Abstract
Pregnancy loss directly impairs reproductive performance in dairy cattle. Here, we investigated genetic factors associated with pregnancy loss following detection of a viable embryo around 42 days of gestation. The objectives of this study were to perform whole-genome scans and subsequent gene-set analyses for identifying candidate genes, functional gene-sets and gene signaling pathways implicated in pregnancy loss in US Holstein cows. Data consisted of about 58,000 pregnancy/abortion records distributed over nulliparous, primiparous, and multiparous cows. Threshold models were used to assess the binary response of pregnancy loss. Whole‐genome scans identified at least seven genomic regions on BTA2, BTA10, BTA14, BTA16, BTA21, BTA24 and BTA29 associated with pregnancy loss in heifers and lactating cows. These regions harbor several candidate genes that are directly implicated in pregnancy maintenance and fetal growth, such as CHST14, IGF1R, IGF2, PSEN2, SLC2A5 and WNT4. Moreover, the enrichment analysis revealed at least seven significantly enriched processes, containing genes associated with pregnancy loss, including calcium signaling, cell–cell attachment, cellular proliferation, fetal development, immunity, membrane permeability, and steroid metabolism. Additionally, the pathway analysis revealed a number of significant gene signaling pathways that regulate placental development and fetal growth, including Wnt, Hedgehog, Notch, MAPK, Hippo, mTOR and TGFβ pathways. Overall, our findings contribute to a better understanding of the genetic and biological basis of pregnancy loss in dairy cattle and points out novel strategies for improving pregnancy maintenance via marker‐assisted breeding.
Similar content being viewed by others
Introduction
Reproductive performance is a major determinant of efficient dairy production. Given its importance, there has been a considerable effort in improving dairy cow reproductive efficiency through nutrition, cow comfort, control of ovulation, and, more recently, inclusion of reproductive traits into breeding objectives and selection programs1. Yet, reproductive performance remains suboptimal in many dairy herds. Pregnancy loss is arguably the major factor contributing to poor reproductive performance in dairy cattle. In fact, fertilization is observed in 70% to 75% of high producing dairy cows subjected to insemination whereas embryonic and fetal losses result in calving rates that range between 30 and 35%2,3. Pregnancy losses during the embryonic period (i.e., first 42 days of gestation) have been estimated around 25% to 40% whereas pregnancy losses during the fetal period (i.e., after 42 days of gestation) range between 2 and 12%4. The cost of a pregnancy loss in dairy cattle ranges from US$90 to $1900 depending on the gestational stage at which pregnancy is lost and is associated with longer calving interval, reduced availability of potential replacement heifers, decreased milk production, increased insemination, veterinary, and labor costs, and premature culling5,6. Although pregnancy losses during the embryonic period are more frequent, pregnancy losses during the fetal period have a much greater economic impact due to extended calving intervals and early culling of productive cows7. Fetal loss may also result in the retention of fetal membranes and the development of endometritis, which further reduces reproductive performance and increases veterinary and labor costs. Therefore, pregnancy loss during the fetal period should not be overlooked in breeding schemes aimed at improving the reproductive performance of dairy cattle.
Numerous factors may cause fetal loss, including infectious diseases, toxic agents, heat stress, and genetic factors. Indeed, previous studies have reported that there is a substantial genetic variation underlying fetal loss in dairy catlle8,9,10. Known genetic factors include lethal recessive alleles, such as complex vertebral malformation (CVM) and deficiency of uridine monophosphate synthase (DUMPS), and an increasing list of recessive haplotypes11. Recently, Gershoni and colleagues identified ABCA9 gene as a candidate gene for early abortions in Israeli dairy cattle10. Interestingly, ABCA9 gene is located within an ATP-binding cassette (ABC) genes cluster on BTA19, and the ABC family is the major class of primary active transporters in the placenta. Overall, current evidence suggests that fetal loss is influenced by genetic factors, and hence it could be improved by genetic means.
