-time polymerase chain reaction (RT-PCR). Briefly, after total RNA was extracted by nucleic acid extraction and separation reagent (Sansure Biotech Inc., Changsha, China), one-step RT-PCR (bioPerfetus Technologies Co., Taizhou, China and Shanghai BioGerm Medical Technology Co., Shanghai, China) was performed using the following conditions: 50 °C for 10 min and 95℃ for 5 min, 40 cycles at 95 °C for 10 s and 55 °C for 40 s. The viral nucleic acid testing for patients was performed in the clinical laboratory of the People's Hospital of Guangxi Zhuang Autonomous Region Yongwu branch. Laboratory findings, including hematological and serological parameters and serum biochemistry, were traced.
Chest CT
Eighty-five patients were imaged with a 64-detector row SOMATOM go. Top (Siemens Healthineers, Germany). The patient was in the supine position, and the images were collected at the end of inspiration. Chest CT was assessed at two time points: on admission and before discharge (or death). All images were independently reviewed by two radiologists with more than 5 years of working experience, and the final results were determined by consensus the two doctors.
Each image was evaluated for the following features: lesion morphology, fibrosis, number of lobes affected, total lung severity score, distribution of lesions, and other findings, including linear consolidation, discrete pulmonary nodules, pulmonary emphysema, bronchiectasis, pleural effusion and pleural incrassation. The total lung severity score was obtained by adding the involvement of each lobe52,53. The corresponding scores of each lobe in the five lobes were 0, 1, 2, 3, and 4, and the corresponding affected areas were 0, 1–25%, 26–50%, 51–75%, and 76–100%.
According to whether there were fibrosis bands on chest CT, the patients were divided into nonpulmonary fibrosis and pulmonary fibrosis groups. The latter group includes the nonprogressive pulmonary fibrosis group and the progressive pulmonary fibrosis group, based on the increased severity of fibrotic pulmonary lobe involvement.
Proteome analysis
Serum samples were placed at 56 °C (water bath) for 30 min to inactivate the SARS-CoV-2 virus and were prepared as previously described with some modifications16. One hundred microliters of serum sample was denatured in 900 μL of L3 buffer containing 7 M urea, 2 M thiourea and 20 mM Tris (pH 8–8.5). The serum protein was extracted by the two following reactions: dithiothreitol (10 mM) at 37 °C (water bath) for 30 min and iodoacetamide at room temperature for 30 min (dark condition). The protein was enriched by a C18 column (Waters, USA) at a flow rate of 3 mL/min, washed with 0.1% formic acid (FA) and 75% ACN, and then dried in 75% ACN. Isolated proteins were digested with 1:20 trypsin overnight at 37 °C.
Peptides were fractionated using an LC-20AB system (Shimadzu, Japan) for high-pH reversed-phase fractionation and separated on a Gemini C18 column (5 μm × 4.6 mm × 250 mm; Waters, USA) using a multistep gradient from 5 to 95% buffer B (95% ACN, pH 9.8) at a flow rate of 1 mL/min for 64 min. Eluting peptides were collected every min, which were combined with a chromatographic elution peak diagram. Then, all the fractions were combined into a total of 10 fractions, which were then frozen and dried.
The fractions were redissolved in buffer A (2% ACN and 0.1% FA) and separated on an UltiMate 3000 UHPLC (Thermo Fisher Scientific) equipped with a C18 column (150 μm i.d, 1.8-μm particles, 35 cm long; Waters, USA). Peptides were eluted by a gradient from 5 to 80% buffer B. For data-dependent acquisition (DDA) mode detection, peptides were ionized by a nanoESI source and were then introduced into a Q-Exactive HF tandem mass spectrometer (Thermo Fisher Scientific, San Jose, CA) at a spray voltage of 1.9 kV. The MS spectra (350–1500 m/z) were acquired at a resolution of 120,000, a maximum injection time (MIT) of 100 ms and an AGC target value of 3 × 106. The MS2 spectra (350–1500 m/z) were acquired at a resolution of 120,000, an MIT of 100 ms and an AGC target value of 1 × 105. The data were identified using the Andromeda integrated by MaxQuant, and then the spectrum library was established.
