Abstract
The Hedgehog (Hh)/Gli signaling pathway controls cell proliferation and differentiation, is critical for the development of nearly every tissue and organ in vertebrates and is also involved in tumorigenesis. In this study, we characterize the oncoprotein SET/I2PP2A as a novel regulator of Hh signaling. Our previous work has shown that the zebrafish homologs of SET are expressed during early development and localized in the ciliated organs. In the present work, we show that CRISPR/Cas9-mediated knockdown of setb gene in zebrafish embryos resulted in cyclopia, a characteristic patterning defect previously reported in Hh mutants. Consistent with these findings, targeting setb gene using CRISPR/Cas9 or a setb morpholino, reduced Gli1-dependent mCherry expression in the Hedgehog reporter zebrafish line Tg(12xGliBS:mCherry-NLS). Likewise, SET loss of function by means of pharmacological inhibition and gene knockdown prevented the increase of Gli1 expression in mammalian cells in vitro. Conversely, overexpression of SET resulted in an increase of the expression of a Gli-dependent luciferase reporter, an effect likely attributable to the relief of the Sufu-mediated inhibition of Gli1. Collectively, our data support the involvement of SET in Gli1-mediated transcription and suggest the oncoprotein SET/I2PP2A as a new modulator of Hedgehog signaling.
Similar content being viewed by others
Introduction
The Hedgehog (Hh) signaling pathway controls cell proliferation and differentiation and is critical for the development of nearly every tissue and organ in vertebrates1. Lack of Hh signaling during development is embryonic lethal, while hyperactivation of the Hh pathway promotes tumor formation and maintenance of a wide range of human malignancies, including medulloblastoma and basal cell carcinoma of the skin2. Several findings suggest that Hh signaling might also modulate the tumor microenvironment, which has important implications for drug screening and therapeutic interventions3. Therefore, Hh signal transduction has emerged as a key pathway for cancer therapy and its pharmacological modulation is of great importance in clinical trials4.
Unlike other signaling pathways involved in development, the vertebrate Hh signaling is dependent on the primary cilium, a microtubule-based membrane protrusion that functions as an antenna in the cells5. The main effectors of the pathway are Gli transcription factors (Gli1, Gli2, Gli3) which belong to the Krüppel family of zinc finger proteins. Gli1 and Gli2 are positive regulators of transcription, whereas Gli3 mainly functions as a transcription repressor6. Upon their transportation into the nucleus, Gli transcription factors activate target genes that participate in the regulation of proliferation and self-renewal7,8. The localization and the activity of Gli transcription factors, control both the amplitude and the outcome of the Hh expression program, especially during development, and are regulated by a complex combination of posttranslational modifications, including phosphorylation8,9, acetylation10 and ubiquitination/proteasomal truncation11.
The oncoprotein SET is mainly a nuclear protein that functions as a histone chaperone12,13, transcription cofactor14,15,16, regulator of histone acetylation17,18, modulator of DNA repair19 and as an epigenetic regulator20. Besides its nuclear function, SET is an inhibitor of protein phosphatase 2A (PP2A)21 and has also been reported as a signaling amplifier of Rac1-dependent cell migration22.
SET was first characterized in acute undifferentiated leukemia23 and is overexpressed in various cancers, including lymphoid malignancies24 and non-small cell lung carcinoma (NSCLC)25. In addition, SET is involved in the epithelial mesenchymal transition (EMT)26 and in the regulation of cancer cell stemness27.
Several studies have focused on SET targeting as a potential therapeutic approach in cancer28,29, by testing the effects of two SET inhibitors, COG11228,30 and FTY72024,31,32,33 on cancer progression. COG112 is an apolipoprotein E (apoE)-mimetic peptide which inhibits SET-Rac1 interaction resulting to decreased cell migration and invasion of cancer cells28,30. FTY720 (Fingolimod) is a sphingosine immunosuppressant drug that also mediates the suppression of several tumors24,31,32,33. Recent work has reported impairment of triple negative breast cancer progression upon blockage of SET function by TD19, a new compound which is a derivative of erlotinib34. Besides the pharmacological inhibition of SET, other studies have shown that knocking down SET in Smo1 primary medulloblastoma cells induces cell death, suggesting a potential function of the oncoprotein in Hh signaling35. In this study, we present several pieces of evidence which support the involvement of SET in Gli1-mediated transcription that plays a crucial role in the Hh signaling cascade.
Results
SET is involved in Hh signaling in zebrafish
Previous work of our lab has shown that the zebrafish homologs of SET oncoprotein are expressed during early zebrafish development and localized in the ciliated organs of zebrafish, suggesting a potential function of SET in signaling cascades coordinated by cilia36. This study also demonstrated that targeting setb gene expression by injections of setb gRNA with Cas9 mRNA or seta/b morpholinos into one-cell stage zebrafish embryos resulted in several morphological abnormalities, including defects in spinal cord and eye development36. A systematic scoring of the eye defects in setb crispants showed that approximately 80% of the larvae with severe phenotype and 33% of the larvae with moderate phenotype exhibited cyclopia (Fig. 1A), a characteristic embryonic patterning defect reported in the Sonic Hedgehog mutant mouse and zebrafish37,38.
Impact of targeting the setb gene on Hedgehog signaling in zebrafish embryos. (A) Zebrafish setb crispants exhibit cyclopia. One- to two-cell stage zebrafish embryos were injected with 4.6 nl of 150 ng/μl of Cas9 mRNA or with 50 ng/μl of setb gRNA and 150 ng/μl of Cas9 mRNA and were examined morphologically at 48 hpf. (Right graph). Phenotypic scoring of the injected embryos (n = 262, n denotes the number of injected embryos). Numbers in X-axis refer to percentage of embryos in each phenotypic group and numbers on the bars refer to the percentage of cyclopic embryos in each phenotypic group. (B) Hh signaling is reduced in Tg12x_Gli transgenic zebrafish embryos injected with setb gRNA/Cas9. One- to two-cell stage Tg12x_Gli transgenic zebrafish embryos were injected with setb gRNA/Cas9 and examined under a confocal fluorescence microscope at 24 hpf. The phenotypic scoring of the injected embryos is shown in the graph (n = 96). The squares show enlargement of the trunk areas that display decreased mCherry fluorescence intensity. Note the disrupted morphology of the somites in the marked regions.