The main objective of this study was to characterize the genetic basis of pregnancy loss in US Holstein cattle. We investigated pregnancy loss following the detection of a viable embryo, and hence, we analyzed direct records of pregnancy loss, rather than indirect indicators of pregnancy loss, such as long insemination intervals. We performed whole-genome scans to identify candidate genes associated with pregnancy loss. Given that association studies detect, in general, only the most significant genetic markers, we also applied SNP-based gene-set enrichment tools to gain additional insight into the biological processes and molecular mechanisms that could be affecting pregnancy loss. Our findings will contribute to a better understanding of the genetic variants and complex biological mechanisms underlying pregnancy loss in dairy cattle, in addition to providing novel tools for improving reproductive efficiency via marker-assisted selection.
Materials and methods
Phenotypic and genotypic data
Records from nulliparous (heifers), primiparous (first lactation cows) and multiparous (second lactation cows) Holsteins diagnosed as pregnant 39 ± 3 days after breeding were compiled from a single dairy herd located in Florida, USA between 2001 and 2019 (Table 1). Records of pregnancy loss were obtained from veterinary pregnancy examinations. Females were assumed to undergo pregnancy loss if a negative veterinary examination was reported in subsequent pregnancy checks, after the initial confirmation of pregnancy at 42 days. Pregnancy loss was recorded as a binary trait, i.e., Y = 1 if the heifer or cow diagnosed as pregnant 39 ± 3 days after breeding was diagnosed as non-pregnant in subsequent examinations, and Y = 0 if the heifer or cow remained pregnant in subsequent examinations. The pedigree file was created by tracing the pedigree of cows back five generations.
Genotype data for 72,444 SNPs across the bovine genome were available for heifers and cows with records, and also sires in the pedigree. The SNP information was updated to the new bovine reference genome ARS-UCD 1.2. Quality control was conducted both at the level of animal and SNP markers. Animals were removed from the analysis if they had a call rate < 95%. The SNPs were removed if they mapped to the sex chromosomes, had a call rate < 95%, minor allele frequency < 1%, or if they failed the Hardy–Weinberg equilibrium test. After quality control, a total of 69,051 SNP markers were retained for subsequent genomic analyses. Summary statistics of the final dataset used for the analyses are presented in Table 1.
Statistical model
The threshold model, also known as probit model, was used to evaluate the incidences of pregnancy loss in each of the three parities, i.e., nulliparous, primiparous and multiparous. This model describes the variation observed at a binary response variable (Y, either 0 or 1) using an underlying random variable \(\mathbf{z}\), known as liability,
where \(\boldsymbol{\upeta }\) is a vector of linear predictors and \(\boldsymbol{\upepsilon }\) is a vector of independent and identically distributed standard normal random variables, ε ~ \(N(0,1)\). Here, pregnancy loss is \(Y=1\) if the underlying liability \((z)\) is greater than zero, i.e., \(\mathbf{Y}={1 \; \text{if} \quad \mathbf{z}>\mathbf{0}}\); 0 otherwise. Therefore, the conditional probability of observing a pregnancy loss event is P \(\left(\mathbf{Y}=\mathbf{1}|\boldsymbol{\upeta }\right)\) = \({\Phi }\)(\(\boldsymbol{\upeta }\)) where \({\Phi }\)(.) is the standard normal cumulative distribution. The likelihood function for the binary response variable then becomes:
where \(\mathbf{Y}\) and \(\boldsymbol{\upeta }\) are the vectors of the binary responses and the linear predictors, respectively.