For data-independent acquisition (DIA) mode detection, the same nano-LC system and gradient was used as DDA analysis. Briefly, the MS spectra (400–1250 m/z) were acquired at a resolution of 120,000, an MIT of 50 ms and an AGC target value of 1 × 106. The ion fragmentation mode was HCD, the MIT was selected as automatic mode, the fragment ions were detected in Orbitrap, and the resolution was set to 30,000. The fragmentation energy was distributed as follows: 22.5, 25, 27.5. The data were analyzed and controlled by the mProphet algorithm.
Metabolome analysis
The metabolites of serum samples were extracted by internal labeling and with a mixture of methanol and ACN at a ratio of 2:1. Ten microliters of supernatant from each sample was mixed into QC quality control samples to evaluate the repeatability and stability of the LC–MS analysis process. A Waters 2D UPLC (Waters, USA) series Q Exactive HF high-resolution mass spectrometer (Thermo Fisher Scientific, USA) was used for the separation and detection of metabolites.
A 5-μL aliquot of sample solution was injected on a BEH C18 column (100 mm × 2.1 mm, 1.7 μm; Waters, USA). The column oven was maintained at 55 °C. The positive ion mode mobile phases were aqueous solution containing 0.1% formic acid (A solution) and 100% methanol containing 0.1% formic acid (B solution). The negative ion mode mobile phase consisted of aqueous solution containing 10 mM ammonium formate (C solution) and 95% methanol containing 10 mM ammonium formate (D solution). The following gradient was used for elution: 0–1 min, 2% B/D solution; 1–9 min, 2–98% B/D solution; 9–12 min, 98% B/D solution; 12–12.1 min, 98–2% B/D solution; 12.1–15 min, 2% B/D solution. The flow rate was 0.35 mL/min. The range of mass spectrometry scanning with mass nucleus ratio was 70–1050, the first-stage resolution was 120,000, the AGC was 3 × 106, and the MIT was 100 ms. According to the parent ion strength, top3 was selected for fragmentation, and secondary information was collected. The secondary resolution was 30,000, the AGC was 1 × 105, and the MIT was 50 ms. The stepped energy was set to 20, 40, and 60 eV. The ion source was set at the following conditions: sheath gas flow rate, 40; aux gas flow rate, 10; spray voltage positive ion mode, 3.8; spray voltage negative ion mode, 3.2; capillary temperature, 320 °C; aux gas heater temperature, 350 °C.
Statistical analysis
Continuous variables are represented as medians with interquartile ranges, while categorical variables are expressed as counts (%). The statistical analyses were performed using SPSS 13.0.
DDA data were identified by MaxQuant 1.5.3.30 (Max Planck Institutes) software and the UniProtKB/Swiss-Prot Homo sapiens database. Identification information that met FDR ≤ 1% was used to build the final library. The parameters were selected as follows: enzyme: trypsin; minimal peptide length: 7; PSM-level FDR: 0.01; protein FDR: 0.01; fixed modifications: carbamidomethyl (C); variable modifications: oxidation (M), acetyl (protein N-term). DIA data were analyzed by Spectronaut (Biognosys) software, which used iRT peptide to correct the retention time. Then, based on the target-decoy model applicable to SWATH-MS, the false-positive control was completed at FDR 1% to obtain significant quantitative results. The R package MSstats (Bioconductor) screened differential proteins according to the fold change > 1.5 and P value < 0.05, while screened differential metabolites according to variable importance for the projection (VIP) ≥ 1, fold change ≥ 1.2 or ≤ 0.83, and P value < 0.05. Meanwhile, differential proteins and differential metabolites were enriched by network pathway analysis database: gene ontology (GO) (http://www.geneontology.org.) and the kyoto encyclopedia of genes and genomes (KEGG). The protein–protein interaction network of the difference proteins was analyzed by STRING database (http://string-db.org). DEPs and differential metabolites were calculated by regularized canonical correlation analysis and were visualized using the plotVar functions in the mixOmics package.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository54 with the dataset identifier PXD024311. All data are fully available without restriction.
References
Wu, Y. et al. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet 395, 949–950 (2020).
WHO. Novel coronavirus disease named COVID-19. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 11 Feb 2020.
WHO. Coronavirus disease (COVID-19) weekly epidemiological update and weekly operational update. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed 25 Oct 2020.
Lurie, N., Saville, M., Hatchett, R. & Halton, J. Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 382, 1969–1973 (2020).
Zhu, F. C. et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 395, 1845–1854 (2020).
Xu, Y. H. et al. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. J. Infect. 80, 394–400 (2020).
Bradley, B. T. et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet 396, 320–332 (2020).