To investigate in detail the potential role of setb gene in Hh signaling we next used the Sonic Hedgehog reporter zebrafish line Tg(12xGliBS:mCherry-NLS), which expresses the mCherry reporter under the transcriptional regulation of Gli139. To this end, setb gRNA was coinjected with Cas9 mRNA in Tg12x_Gli embryos and the transgene expression was recorded at 24 hpf. As shown in Fig. 1B, setb crispants displayed a decrease of the mCherry reporter expression, mainly in the trunk region. Importantly, the somite development, a process that depends on the Hh signaling pathway, was disturbed in the injected embryos, as shown by the defective segmentation of the somites (Fig. 1B). Similar results were obtained upon targeting both seta and setb genes in the transgenic embryos with a translation blocking morpholino (MOab)36 (Fig. 1S).
In parallel, we performed knockdown experiments to block the setb function by injecting one-cell stage Tg12x_Gli zebrafish embryos with antisense morpholino oligonucleotides targeting the setb gene. setb morphants exhibited reduced mCherry reporter activity compared to age-matched siblings, indicating lower levels of Hh signaling activation. In accordance with the setb crispants, the morphants appear to show decreased mCherry fluorescence in both the brain and the trunk area (ventral neural tube and conformation of somites) (Fig. 2A). In order to analyze at cellular resolution the reduction of Hh signaling at the neural tube of setb morphants, 48 hpf larvae were sectioned transversally. Hh-activated cells (mCherry+) both at the floor plate of neural tube and the slow fibers adjacent to the notochord exhibited significant signal reduction compared to the control siblings (Fig. 2B). These data suggest the involvement of set genes in the regulation of Hh signaling during zebrafish development.
Activation of Hh signaling is reduced in setb morphants. (A) Fluorescent and brightfield images of 48 hpf setb-morpholino injected and control sibling Tg12x_Gli embryos. Embryos are placed anterior to left, dorsal up. Scale bars 100 μm. Quantification of pixel intensity at 48 hpf of the morphologically mildly affected embryos (***p < 0.001). Data are expressed as mean ± SEM (n = 129, after exclusion of the injected embryos developing apoptotic tissue at 24 hpf). Percentage of phenotypic scoring of 48 hpf morphants is presented with mean ± SEM and the numbers of total embryos per group is indicated. At the brain region: cerebellum (c, white arrow), mid-hindbrain boundary (MHB, yellow arrow). At the trunk region: floor plate (FP, yellow arrow), slow muscle pioneer cells (MP, white arrow, high mCherry signal), superficial slow fibers (SSF, white arrows, mid mCherry signal). (B) Confocal images of cross-sections throughout the trunk region of 48 hpf setb morphants and age-matched siblings. Note the reduction of mCherry+ cells at the ventral side of neural tube (yellow arrows) and at the slow fibers surrounding notochord (white arrows) of setb morphants; indication of lower activation levels of the Hh pathway. Sections were stained with phalloidin-633 (blue) and DAPI (grey). NT: neural tube, nc: notochord. Scale bars 50 μm.
Targeting SET in NIH 3T3 cells inhibits Hh signaling
To further study the role of the oncoprotein SET in the Hh pathway, we used NIH 3T3 cells, a well validated Hh-responsive cell line. To this end, we established SET knock down (SETKD) cell lines by utilizing the CRISPR/Cas9 technology. A plasmid expressing a small guide RNA targeting exon 4 of the mouse Set gene was cotransfected with Cas9 mRNA. Set mRNA and protein levels were analyzed in different clones to verify the ablation of the protein. As shown in Fig. 3A, H4 and H11 clones exhibited reduced SET levels (lanes 2, 3), while the G5 clone displayed SET expression comparable to control cells (lane 4). These results were verified by qPCR analysis of SETKD clones which showed that Set gene expression was knocked down in the H4 and H11 clones. The generation of multiple Set knockdown clones and no clone with complete SET loss in our CRISPR experiment may indicate that mouse Set genes are essential and therefore their complete knockout was not feasible. Nevertheless, SETKD cell lines represent a cell-based experimental system complementary to zebrafish morphants and crispants that also express reduced protein levels of the zebrafish Seta/b proteins36.
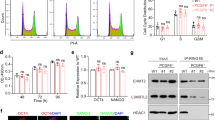
We next asked whether the stimulated expression of Gli1 transcription factor, the constitutive activator of Hh signaling, was affected in the H4 and H11 clones. Upon treatment of control NIH 3T3 cells with purmorphamine, a specific agonist of Smo, the expression of Gli1 was increased, as expected (Fig. 3C, lanes 1, 2). Interestingly, the decrease of SET protein levels in H4 and H11 SETKD cells attenuated the stimulated Gli1 expression (Fig. 3C, lanes 4, 8), in agreement with the in vivo experiments in zebrafish (Figs. 1, 2). On the contrary, the stimulated expression of Gli1 in the G5 clone was similar to Gli1 levels of control cells (Fig. 3C, lane 6). Ciliation analysis of H4 and H11 cells showed that cilia number, a crucial structural component of Hh signaling5, was not affected upon the decrease of SET protein levels, indicating a direct effect of SET in Hh signaling (Fig. 3D).
CRISPR/Cas9-mediated SETKD is linked with a decrease in Gli1 expression. (A) SET protein levels in NIH 3T3 control cells and SETKD clones. Western blot analysis of control NIH 3T3, H4, H11 and G5 cell extracts using the anti-I2PP2A and anti-α-tubulin antibodies (loading control). Original uncropped Western Blot images are shown in Supplementary Fig. 2S. (B) Comparison of the relative expression differences for Seta and Setb, the two splicing forms of mouse Set gene, in control NIH 3T3, H4, H11 and G5 cells. The fold changes in qPCR were normalized to Gapdh. Data are expressed as mean ± SEM (n = 3 ) **p < 0.01***p < 0.001. (C) Gli1 expression levels in control NIH 3T3 and SETKD cells. Western blot analysis of extracts obtained from control NIH 3T3 cells and H4, H11 and G5 cells after incubation at 0.5% serum with DMSO (lanes 1, 3, 5, 7) or 2 μM purmorphamine (lanes 2, 4, 6, 8) using the anti-Gli1 and anti-α-tubulin antibodies. Original uncropped Western Blot images are shown in Supplementary Fig. 2S. (D) Ciliation analysis of control NIH 3T3 cells and H4, H11 clones after 24 h of serum starvation. 100–150 cells were analyzed per experiment (n = 3).