For pregnancy loss incidence, the linear predictor \(\boldsymbol{\upeta }\) has the following form:
where \(\boldsymbol{\upbeta }\) is a vector for fixed effects of year-season of breeding (39 levels), type of breeding (2 levels; insemination or embryo transfer), days in milk (DIM) at breeding (3 levels; only for lactating cows), and occurrence of uterine diseases (binary trait, only for lactating cows), \(\mathbf{a}\) is a vector of random additive genetic effects, and \(\mathbf{s}\mathbf{s}\) is a vector of random service sire effects. The matrices \(\mathbf{X}\), \({\mathbf{Z}}_{1}\), and \({\mathbf{Z}}_{2}\) are the incidence matrices relating phenotypic records to fixed, animal, and sire effects, respectively. Random effects were assumed to follow a multivariate normal distribution,
where \(\mathbf{a}\) and \(\mathbf{ss}\) are the vectors of animal and service sire effects respectively; \({\sigma}_{a}^{2}\) and \({\sigma}_{ss}^{2}\) are animal and service sire variances, respectively. Here, the classical pedigree relationship matrix \(\mathbf{A}\) is replaced by \(\mathbf{H}\) which combines pedigree and genotypic information. This method is known as single-step genomic best linear unbiased prediction (ssGBLUP). The matrix \({\mathbf{H}}^{-1}\) was calculated as follows,
where \({\mathbf{G}}^{-1}\) is the inverse of the genomic relationship matrix and \({\mathbf{A}}_{22}^{-1}\) is the inverse of the pedigree-based relationship matrix for genotyped animals. Here, G matrix was obtained using the observed allele frequency of the genotypes. The G was created considering heifers and cows with both genotypes and abortion records, plus genotyped sires in the pedigree. The pedigree relationship A matrix was created using a five-generation pedigree file obtained from the Council of Dairy Cattle Breeding (Table 1).
Model implementation
Threshold models were implemented in a Bayesian framework using THRGIBBS1F90 (version 1.93)12. A Monte Carlo Markov chain approach through Gibbs sampling was used to obtain features of the posterior distribution. Inferences were based on 500,000 samples obtained after discarding the first 100,000 samples as burn in. A thinning interval of 100 was used to compute statistics of the posterior distribution. Convergence diagnostics of Markov chain Monte Carlo sampling output were carried out by visual inspection of trace plots of key parameters such as variance components.
Genome-wide association mapping
Candidate genomic regions associated with pregnancy loss in each parity under study were identified based on the amount of genetic variance explained by 2.0 Mb moving windows of adjacent SNPs. The SNP effects were estimated as \(\widehat{\boldsymbol{s}}\) = \(\boldsymbol{D}\boldsymbol{M}\boldsymbol{^{\prime}}\left[\boldsymbol{M}\boldsymbol{D}{\left.\boldsymbol{M}\boldsymbol {^{\prime}} \right]}^{-1}\right.{\widehat{\boldsymbol{a}}}_{\boldsymbol{g}}\), where \(\widehat{\boldsymbol{s}}\) is the vector of SNP marker effects, \(\boldsymbol{D}\) is a diagonal matrix of weights of SNPs, \(\boldsymbol{M}\) is a matrix relating genotype of each SNP marker to observations and \({\widehat{\boldsymbol{a}}}_{\boldsymbol{g}}\) is the vector of GEBVs for genotyped individuals13. The percentage of genetic variance explained by a given 2.0 Mb region was then calculated as
where \({u}_{i}\) is the genetic value of the \({i}\)th genomic region under consideration, B is the total number of adjacent SNPs within 2.0 Mb region, \({M}_{j}\) is the genotype code of \({j}\)th marker, \({S}_{j}\) is the marker effect of the \({j}\)th SNP within the \({i}\)th region. In this study, all SNPs were equally weighted. All these ssGBLUP calculations were performed using POSTGSF90 (version 3.08) of BLUPF90 family of programs14.
Gene-set enrichment analysis
Mapping SNPs to genes
The first step in gene-set enrichment analysis is to map SNPs to genes. The Bioconductor R package biomaRt15 and the latest bovine reference genome ARS-UCD 1.2. were used to map SNPs to genes. Specifically, SNPs were assigned to genes if they were located within the genomic sequence of an annotated gene or within 15 kb either upstream or downstream the gene. The distance of 15 kb was used to capture proximal regulatory regions and other functional sites that may lie outside (e.g., promoter region) but close to each gene. If a SNP was found to be located within or close to more than one gene, all these genes were included in subsequent analyses. A 5% threshold of the SNP effects was used to define relevant SNPs; hence, a gene was associated with pregnancy loss if it contained at least one SNP whose effect was in the top 5% of the distribution.