George, P. M., Patterson, C. M., Reed, A. K. & Thillai, M. Lung transplantation for idiopathic pulmonary fibrosis. Lancet Respir. Med. 7, 271–282 (2019).
Li, F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 89, 1954–1964 (2015).
Qiao, B. & de la Olvera, C. M. Enhanced binding of SARS-CoV-2 spike protein to receptor by distal polybasic cleavage sites. ACS Nano 14, 10616–10623 (2020).
Ganter, M. T. et al. Interleukin-1beta causes acute lung injury via alphavbeta5 and alphavbeta6 integrin-dependent mechanisms. Circ. Res. 102, 804–812 (2008).
Shimbori, C. et al. Mechanical stress-induced mast cell degranulation activates TGF-β1 signalling pathway in pulmonary fibrosis. Thorax 74, 455–465 (2019).
Li, M. et al. Epithelium-specific deletion of TGF-β receptor type II protects mice from bleomycin-induced pulmonary fibrosis. J. Clin. Invest. 121, 277–287 (2011).
Azuma, A. et al. Interferon-β inhibits bleomycin-induced lung fibrosis by decreasing transforming growth factor-β and thrombospondin. Am. J. Respir. Cell Mol. Biol. 32, 93–98 (2005).
King, T. E. Jr. et al. Effect of interferon gamma-1b on survival in patients with idiopathic pulmonary fibrosis (INSPIRE): a multicentre, randomised, placebo-controlled trial. Lancet 374, 222–228 (2009).
Shen, B. et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 182, 59–72 (2020).
Bojkova, D. et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 583, 469–472 (2020).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Antonio, G. E. et al. Thin-section CT in patients with severe acute respiratory syndrome following hospital discharge: preliminary experience. Radiology 228, 810–815 (2003).
Geyer, P. E., Holdt, L. M., Teupser, D. & Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 13, 942 (2017).
Bensadoun, E. S., Burke, A. K., Hogg, J. C. & Roberts, C. R. Proteoglycan deposition in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 154, 1819–1828 (1996).
Li, Y. et al. Hyaluronan synthase 2 regulates fibroblast senescence in pulmonary fibrosis. Matrix Biol. 55, 35–48 (2016).
Liang, J. et al. Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nat. Med. 22, 1285–1293 (2016).
Cavallaro, U. & Dejana, E. Adhesion molecule signalling: not always a sticky business. Nat. Rev. Mol. Cell Biol. 12, 189–197 (2011).
Harjunpää, H., Llort, A. M., Guenther, C. & Fagerholm, S. C. Cell adhesion molecules and their roles and regulation in the immune and tumor microenvironment. Front. Immunol. 10, 1078 (2019).
Marelli-Berg, F. M., Clement, M., Mauro, C. & Caligiuri, G. An immunologist’s guide to CD31 function in T-cells. J. Cell Sci. 126, 2343–2352 (2013).
Privratsky, J. R., Newman, D. K. & Newman, P. J. PECAM-1: conflicts of interest in inflammation. Life Sci. 87, 69–82 (2010).
Villar, J., Zhang, H. & Slutsky, A. S. Lung repair and regeneration in ARDS: role of PECAM1 and wnt signaling. Chest 155, 587–594 (2019).
Watson, W. H., Ritzenthaler, J. D. & Roman, J. Lung extracellular matrix and redox regulation. Redox Biol. 8, 305–315 (2016).
Cox, N., Pilling, D. & Gomer, R. H. Distinct Fcγ receptors mediate the effect of serum amyloid p on neutrophil adhesion and fibrocyte differentiation. J. Immunol. 193, 1701–1708 (2014).
Pilling, D., Buckley, C. D., Salmon, M. & Gomer, R. H. Inhibition of fibrocyte differentiation by serum amyloid P. J. Immunol. 171, 5537–5546 (2003).
Song, J. W. et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 11, 3410 (2020).
Bournazos, S. et al. Copy number variation of FCGR3B is associated with susceptibility to idiopathic pulmonary fibrosis. Respiration 81, 142–149 (2011).
Castaño, A. P. et al. Serum amyloid P inhibits fibrosis through Fc gamma R-dependent monocyte-macrophage regulation in vivo. Sci. Transl. Med. 1, 5ra3 (2009).
Satoh, T. et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 541, 96–101 (2017).
Oh, J. S. et al. NK cells lacking FcεRIγ are associated with reduced liver damage in chronic hepatitis C virus infection. Eur. J. Immunol. 46, 1020–1029 (2016).