In a growing number of studies, pharmacological inhibition of SET has been achieved by means of two well-studied inhibitors of SET, FTY72024,31,32,33 and COG11228,40. FTY720 is a sphingosine immunosuppressant drug which has been shown to have anti-tumor properties in different human malignancies31,32, including Hh-dependent medulloblastoma33. The interaction of FTY720 with SET is direct and has been studied in detail by distinct and complementary approaches, including molecular modelling31, biochemical binding assays31 and recently by NMR experiments41. To analyze the role of FTY720 in Hh signaling, we checked the effect of this compound in the stimulated levels of Gli1 by purmorphamine. Our results showed that treatment of the cells with 5 μΜ FTY720 prevented the increase of the stimulated levels of Gli1 (Fig. 4A, lane 4). Interestingly, treatment of the cells with COG112, an apolipoprotein E peptide 28,40 resulted to similar inhibition of Gli1 protein levels (Fig. 4A, lane 3). To account for differences in the temporal transcriptional response of genes involved in the Hedgehog pathway, in the presence of SET inhibitors, we performed quantitative RT-PCR analysis. Cells were cultured with 2 μΜ purmorphamine in the presence of 5 μΜ FTY720 or 3 μΜ COG112 and mRNA was isolated. As expected, upon treatment of the cells with the Hh agonist, the expression of Gli1 and Ptch1, key genes that are directly stimulated by Hh signaling, was increased relative to control (Fig. 4B, C). Furthermore, the expression of the cell cycle regulator cyclin D1 (Ccnd1), a Gli1 target gene, was also increased, as expected (Fig. 4B, C). Importantly, upon treatment of the cells with FTY720 or COG112, the abundance of Gli1 and Ptch1 mRNA was reduced in both cases, while Ccnd1 was downregulated only in the presence of FTY720 (Fig. 4B, C). These results, taken together, suggest the involvement of SET in Hh signaling, in agreement with the data obtained from the zebrafish model system.
SET inhibitors decrease Gli1 expression. (A) The effect of FTY720 and COG112 on Gli1 protein expression. Western blot analysis of extracts obtained from NIH 3T3 cells after incubation at 0.5% serum with DMSO (lane 1) or 2 μM purmorphamine (lane 2) in the presence of 3 μM COG112 (lane 3) or 5 μM FTY720 (lane 4). Proteins were resolved in 10% SDS-PAGE and the blots were probed with the anti-Gli1, anti-SET and anti-α-tubulin antibodies (loading control). Original uncropped Western Blot images of panel A are shown in Supplementary Fig. 2S. (B, C). The effect of FTY720 and COG112 on the mRNA levels of selected genes related to Hedgehog signaling. Comparison of relative expression differences for Gli1, Ptch1, and Ccnd1 in NIH 3T3 cells cultured at 0.5% serum after incubation with 2 µM purmorphamine in the presence of 5 μΜ FTY720 or 3 μΜ COG112, as determined by qPCR analysis. The mRNA levels of the genes were normalized to Gapdh. The experiments were repeated three times. Data are expressed as mean ± SEM ***p < 0.001.
SET affects Gli1-mediated transcription
Several pieces of evidence have demonstrated distinct roles of SET in gene regulation, as a transcription cofactor through interaction with CBP/p30014,16 and as regulator of histone acetylation17,18. We therefore hypothesized that SET may regulate Hh target gene expression by affecting the transcriptional activity of Gli proteins, rather than by targeting upstream pathway components, such as Patched or Smo. To test this hypothesis, we first asked whether SET could affect Gli1-mediated transcription in the absence of upstream Hh pathway activation. To this end, HEK 293 cells were transfected with a 12xGliBS luciferase construct that contains twelve copies of Gli-binding sites42 together with Gli1 and Flag-SET. As shown in Fig. 5A, transcription driven by the luciferase reporter was activated by Gli1, as expected. Interestingly, SET overexpression increased Gli1-mediated transcription (Fig. 5A).
SET overexpression increases Gli1-mediated transcription. (A) The effect of SET on Gli1-mediated transcription. To determine Gli1-mediated transcription upon SET overexpression, 12xGliBS-luc and Gli1 expression plasmids were cotransfected with a human Flag-SET expressing construct in HEK 293 cells. (B) The effect of SET on Sufu inhibition of Gli1-mediated transcription. HEK 293 cells were transfected with 12xGliBS-luc reporter with the indicated amounts of plasmids encoding Gli1 (100 ng), Flag-SET (100 and 200 ng) and Sufu (10 ng). pRL-TK expresses Renilla luciferase and was included in all samples to normalize the transfection efficiency. Experiments were repeated three times and samples were analyzed in triplicates, *p < 0.05, **p < 0.01, ***p < 0.001. (C) Expression of GST-SET and GST-SAP18 recombinant proteins. Bacterially expressed glutathione-S-transferase-tagged SAP18 (GST-SAP18), GST protein and baculovirus expressed GST-SET (approximately 2–3 μg each) were immobilized on glutathione-Sepharose beads and analyzed by 12% SDS-PAGE. An original uncropped image of panel C is shown in Supplementary Fig. 2S. (D) GST-SET pull down experiments. GST-SET or GST was immobilized on glutathione-Sepharose beads and incubated with cell extracts expressing GFP-SAP18 or GFP. (E) GST-SAP18 pull down experiments. GST-SAP18 or GST was immobilized on glutathione-Sepharose beads and incubated with cell extracts expressing GFP-SET or GFP. The beads and the supernatant of the pull down assays were analyzed by western blot using the anti-GFP antibody. Original uncropped Western Blot images of panels D and E are shown in Supplementary Fig. 2S.
It is well documented that Gli1 function is regulated by Sufu, an essential negative modulator of Hh signaling that affects both the nucleocytoplasmic localization of Gli17,43,44 and the transcriptional activity of Gli1 in the nucleus, by distinct mechanisms45,46,47,48.