Assignment of genes to functional categories
Different databases including GO, MeSH, Reactome, InterPro and MsigDB were used to define functional sets of genes. Genes assigned to the same functional category can be regarded as more closely related in terms of biology or function than random sets of genes. Only gene-sets, biological processes, molecular mechanisms, or gene signaling pathways with 10 or more genes were considered in these analyses.
Association analysis between functional categories and pregnancy loss
The significant association of a given functional term with pregnancy loss was analyzed using the Fisher’s exact test, a test of proportions based on the cumulative hypergeometric distribution16. This test was performed to search for an overrepresentation of relevant genes in each functional category. The P-value of observing \(g\) relevant genes in the term was calculated by
where \(S\) is the total number of relevant genes associated with pregnancy, \(N\) is the total number of genes that were analyzed, and \(K\) is the total number of genes in the term considered. The gene-set enrichment analysis was performed using the R package EnrichKit, developed by Lihe Liu and Francisco Peñagaricano, available at https://github.com/liulihe954/EnrichKit.
Results and discussion
Improving reproductive efficiency is one of the most important goals for the dairy industry worldwide. Yet, reproductive efficiency remains suboptimal in most of the dairy herds. Pregnancy loss is recognized as one of the most important factors that reduces reproductive performance and dairy farm profitability. In fact, pregnancy loss is expensive, and the cost increases rapidly as the gestation progresses. Most genetic studies have focused on early pregnancy loss during the embryonic period, whereas late pregnancy loss during the fetal period has been largely overlooked. Despite having a smaller incidence, late fetal losses have a much greater economic impact compared with earlier embryonic losses5,6. As such, this study was conducted to unravel individual genes, functional gene-sets, biological processes, and gene signaling pathways underlying pregnancy losses during the fetal stage in US Holstein cattle.
Gene mapping
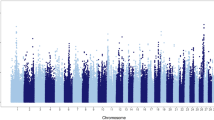
Single-step genomic BLUP methodology was utilized to identify genomic regions and putative candidate genes affecting pregnancy loss in nulliparous, primiparous and multiparous cows. This method combines all available phenotypic, pedigree, and genotypic data and fits all SNPs simultaneously. Figure 1 displays Manhattan plots for pregnancy loss across the three parities. Genomic regions and candidate genes were identified based on the amount of genetic variance explained by 2.0 Mb SNP-windows across the entire bovine genome. Note that most of the genomic regions associated with pregnancy loss are parity specific. In other words, there is a remarkable genotype-by-parity interaction, regions and major genes associated with pregnancy loss vary across lactations. The low genetic correlations (calculated as correlations between SNP effects) across parities support these findings (Table 2).
Whole-genome scans for pregnancy loss across the first three parities in Holstein dairy cows: Percentage of additive genetic variance explained by 2.0 Mb single nucleotide polymorphism (SNP)-windows across the genome. Genes directly implicated in pregnancy maintenance and fetal growth are highlighted in green.
Genomic scan identified peaks on BTA16, BTA24, and BTA29 associated with pregnancy loss in nulliparous heifers. Interestingly, these genomic regions harbor putative candidate genes, such as PSEN2 and KIF26B (BTA16, 29.68–31.66 Mb), IER3IP1 (BTA24, 45.18–47.18 Mb), and IGF2, PHLDA2, and TOLLIP (BTA29, 48.33–50.31 Mb). The PSEN2 gene is associated with notch signaling pathway and is involved in cellular proliferation, differentiation and placental angiogenesis17. Knock-out of PSEN2 in mice exhibits abnormal tissue development, including defective somite formation leading to intrauterine growth restrictions and fetal death18. The KIF26B gene of the kinesin family is involved in fetal kidney development and many crucial cellular processes, including cell proliferation, differentiation, migration19. The IER3IP1 gene is highly expressed in the fetal brain cortex and regulates fetal brain development20. The IGF2 gene is involved in fetal growth as it controls the supply of oxygen nutrients available to the fetus through placental circulation21. The IGF2 gene affects the metabolism, mitogenesis and differentiation of many cells including the placenta, while mutations in this gene cause fetal death by altering the supply of oxygen and nutrients22. Maternal expression of the PHLDA2 gene has been linked to fetal growth restrictions resulting in poor perinatal outcomes such as fetal distress and fetal death thereby supporting a role of the PHLDA2 gene in fetal growth and placental development23. In mice, maternal expression of the PHLDA2 gene results in the termination of pregnancy24. The TOLLIP gene is a Wnt signaling associated gene involved in the immune regulation at the maternal–fetal interface. Activation of TOLLIP gene at the maternal–fetal interface produces Tumor necrosis factor-α (TNF-α) that stimulates apoptosis of villi-trophoblast cells, which suggests that TOLLIP gene may be a potential risk factor for pregnancy failure25. Our results confirm a genetic basis of pregnancy loss in nulliparous heifers and mapping of potential candidate genes at associated loci identified several genes (PSEN2, KIF26B, IER3IP1, IGF2, PHLDA2, and TOLLIP) with a plausible biological role in placental biology, fetal growth, and immune modulation.