Asai, Y. et al. Aberrant populations of circulating T follicular helper cells and regulatory B cells underlying idiopathic pulmonary fibrosis. Respir. Res. 20, 244 (2019).
Gross, B., Pawlak, M., Lefebvre, P. & Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. Endocrinol. 13, 36–49 (2017).
Kheirollahi, V. et al. Metformin induces lipogenic differentiation in myofibroblasts to reverse lung fibrosis. Nat. Commun. 10, 2987 (2019).
Bargagli, E. et al. Metabolic dysregulation in idiopathic pulmonary fibrosis. Int. J. Mol. Sci. 21, 5663 (2020).
Landi, C. et al. A system biology study of BALF from patients affected by idiopathic pulmonary fibrosis (IPF) and healthy controls. Proteomics Clin. Appl. 8, 932–950 (2014).
Avouac, J. et al. Pan-PPAR agonist IVA337 is effective in experimental lung fibrosis and pulmonary hypertension. Ann. Rheum. Dis. 76, 1931–1940 (2017).
El Agha, E. et al. Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell 20, 261–273 (2017).
Jain, M. et al. Leptin promotes fibroproliferative acute respiratory distress syndrome by inhibiting peroxisome proliferator-activated receptor-γ. Am. J. Respir. Crit. Care Med. 183, 1490–1498 (2011).
Steinritz, D., Stenger, B., Dietrich, A., Gudermann, T. & Popp, T. TRPs in tox: involvement of transient receptor potential-channels in chemical-induced organ toxicity-a structured review. Cells 7, 98 (2018).
Grace, M. S., Baxter, M., Dubuis, E., Birrell, M. A. & Belvisi, M. G. Transient receptor potential (TRP) channels in the airway: role in airway disease. Br. J. Pharmacol. 171, 2593–2607 (2014).
Moncada, S. & Higgs, A. The L-arginine-nitric oxide pathway. N. Engl. J. Med. 329, 2002–2012 (1993).
Mehta, S., Stewart, D. J., Langleben, D. & Levy, R. D. Short-term pulmonary vasodilation with L-arginine in pulmonary hypertension. Circulation 92, 1539–1545 (1995).
Gao, L. et al. Combination of L-Arginine and L-Norvaline protects against pulmonary fibrosis progression induced by bleomycin in mice. Biomed. Pharmacother. 113, 108768 (2019).
Ellul, M. A. et al. Neurological associations of COVID-19. Lancet Neurol 19, 767–783 (2020).
China NHCO. New coronavirus pneumonia diagnosis and treatment scheme (6th edn). https://www.gov.cn/zhengce/zhengceku/2020-02/19/content_5480948htm. Accessed 18 Feb 2020.
Chung, M. et al. CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology 295, 202–207 (2020).
Bernheim, A., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology 295, 200463 (2020).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res 47, D1211–D1217 (2019).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81960246, 81701089, 81560044, 30860113, 81960026 and 82003967), the Guangxi Natural Science Foundation (2017GXNSFBA198010 and 2020GXNSFAA238003), the Guangxi Medical and Health Appropriate Technology Research and Development Project (S2020076, S201422-01 and S2019087), and the Shanxi Health Research Project (2019165).
Author information
Authors and Affiliations
Contributions
J.R.Y., C.X.C., W.C., W.S.L. and M.Z. designed the research study and had full access to all data in this study. C.X.C., W.C., L.Y.H., Z.F., K.Y., and L.W.L. collected the data. J.R.Y., C.X.C., and W.C. drafted the manuscript and summarized all data. Z.H.N. and X.Z. analyzed the data. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, J., Chen, C., Chen, W. et al. Proteomics and metabonomics analyses of Covid-19 complications in patients with pulmonary fibrosis. Sci Rep 11, 14601 (2021). https://doi.org/10.1038/s41598-021-94256-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-94256-8
This article is cited by
-
Gene expression profiling of host lipid metabolism in SARS-CoV-2 infected patients: a systematic review and integrated bioinformatics analysis
BMC Infectious Diseases (2024)
-
Plasma Metabonomics of Human Adenovirus-infected Patients with Pneumonia and Upper Respiratory Tract Infection
Current Medical Science (2024)
-
Multi-omics Approach in Kidney Transplant: Lessons Learned from COVID-19 Pandemic
Current Transplantation Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.