Based on this knowledge, we then asked whether SET could affect Sufu-mediated repression of Gli1 transcriptional activity. As shown in Fig. 5B, upon expression of Sufu, Gli1-mediated transcription was markedly inhibited, as previously reported45. Interestingly, coexpression of Flag-SET significantly relieved SuFu-mediated inhibition of Gli1 transcriptional activity (Fig. 5Β). A possible explanation for these results is that SET may modulate Sufu protein interactions, leading to increased Gli1 transcriptional activity. We tested this hypothesis by analyzing protein interactions of GFP-SET or GFP by immunoprecipitation experiments using the anti-GFP antibody, followed by mass spectrometry (MS) analysis of the immunoprecipitated proteins. This experimental approach identified SAP18 as an interactor of SET, a component of the mSin3 and histone deacetylase complex48,49 that cooperates with Sufu to repress Gli1-mediated transcription45,46. The MS analysis also revealed two known interactors of SET, glyceraldehyde-3-phosphate dehydrogenase50 and T-complex protein 1 subunit delta51. The interaction between SET and SAP18 was verified biochemically by glutathione S-transferase (GST) pull-down assays using GST-SET as bait in cell extracts expressing GFP-SAP18 or GFP protein (Fig. 5C). Consistent with the results of the mass spectrometry analysis, GFP-SAP18 was bound specifically to bead-immobilized GST-SET (Fig. 5D). The interaction of SET with SAP18 was further validated by GST-SAP18 pull down experiments in cell extracts expressing GFP-SET or GFP (Fig. 5E). These experiments provide evidence that SET interacts specifically with SAP18. Further studies should investigate the functional significance of SET-SAP18 binding with respect to Gli1 transcription at the level of Gli1-Sufu complex, taking into consideration the dynamic nature of these interactions with respect to space and time and their dependence on the cellular context.
Discussion
The remarkable network of gene activity regulated by Hh signaling can govern cell proliferation, survival and fate, alternatives that should be fine-tuned depending on cell context and time1. The Hh-dependent genes involved in cell proliferation, survival and differentiation are mostly activated and correspond to a wide variety of molecular functions52,53. In the present study, using in vivo experiments and cell-based assays we present several pieces of evidence which demonstrate that the oncoprotein SET is involved in the Hedgehog pathway through modulation of Gli1-mediated transcription.
In our work, using the zebrafish model organism, we have shown that the setb crispant larvae exhibited cyclopia, a characteristic embryonic patterning defect, first reported in the Shh mutant mouse37 and later in zebrafish38. Besides the craniofacial and eye malformations, the crispants also displayed characteristic patterning defects in the spinal cord and somite development. These results are in agreement with genetic studies in zebrafish which have shown that loss of Hh signaling leads to ventral curvature of the body, deficiencies in ventral forebrain specification and spinal cord defects6, 54. Further evidence for the involvement of SET in the Hh pathway was obtained using the Tg12x_Gli transgenic embryos that express the mCherry reporter under the transcriptional regulation of Gli142. Upon blocking the expression of setb gene using two different morpholinos, the setb splice blocking morpholino (MOb) and the translation blocking morpholino (MOab) that targets both seta and setb36, the transgene expression was markedly reduced. Importantly, the same effect on the transgene expression was observed in setb crispants. These experiments also revealed defects in somite development of both set morphants and setb crispants. The reduction of Hh signaling activation was observed in Tg12x_Gli embryos also when the overall morphology of the morphants or crispants was not severely affected, indicating that the modulation of the Hh pathway by Seta/b is one of its primary targets.
The somite morphology is an Hh-dependent process55 and is considered as a suitable way to diagnose impaired Hh signaling in living zebrafish embryos56. The first somites appear at 11 hpf on both sides of the midline and at 16 hpf the formed somites reach the tail tip and acquire the characteristic chevron shape. In contrast to wild-type embryos, Hh mutants exhibit unshaped and rounded somites56. Our data are also in agreement with previous studies which showed similar developmental defects upon loss of Hh activation and identified Gli1 as the main activator of Hh signaling in zebrafish54.
At the cellular level, and in agreement with the in vivo data from the zebrafish, the decrease of SET protein levels in SETKD cells attenuated the stimulated Gli1 expression. Similar results on Gli1 expression levels were obtained upon blocking SET function in NIH 3T3 cells with FTY720, a known inhibitor of SET28,31,32. Saddoughi et al.31 demonstrated that FTY720 directly binds SET with a Kd = 11 nM, as determined by surface plasmon resonance (SPR). Recently, the SET-FTY720 complex was characterized by NMR experiments revealing important structural information for the design of new SET inhibitors41. Consistent with the effect of FTY720 on Gli1 expression, upon treatment of the cells with COG112, an apolipoprotein E peptide that inhibits SET28,40, Gli1 expression after purmorphamine stimulation was also decreased. Future studies are required to test whether SET inhibitors also affect Hh signaling in the context of cancer.
Nowadays, ongoing research has focused on targeting the Hh pathway downstream of Smo by studying molecules that modulate Hh signaling at the level of Gli1 transcription factor57,58. Therefore, unraveling the underlying mechanisms by which SET affects Gli1 transcription and Hh signaling is an important task. SET is an intrinsic protein inhibitor of Protein phosphatase 2A (PP2A)21 and thus may regulate PP2A activity participating in the complex interplay of kinases, phosphatases and associated factors that function at different levels in the Hh pathway8,9,11. Genetic studies in Drosophila have shown that PP2A is involved in the regulation of Hh signaling59. In mammalian cells, an increase of PP2A activity in cancer cells led to retention of full-length Gli3 in the cytoplasm and consequently Gli3 decreased transcription activity, while Gli1 and Gli2 activities were unaffected60. Our preliminary experiments have shown that the protein levels of PP2A and its enzymatic activity in NIH 3T3 cells were not significantly affected upon purmorphamine activation of the Hh pathway. Therefore, future work should investigate whether SET-PP2A interaction plays a role in regulating the phosphorylation status of the Gli1 transcription factor.