Two genomic regions on BTA2 and BTA10 were strongly associated with pregnancy loss in both primiparous and multiparous cows. These genomic regions harbor strong candidate genes implicated in cellular functions related to placental development and fetal growth, such as PAFAH2, CLIC4, and WNT4 (BTA2, 126.70–128.26 Mb), and CHAC1, CHST14, and DLL4 (BTA10, 33.62–35.56 Mb). The PAFAH2 gene has been implicated in various stages of reproduction, including implantation, fetal development, and parturition26. The PAFAH2 gene stimulates the formation of IP3 and DAG and increases intracellular calcium27. Calcium is involved in bone formation and fetal mineralization. The CLIC4 gene is implicated in mid gestational brain differentiation and neurogenesis28. The WNT4 gene, a member of the Wnt signaling pathway, is implicated in the development of organ systems and extra embryonic tissues, particularly vascularization of the placenta. It promotes placental development through trophoblast lineage determination, chorioallantois fusion, and placental branching morphogenesis29. Failure in proper placental development creates an anaerobic environment and causes oxidative stress and fetal death30. The CHAC1 gene is an endoplasmic reticulum stress gene that is involved in apoptosis initiation and execution31. The CHST14 gene is implicated in dermatan sulfate biosynthetic process. In mice, knock-out of CHST14 gene produces changes in the placenta, including reduced weight, alterations in the vascular structure, and ischemic and/or necrotic-like changes, indicating this gene is essential for placental vascular development and perinatal survival of the fetus32. In brief, whole-genome scans revealed two common genomic regions on BTA2 and BTA10 as associated with pregnancy loss in primiparous and multiparous cows, suggesting that genes important for pregnancy loss are common in lactating cows. These genomic regions harbor candidate genes important for fetal development, calcium regulation, fetal-maternal circulation, and immunity.
For primiparous cows, one genomic region located on BTA14 explained almost 1.0% of additive genetic variance for pregnancy loss. This genomic region harbors several genes, including SLC2A5 (BTA14, 81.94–83.93 Mb). The SLC2A5 gene, a solute carrier transporter, is expressed in uterine-placental interface during mid to late pregnancy stage. The SLC2A5 gene transports fructose from placenta to fetus and hence, supports the growth and development of fetus33. For multiparous cows, two different genomic regions on BTA21 (72.83–92.67 Mb) and BTA29 (43.42–45.42 Mb) explained more than 0.5% of the additive genetic variance for pregnancy loss. The genomic region on BTA21 harbors several genes including IGF1R which is involved in fetal growth and development. The IGF1R gene is implicated in the transfer of nutrients to the fetus and promotes anabolic events during the fetal stage of development34. The RAD9A gene, a key gene of the DNA damage response pathway, is involved in genomic stability and embryo-fetal development; deletion of mouse RAD9A gene results in post-zygotic loss and reduced fertility35. The GSTP1 gene on BTA29 is important for detoxification of toxic compounds that pass the placental barrier and protects the placenta and fetus from toxic products36.
Overall, our whole-genome scans have detected several genomic regions and candidate genes associated with pregnancy loss in both non-lactating heifers and lactating primiparous and multiparous cows. Interestingly, these genomic regions harbor candidate genes that have diverse biological roles in placental development, fetal growth, pregnancy maintenance, immune modulation, calcium signaling, vascularization and organogenesis. These genomic regions are excellent candidates for future research to identify functional mutations associated with pregnancy loss in dairy cattle.
Gene-set analysis
Whole-genome association mapping evaluated 72,444 SNP markers, however, only SNPs located within annotated genes or at most 15 kb upstream or downstream from annotated genes were used for the gene-set analysis. This set of SNPs defined a total of 20,087 genes in the new ARS-UCD 1.2. bovine reference genome. A subset of 1903 genes in nulliparous heifers, 1892 genes in primiparous, and 1882 genes in multiparous cows were considered as significantly associated with pregnancy loss, i.e., set of genes flagged by at least one SNP located in the top 5% of the SNP effect distribution.
Figure 2 shows the most relevant biological terms and pathways associated with pregnancy loss. Note that our gene-set analysis used different public annotation databases, including GO, MeSH, InterPro, Reactome, and MSigDB. Across these annotation databases, genome-wide association signals for pregnancy loss were highly enriched in at least seven groups of gene-sets, namely calcium signaling, steroid metabolism, fetal development, immunity, cellular proliferation, membrane permeability and cell–cell attachment. Supplementary Tables S1–3 report the full list of significant biological terms, including term ID, term name, total number of genes, number of significant genes and Fisher’s P‐value.
Functional terms and pathways significantly enriched with genes associated with pregnancy loss. Several annotation databases were analyzed, including GO, Medical Subject Headings, InterPro, Reactome and MSigDB. The y-axis displays the names of gene-sets associated with pregnancy loss, and letters within parenthesis represent the three parities (N: Nulliparous; P: Primiparous and M: Multiparous). The size of the dots represents the significance of enrichment (− log10 P-Value, Fisher’s exact test) and x-axis represents the percentage of significant genes in each functional term.
Noticeably, some of the enriched terms are directly involved in fetal growth and organogenesis, such as post-embryonic development (GO:0009791), limb development (GO:0060173), skeletal system development (GO:0001501), central nervous system development (GO:0007417), positive regulation of neurogenesis (GO:0050769), heart development (GO:0007507), developmental biology (R-BTA-1266738), and developmental pluripotency-associated protein 2/4, C-terminal domain (R025891). These significant terms contain relevant genes, such as BMPR1B, BMPR1, MAP2K1, and DLG4, all of which are involved in embryogenesis, development of multiple organs systems, and maintenance of tissues homeostasis37,38,39.
The significant terms enriched in genes involved in calcium signaling include positive regulation of calcium-mediated signaling (GO:0050850), calcium channel activity (GO:0005262), release of sequestered calcium ion into cytosol (GO:0051209), calcium channels, T-type (D020747) and calcium signaling (D020013). The calcium ion is an important secondary messenger involved in proper placental development and function, critical for the growth and survival of the fetus. These calcium‐related gene‐sets contained several significant genes, namely ITPR2, ITPR3, and PLD1, which are associated with cell proliferation, survival, differentiation, cytoskeletal organization, and maintenance of the fetal-maternal connections and viability of the fetus40,41.
Steroid metabolism was identified as a biological process significantly enriched with genes implicated in pregnancy maintenance. Functional terms or pathways associated with steroid metabolism include positive regulation of phospholipid translocation (GO:0061092), glycerol-lipid metabolic process (GO:0046486), glycero-lipid metabolism (bta00561), metabolism of steroids (R-BTA-8957322) and steroid hormone biosynthesis (bta00140). These terms were enriched with relevant genes, including DGKG, DGAT2, GPAT3, and ATP8A1, which maintain the steroid hormones, namely progesterone, estrogens, gluco- and mineralocorticosteroids between the mother, fetus, and placenta. Steroid hormones are involved in several events during pregnancy, including proliferation of trophoblasts and healthy progression of pregnancy42,43.