At the transcription level and in the absence of upstream Hh pathway activation, SET overexpression in HEK 293 cells resulted to an increase of Gli1-mediated transcription. This result was not surprising, since several studies have shown that SET modulates chromatin status in different ways and at different levels, acting as a histone chaperone12,13, regulator of acetylation/deacetylation17,18, epigenetic cofactor20 and transcription coactivator through interaction with CBP/p30014,16. Concerning the regulation of Gli transcription factor activity, genetic studies, initially reported in Drosophila and later in mammalian cells, have identified CBP as an essential cofactor for Ci and Gli transcription factors61. Since CBP possesses histone acetyltransferase (HAT) activity, its interaction with Gli transcription factors possibly leads to epigenetic changes of Gli targets, facilitating the accessibility of other regulators to chromatin. In this context, SET may co-operate with CBP to enhance Gli1 mediated transcription of different Hh target genes. Consistent with this hypothesis, previous work has shown that the acetyltransferase PCAF interacts with Gli1 and regulates acetylation of histone H3 on Hh target gene promoters62. Further evidence on the epigenetic regulation of Gli1 transcriptional activity comes from studies which have documented the recruitment of SAP18, a component of the Sin3 histone deacetylase complex47,48,49, through Sufu to Gli145,48. SAP18 enhances mSin3-HDAC1-mediated transcriptional repression, directing the formation of a repressive complex when tethered on promoters48. Besides its involvement in the Hh pathway, SAP18 also functions as a scaffold protein for the formation of a splicing complex and the assembly of other multiprotein complexes via its ubiquitin-like fold63. Interestingly, our proteomic analysis identified SAP18 as a new interactor of SET. Furthermore, gene reporter assays showed that SET overexpression relieved the Sufu-mediated inhibition of Gli1 transcriptional activity. Based on these results, we could propose the involvement of SET in the epigenetic network that fine tunes the activity of Gli proteins in response to the Hh signaling gradient through its interaction with SAP18. Further experiments are required to shed light into the underlying mechanisms of SET function in the Hh pathway, facing the major challenges associated with the dynamic nature of Gli-Sufu-SAP18 interactions in the cytoplasm and the nucleus.
Methods
Zebrafish experiments
Breeding and maintenance of wild-type zebrafish and 12xGliBS:mCherry-NLS transgenic line were carried out as previously described36 in accordance with the European Directive 2010/63 for the protection of animals used for scientific purposes and the Recommended Guidelines for Zebrafish Husbandry64 . The experimental protocols described in this study were carried out with zebrafish larvae up to 48 h post fertilization and were approved by the Foundation for Research and Technology-Hellas (FORTH) Ethics Committee. Reverse genetics experiments involving CRISPR/Cas9 were performed and analyzed during embryonic stages of zebrafish development, as described36. The study was carried out in compliance with the ARRIVE guidelines (https://arriveguidelines.org).
Zebrafish embryo microinjections, imaging and analysis
Microinjections of setb morpholino (5–7 ng/embryo) were conducted at the one-cell stage of Tg12x_Gli embryos. Monitoring of setb morphants and controls was done under stereoscope for scoring phenotypic abnormalities, and brightfield images were obtained at 24 and 48 hpf with DFK2BUC03 camera from The Imaging Source under SMZ1000 stereoscope. mCherry fluorescence (and therefore Hh activity) of Tg12x_Gli injected and siblings embryos was monitored at 48 hpf and brightfield and fluorescent images were acquired from injected and control embryos under microscope inverted Leica DMIRE2 with a mounted Hamamatsu ORCA-Flash4.0 camera. In total, 150 embryos were injected. For quantitative image analysis of microscopy images, the open-source software Image J/FiJi (ImageJ 1.52p) (http://imagej.nih.gov/ij)65 was used. Pixel intensity was calculated from the captured images by: mean of Region of Interest (ROI)-mean of unstained area (same size as ROI) for each embryo at the region of the brain and the trunk.
Immunohistochemistry and confocal microscopy
48 hpf embryos were fixed with 4% paraformaldehyde overnight at 4℃. Whole-mount embryos were embedded in 4% agarose and cut in 150 μm thick sections with Leica VT1OOOS Vibratome. Sections were washed 3 times for 15 min with PBT (0.8% Triton X-100 in PBS) and then incubated for 3 h at room temperature in phalloidin-633 (1:300 in PBT) for filamentous actin staining and 20 min at room temperature for DAPI-stained nuclei. Imaging was performed using Leica TCS SP5II upright confocal microscope. Images were capture with LAS AF Software, analyzed in ImageJ Software and presented as maximum projection of a set of z-stacks for each stained tissue section.
Cell culture and drug treatments
Mouse embryonic fibroblast cells NIH 3T3 and HEK 293 cells were cultured in DMEM (Dulbecco’s Modified Eagle Medium) supplemented with 10% heat inactivated FBS, 100 units/ml penicillin, 100 units/ml streptomycin, 1000 mg/lt glucose and 1 mM L-glutamine. Cells were maintained at 37 °C in a humidified incubator with 5% CO2 in the air atmosphere. FTY720 (SML0700, Sigma-Aldrich) was resuspended in sterile H2O and used in final concentrations of 5, 7.5 and 10 μM. COG112 (acetyl-RQIKIWFQNRRMKWKKCLRVRLASHLRKLRKRLL-amide) peptide, which was synthesized and purchased from Davids Biotechnologie, was dissolved in PBS and used at a working concentration of 3 μM. The Smo agonist, purmorphamine (Sigma-Aldrich) was dissolved in DMSO and used at a final concentration of 2 μΜ. Pharmacological experiments involving the exposure of cells to SET inhibitors, FTY720 and COG112, were performed in the context of activated Hedgehog signaling. More specifically, NIH 3T3 cells were cultured in DMEM and when cells reached about 60% confluency, the full serum medium was replaced with low serum medium (0.5% FBS). Then, FTY720 or COG112 were added to the medium at different concentrations, and one hour later purmorphamine (2 μΜ) was also added. Cells were collected after 24 h. Protein or total RNA extracts were prepared for further analyses.
Cilia number measurements
Control NIH 3T3, H4 and H11 cells were serum starved in DMEM containing 0.5% FBS for 24 h and then stained for acetylated tubulin using mouse anti-acetyl-alpha tubulin antibody (clone 6-11B-1 Sigma-Aldrich, 1:2000). The secondary donkey anti-mouse antibody-Alexa Fluor 488 (1:200) was obtained from Invitrogen. The number of the cilia was determined by the acetylated alpha tubulin signals by randomly counting cilia number in three different fields each containing 70–120 cells. For each clone, the experiments were performed in triplicates. Ciliated cells were counted using the open-source software Image J/FiJi (ImageJ 1.52p)65.