There were also some relevant functional terms related to immunity, including immunoglobulin secretion (GO:0048305), positive regulation of B cell activation (GO:0050871), immunoglobulin production (GO:0002377), positive regulation of B cell proliferation (GO:0030890) and Fc epsilon RI signaling pathway (bta04664) and Fc gamma R-mediated phagocytosis (bta04666). These functional gene-sets harbor many relevant genes, such as CD81, CD40, CLCF1, and MAPK12, which establish immune tolerance at the fetal-maternal interface and allow allogeneic fetal trophoblasts to invade maternal tissues, which in turn permits fetal and placental development44,45.
Cell–cell attachment and cellular proliferation were identified as two important biological processes significantly enriched with genes implicated in pregnancy loss. Functional terms associated with cell–cell attachment include adherens junction (GO:0005912), cell–cell adhesion (GO:0098609), cell adhesion (GO:0007155), focal adhesion (bta04510), cell adhesion molecules (D015815), gap junction (bta04540), and adherens junction (bta04520). Functional terms associated with cellular proliferation include microtubule associated protein (IPR026074), neuroepithelial cell differentiation (GO:0060563), establishment of epithelial cell polarity (GO:0090162), positive regulation of epithelial cell migration (GO:0010634), microtubule organizing center organization (GO:0031023), and blood vessel development (GO:0001568). These biological processes are involved in many stages of reproduction including conceptus attachment to uterine epithelium, proliferation, and differentiation of trophoblast cells for development of placenta, and proliferation of endothelial cells to establish fetal-maternal circulation46,47.
Finally, membrane permeability was another term associated with many significant gene-sets implicated in pregnancy loss, including potassium channel, voltage dependent, EAG (IPR003949), positive regulation of potassium ion transmembrane transport (GO:1901381), glucose transporter type 1 (DO51272), regulation of glucose transmembrane transport (GO:0034765), regulation of ion transmembrane transport (GO:0034765), membrane potentials (D008564), potassium ion transmembrane transport (GO:0071805). Membrane permeability plays a crucial role in signal transduction, intercellular communication for exchange of gas and nutrients, and controls embryogenesis and growth48. Voltage gated potassium channels are also expressed in uterine smooth muscle and play a significant role in modulating uterine contractility during pregnancy49.
Overall, all these enriched gene-sets play crucial roles in pregnancy maintenance, placental and fetal development, via cell attachment, proliferation, changes in membrane permeability, immunological modulation at the fetal-maternal interface, and steroid metabolism. Dysregulation of biological and molecular processes associated with these gene-sets might impair placenta function and fetal growth, causing pregnancy loss.
Gene signaling pathways
Pregnancy maintenance, placental, and fetal development are characterized by cellular processes such as cell proliferation, migration, differentiation, transformation, and apoptosis. These cellular processes are genetically controlled and depend on the activities of gene signaling pathways, which coordinate the cell activities leading to organogenesis50. Our enrichment analysis revealed a number of significant gene signaling pathways, including Integrin, DAG & IP3, Wnt, Notch, Hedgehog, Hippo, MAPK, Rap1, SREBP, TGFβ and mTOR, that are all important for placental and fetal development (Fig. 3). These developmental signaling pathways are involved in cellular functions needed for morphogenesis, tissues homeostasis and organogenesis. For instance, the Wnt pathway comprises a family of ligands that bind to multiple receptor complexes triggering several downstream signaling cascades, including Wnt/β-catenin dependent signaling pathways, non-canonical Wnt/planar cell polarity (PCP), and the Wnt/Ca2+ pathways51. The Wnt signaling pathways are involved in chorion–allantois fusion, placental angiogenesis, and trophoblast differentiation52. As a result, perturbations in the Wnt signaling pathways may lead to failure in chorioallantois fusion or defects in the formation of placental vessels, leading to intrauterine growth restrictions and pregnancy loss53.