Western blot analysis
Cells were cultured in 6-well plates, washed with PBS and resuspended in RIPA buffer (20 mM Tris, 150 mM NaCl, 0.1% Triton X-100, pH 7.4) supplemented with Protease Inhibitor Cocktail (Roche). Protein concentrations were determined by the Bradford Assay (Bio-Rad). For the detection of SET and Gli1, proteins were separated on 10% SDS-PAGE and transferred to nitrocellulose membranes (0.45 μm, GE Healthcare) for 70 min (200 mA). The membranes were cut into two pieces at the 75 kD marker for the detection of Gli1 (upper part) and SET (lower part). The membranes were rinsed with PBS-0.1% Tween 20 (PBST ) and incubated with blocking buffer (5% non-fat milk in PBST) for 1 h at RT and incubated overnight at 4 °C with mouse monoclonal anti-Gli1 (L42B10, Cell Signaling, 1:1000) or for 2 h at RT with mouse monoclonal anti-I2PP2A (F-9 sc-133138 Santa Cruz, 1:2000) or mouse monoclonal anti-α-Tubulin (B-5–1-2 Sigma-Aldrich, 1:4000) to verify equal loading. Membranes were then washed with PBST and incubated with the monoclonal HRP conjugated secondary antibody (1:3000). Proteins were detected using the enhanced chemiluminescence method (ECL, Millipore).
Luciferase reporter assays
To determine Gli1-mediated transcription, HEK 293 cells were seeded in 24‐well plates and transfected with 360 ng of 12xGli binding site luciferase reporter plasmid (12xGliBS)42 and 40 ng of Renilla control reporter plasmid (pPRL-TK), together with 100 ng of pCMV5‐Gli1‐Flag expression construct using jetPEI. The total amount of plasmid DNA in each well, 700 ng, was adjusted with the addition of the pcDNA3.1 vector. After 24 h of plasmid transfection, the cells were lysed and the luciferase activity was measured using Dual-Luciferase Reporter Assay. All experiments were repeated three times and samples were analyzed in triplicates.
Glutathione S-Transferase (GST) pull down assays
Baculovirus expressed glutathione-S-transferase-tagged SET was prepared according to Bac-To-Bac baculovirus expression system procedure (Invitrogen), as described15. To generate GST fusion SAP18, the coding sequence of human SAP18 (isoform 2) was subcloned into EcoRI/XhoI sites of pGEX-6P-3 vector (Pharmacia) in frame with glutathione S-transferase (GST) using the following primers: forward 5′-CCCGAATTCCATGCTCGCTGCAGGGGTCG-3′ and reverse 5′-GGGCTCGAGTTAATATGGTCTCATGCGCCC-3′. The GST-SAP18 protein was expressed in E. coli and purified according to standard procedures. For the preparation of cell extracts, HEK 293 cells were transfected with GFP-SET, GFP-SAP18 or GFP expression constructs using jetPEI, as described. 8 × 106 cells were lysed with 150 μl of 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5 mM PMSF (phenylmethylsulfonyl fluoride) supplemented with Protease Inhibitor Cocktail (Roche) and incubated on ice for 5 min. To perform GST-pull down experiments, GST-SET, GST-SAP18 or GST used as control (approximately 2–3 μg each) were immobilized on glutathione-Sepharose beads and the beads were incubated with 150 μg of cell extracts in a total volume of 100 μl for 4 h at 4 °C. Then, the beads were harvested, washed three times with lysis buffer (5 ml total) and resuspended in 25 μl of SDS sample buffer. Bound proteins and the supernatants of the pull down assays were fractionated on 12% SDS–polyacrylamide gels and analyzed by western blotting. GFP-SET and GFP-SAP18 were detected using the goat anti-GFP antibody (SICGEN 1:2000).
CRISPR/Cas9 technology
Identification and selection of the guide sequences to target Set gene, were performed using an online CRISPR Design Tool (http://tools.genome-engineering.org). The search for the most suitable target sites on the genomic sequence of mouse Set (Gene ID: 56086, NC_000068.7:30061 996 30072577) was based on the fact that two splice variants, Seta and Setb, are expressed from this gene. Two gRNAs were selected, located in exon 4, the first common exon for both variants. Set gRNA1, forward 5′-CACCGCAACTCAGTTCAGTTCAAGT-3′ and reverse 5′-AAACACTTGAACTGAACTGAGTTGC-3′; Set gRNA2, forward 5′-CACCGTCTAGTGTCTGCACTGCTTG-3′ and reverse 5′-AAACCAAGCAGTGCAGACACTAGAC-3′. The oligos were synthesized, annealed and cloned into PX459 (Addgene 48139) and pSpCas9(BB)-2A-GFP (PX458, Addgene 48138), respectively. To generate a stable cell line, CRISPR plasmids were introduced into NIH 3T3 cells by transfection with polyethylenimine (PEI). Transfection efficiency was assessed the next day based on GFP expression under a fluorescent microscope. For antibiotic selection, 1.5 μg/ml puromycin was added to the medium and renewed daily. Clonal isolation and expansion were achieved by seeding individual cells in a 96-well plate. Cell growth was monitored and once the formation of single colonies was observed, each monoclonal population was allowed to expand and then transferred to larger culture dishes. To screen for indels genomic DNA was isolated, PCR amplified using primers spanning the target region and subjected to Sanger sequencing (forward 5′-AGTCTGTTTGCAGTTGGTGG-3′ and reverse 5′-CACCTATCAACTGTCTGACTTCAT-3′). Validation of successful gene-editing in selected clones was performed by assessing SET expression levels by western blot analysis using a commercially available antibody (anti-mouse I2PP2A, F-9, sc-133138 Santa Cruz).
Total RNA isolation and cDNA synthesis
Total RNA was extracted from NIH 3T3 cells (control, treated or CRISPR edited) in three biological replicates using the RNeasy Mini Kit (QIAGEN), according to manufacturer’s protocol. To eliminate genomic DNA contamination, all RNA samples were treated with DNase I (RNase-Free DNase Set). The concentration of the isolated RNA was measured with NanoDrop 2000 spectrophotometer, while its quality and integrity were assessed using the Agilent 2100 Bioanalyzer. One μg of total RNA was used as a template for cDNA synthesis using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time, TaKaRa), following the manufacturer’s instructions.
Quantitative real-time PCR
Quantitative Real Time PCR analyses were carried out on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) with the use of KAPA SYBR FAST qPCR Master Mix (2X) Universal. The reaction mixture constituted of 5 µl of SYBR Green Master Mix (2x), 0.25 µl of each primer (10 μM), 2 µl of diluted cDNA template and 2.5 µl of Nuclease-free water. All biological samples were run in technical triplicates. Cycling conditions consisted of enzyme activation at 95 °C for 3 min and 40 cycles of denaturation at 95 °C for 3 s, annealing at 60 °C for 30 s and extension at 72 °C for 15 s, followed by a melting curve analysis. Mouse Gapdh was used as a reference gene. qPCR data analysis, including normalization and relative quantification of gene expression, was performed with the qbasePLUS software (Biogazelle). All primers listed below were designed based on Mus musculus nucleotide sequences found in NCBI. Gli1, forward 5′-CCAAGCCAACTTTATGTCAGGG-3’ and reverse 5′-AGCCCGCTTCTTTGTTAATTTGA-3′; Ptch1, forward 5′-TGCTGTGCCTGTGGTCATCCTGATT-3′ and reverse 5′-CAGAGCGAGCATAGCCCTGTGGTTC-3′; Ccnd1, forward 5′-GTTCATTTCCAACCCACCCTC-3′ and reverse 5′-AGAAAGTGCGTTGTGCGGTAG-3′; Seta, forward 5′-AGTCTGCGATCCTGCCTCA-3′ and reverse 5′-TGTTCAATTGCTTCTTGCTGTTCT-3′; Setb, forward 5′-CCGACGAGACCTCAGAAAAAGA-3′ and reverse 5′-AAAATGGTTGGCGGAGTTTG-3′. Gapdh, forward 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse 5′-GGGGTCGTTGATGGCAACA-3′.
Statistical analysis
Statistically significant changes in gene expression were determined using the One-way ANOVA test followed by Tukey–Kramer post hoc analysis for pairwise comparisons, considering *p < 0.05, **p < 0.01, ***p < 0.001. Statistically significant difference of pixel intensity was determined using two-tailed Student’s t-test, using GraphPad Prism 5.0. p value was considered significant ***p < 0.001.
References
McMahon, A. P., Ingham, P. W. & Tabin, C. J. Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53, 1–114 (2003).
Ruiz-i-Altaba, A., Sánchez, P. & Dahmane, N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat. Rev. Cancer 2, 361–372 (2002).
Sari, I. N. et al. Hedgehog signaling in cancer: a prospective therapeutic target for eradicating cancer stem cells. Cells 7, 208 (2018).
Petrova, E., Rios-Esteves, J., Ouerfelli, O., Glickman, J. F. & Resh, M. D. Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat. Chem. Biol. 9, 247–249 (2013).
Singla, V. & Reiter, J. F. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science 313, 629–633 (2006).
Ingham, P. W. & McMahon, A. P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 (2001).
Humke, E. W., Dorn, K. V., Milenkovic, L., Scott, M. P. & Rohatgi, R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670–682 (2010).
Niewiadomski, P. et al. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell Rep. 6, 168–181 (2014).
Chen, Y. & Jiang, J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 23, 186–200 (2013).
Canettieri, G. et al. Histone deacetylase and Cullin3-Ren (Kctd11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat. Cell Biol. 12, 132–142 (2010).
Niewiadomski, P. et al. Gli proteins: regulation in development and cancer. Cells 8, 147 (2019).
Okuwaki, M. & Nagata, K. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J. Biol. Chem. 273, 34511–34518 (1998).
Karetsou, Z. et al. Identification of distinct Set/Taf-Ibeta domains required for core histone binding and quantitative characterisation of the interaction. BMC Biochem. 10, 10 (2009).
Shikama, N. et al. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 20, 8933–8943 (2000).
Karetsou, Z. et al. Prothymosin alpha associates with the oncoprotein SET and is involved in chromatin decondensation. FEBS Lett. 577, 496–500 (2004).
Karetsou, Z., Martic, G., Sflomos, G. & Papamarcaki, T. The histone chaperone SET/TAF-Ibeta interacts functionally with the CREB-binding protein. Biochem. Biophys. Res. Commun. 335, 322–327 (2005).
Seo, S. B. et al. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104, 119–130 (2001).
Almeida, L. O. et al. SET oncoprotein accumulation regulates transcription through DNA demethylation and histone hypoacetylation. Oncotarget 8, 26802–26818 (2017).
Kalousi, A. et al. The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention to chromatin. Cell Rep. 11, 149–163 (2015).
Cervoni, N., Detich, N., Seo, S. B., Chakravarti, D. & Szyf, M. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J. Biol. Chem. 277, 25026–25031 (2002).
Li, M., Makkinje, A. & Damuni, Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J. Biol. Chem. 271, 11059–11062 (1996).
ten Klooster, J. P., Leeuwen, I., Scheres, N., Anthony, E. C. & Hordijk, P. L. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 26, 336–345 (2007).
Adachi, Y., Pavlakis, G. N. & Copeland, T. D. Identification of in vivo phosphorylation sites of SET, a nuclear phosphoprotein encoded by the translocation breakpoint in acute undifferentiated leukemia. FEBS Lett. 340, 231–235 (1994).
Christensen, D. J. et al. SET oncoprotein overexpression in B-cell chronic lymphocytic leukemia and non-Hodgkin lymphoma: a predictor of aggressive disease and a new treatment target. Blood 118, 4150–4158 (2011).
Liu, H. et al. Overexpression of PP2A inhibitor SET oncoprotein is associated with tumor progression and poor prognosis in human non-small cell lung cancer. Oncotarget 6, 14913–14925 (2015).
Mody, H. R. et al. SET contributes to the epithelial-mesenchymal transition of pancreatic cancer. Oncotarget 8, 67966–67979 (2017).
Enjoji, S. et al. Stemness Is enhanced in gastric cancer by a SET/PP2A/E2F1 axis. Mol. Cancer Res. 16, 554–563 (2018).
Switzer, C. H. et al. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene 30, 2504–2513 (2011).
Hung, M. H. & Chen, K. F. Reprogramming the oncogenic response: SET protein as a potential therapeutic target in cancer. Expert. Opin. Ther. Targets 21, 685–694 (2017).
Cristóbal, I. et al. Deregulation of the PP2A inhibitor SET shows promising therapeutic implications and determines poor clinical outcome in patients with metastatic colorectal cancer. Clin. Cancer Res. 21, 347–356 (2015).
Saddoughi, S. A. et al. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Mol. Med. 5, 105–121 (2013).
Pippa, R. et al. Effect of FTY720 on the SET-PP2A complex in acute myeloid leukemia; SET binding drugs have antagonistic activity. Leukemia 28, 1915–1918 (2014).
Garner, E. F. et al. FTY720 decreases tumorigenesis in group 3 medulloblastoma patient-derived xenografts. Sci. Rep. 8, 6913 (2018).
Liu, C. Y. et al. Targeting SET to restore PP2A activity disrupts an oncogenic CIP2A-feedforward loop and impairs triple negative breast cancer progression. EBioMedicine 40, 263–275 (2019).
Wei, Y. et al. p53 function is compromised by inhibitor 2 of phosphatase 2A in sonic hedgehog medulloblastoma. Mol. Cancer Res. 17, 186–198 (2019).
Serifi, I. et al. The zebrafish homologs of SET/I2PP2A oncoprotein: expression patterns and insights into their physiological roles during development. Biochem. J. 473, 4609–4627 (2016).
Chiang, C. et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413 (1996).
England, S. J., Blanchard, G. B., Mahadevan, L. & Adams, R. J. A dynamic fate map of the forebrain shows how vertebrate eyes form and explains two causes of cyclopia. Development 133, 4613–4617 (2006).
Corallo, D. et al. Emilin3 is required for notochord sheath integrity and interacts with Scube2 to regulate notochord-derived Hedgehog signals. Development 140, 4594–4601 (2013).
Zhang, W. et al. SET protein overexpression contributes to paclitaxel resistance in MCF-7/S cells through PI3K/Akt pathway. J. Drug Target 25, 255–263 (2017).
De Palma, R. M. et al. The NMR-based characterization of the FTY720-SET complex reveals an alternative mechanism for the attenuation of the inhibitory SET-PP2A interaction. FASEB J. 33, 7647–7666 (2019).
Mao, J. et al. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J. Biol. Chem. 277, 35156–35161 (2002).
Kogerman, P. et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1, 312–319 (1999).
Merchant, M. et al. Suppressor of fused regulates Gli activity through a dual binding mechanism. Mol. Cell. Biol. 24, 8627–8641 (2004).
Cheng, S. Y. & Bishop, J. M. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc. Natl. Acad. Sci. U. S. A. 99, 5442–5447 (2002).
Paces-Fessy, M., Boucher, D., Petit, E., Paute-Briand, S. & Blanchet-Tournier, M. F. The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem. J. 378, 353–362 (2004).
Zhang, Z. et al. Suppressor of fused chaperones Gli proteins to generate transcriptional responses to sonic hedgehog signaling. J. Mol. Cell. Biol. https://doi.org/10.1128/MCB.00421-16 (2017).
Zhang, Y., Iratni, R., Erdjument-Bromage, H., Tempst, P. & Reinberg, D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89, 357–364 (1997).
de Castro, I. J., Amin, H. A., Vinciotti, V. & Vagnarelli, P. Network of phosphatases and HDAC complexes at repressed chromatin. Cell Cycle 16, 2011–2017 (2017).
Carujo, S. et al. Glyceraldehyde 3-phosphate dehydrogenase is a SET-binding protein and regulates cyclin B-cdk1 activity. Oncogene 25, 4033–4042 (2006).
Vera, J. et al. Proteomic analysis of SET-binding proteins. Proteomics 7, 578–587 (2007).
Breslow, D. K. et al. CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat. Genet. 50, 460–471 (2018).
Pusapati, G. V. et al. CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen sonic hedgehog. Dev. Cell 44, 271 (2018).
Karlstrom, R. O. et al. Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development 130, 1549–1564 (2003).
Shi, Y. et al. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 8, 2043 (2017).
Tena, T. C. & Philipp, M. Assessing Smoothened-mediated Hedgehog signaling in zebrafish. Methods Cell Biol. 132, 147–164 (2016).
Yang, L., Xie, G., Fan, Q. & Xie, J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene 29, 469–481 (2010).
Infante, P. et al. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. EMBO J. 34, 200–217 (2015).
Jia, H., Liu, Y., Yan, W. & Jia, J. PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development 136, 307–316 (2009).
Krauss, S., Foerster, J., Schneider, R. & Schweiger, S. Protein phosphatase 2A and rapamycin regulate the nuclear localization and activity of the transcription factor GLI3. Cancer Res. 68, 4658–4665 (2008).
Akimaru, H. et al. Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signaling. Nature 386, 735–738 (1997).
Malatesta, M. et al. Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res. 73, 6323–6333 (2013).
Singh, K. K. et al. Human SAP18 mediates assembly of a splicing regulatory multiprotein complex via its ubiquitin-like fold. RNA 16, 2442–2454 (2010).
Aleström, P. et al. Housing and husbandry recommendations. Lab. Anim. 54, 213–224 (2020).
Schneider, C., Rasband, W. & Eliceiri, K. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Acknowledgements
We would like to thank Dr Natascia Tiso and Dr Enrico Moro (University of Padova) for the Tg12x_Gli zebrafish transgenic line, Prof. Peter Zaphiropoulos (Karolinska Institute) for the 12xGliBS-luc and pCMV5‐Gli1-Flag constructs, Dr Shen Yue (Nanjing Medical University) for the Myc-tagged Sufu plasmid and Dr. Paola Vagnarelli for the GFP-SAP18 expression construct. We are grateful to Dr. Michalis Aivaliotis for Mass spectrometry analysis at the Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology-Hellas. We also thank Dr Katerina Soupsana for her help with the statistical analysis. We thank Maria Klimopoulou and Vasiliki Xergia for their contribution during the early stages of this project.
Funding
We acknowledge support of this work by the project “Advanced Research Activities in Biomedical and Agroalimentary Technologies” (MIS 5002469) which is implemented under the “Action for the Strategic Development on the Research and Technological Sector”, funded by the Operational Program "Competitiveness, Entrepreneurship and Innovation" (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund). P.N. was funded by the Grant OPUS 2017/27/B/NZ3/01163 from the National Science Centre (Poland).
Author information
Authors and Affiliations
Contributions
I.S., S.B and P.G. performed experiments. Z.K. performed experiments and interpreted the data. S.B. and Z.K. contributed equally. D.B. provided reagents, analysed data and edited the manuscript. P.N. provided reagents, advice and edited the manuscript. T.P. conceived the study, designed the experiments, and interpreted the data. T.P. and Z.K. wrote the main manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Serifi, I., Besta, S., Karetsou, Z. et al. Targeting of SET/I2PP2A oncoprotein inhibits Gli1 transcription revealing a new modulator of Hedgehog signaling. Sci Rep 11, 13940 (2021). https://doi.org/10.1038/s41598-021-93440-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-93440-0
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.