Notch is another significant signaling pathway that mediates cell–cell communication. Notch signaling is involved in many organ formations, including vertebrate segmentation and skeleton development during fetal development54. Another important signaling pathway is Hedgehog signaling which is involved in trophoblast syncytialization, a critical process for placental development and maturation55. Additionally, Hedgehog signaling regulates fetal growth via IGF1R pathway in the placenta56. Moreover, Hippo signaling pathway is involved in angiogenesis for establishing feto-maternal circulation and regulating fetal growth. The Hippo pathway has essential roles in organ size control, tissues regeneration, cell fate decision and differentiation during fetal development57. The MAPK signaling pathway is involved in complex cellular processes like proliferation, differentiation, transformation, and apoptosis. Additionally, MAPK interacts with other intracellular signaling pathways such as steroid receptors for uterine cell proliferation58 and is involved in embryonic and yolk sac angiogenesis during feto-placental development59.
The Transforming growth factor β (TGFβ) is another significant signaling pathway implicated in cell growth and differentiation. The TGFβ signaling pathway regulates a variety of reproductive processes, including pregnancy, uterine growth, and fetal development60. The mTOR signaling pathway, another developmental signaling pathway, has critical roles in cell growth, survival, and metabolism in response to nutrients, growth factors, energy, and stress signals. The mTOR signaling pathway regulates placental growth, including oxygen and nutrient transport. Inhibition of placental mTOR signaling results in intra-uterine growth restrictions characterized by adverse perinatal outcomes such as neurodevelopmental dysfunction and fetal loss61. Furthermore, the Integrin signaling pathway is implicated in cellular differentiation and tissue assembly during embryogenesis and fetal growth as integrins mediate both signaling and adhesion62. Integrins connect extracellular matrix (ECM) components with the actin cytoskeleton and form ECM-integrin-cytoskeleton signaling axis which is important for proper development and function of fetal skeleton63. The DAG & IP3 is another important developmental signaling pathway that regulates cellular calcium signaling system which is involved in several cellular processes such as proliferation, growth, differentiation, and contraction of uterine smooth muscles during pregnancy64. The SREBP signaling pathway is involved in cholesterol and fatty acid biosynthesis, and cholesterol is required for fetal development65. The Rap1 signaling is implicated in several basic cellular functions, including cell–cell interactions, cell–matrix adhesion, proliferation, and regulation of cellular polarity during fetal development. The Rap1 signaling is also implicated in maintaining epithelial and endothelial cell junction integrity for maintaining the viability of fetus66.
Overall, all these significantly enriched gene signaling pathways play critical roles in pregnancy maintenance, placental, and fetal development. Interestingly, many of these gene signaling pathways are also involved in pregnancy establishment and early embryo development, suggesting that some key pathways are relevant throughout different stages of gestation.
Conclusions
In this study, we performed an integrative genomic analysis to understand the genetic and biological basis of pregnancy loss in dairy cattle. Our study focused on pregnancy loss during the fetal period using data from cows that were first confirmed pregnant and subsequently diagnosed as non-pregnant in later pregnancy checks. Whole-genome scans identified at least seven genomic regions located on BTA2, BTA10, BTA14, BTA16, BTA21, BTA24, and BTA29 that explained more than 0.5% of the additive genetic variances for fetal loss. Interestingly, these genomic regions harbor candidate genes that have diverse biological roles in feto-placental growth, immune modulation, calcium signaling, vascularization, and organogenesis. Moreover, the enrichment analysis revealed at least seven relevant processes, namely cell–cell attachment, cellular proliferation, membrane permeability, immunity, calcium signaling, steroid metabolism and fetal development as significantly enriched with genes associated with pregnancy loss. Additionally, the enrichment analysis revealed a number of important gene signaling pathways, including Integrin, DAG & IP3, Wnt, Notch, Hedgehog, Hippo, MAPK, Rap1, SREBP, TGFβ, and mTOR. These signaling pathways have diverse roles in pregnancy maintenance, placental development, and fetal growth. Overall, this comprehensive study contributes to a better, deeper understanding of the genetic architecture of fetal loss in dairy cattle by unraveling genetic variants, individual genes and complex biological and physiological pathways responsible for pregnancy maintenance in dairy cattle. In addition, these findings can provide opportunities for improving pregnancy success in dairy cattle via marker-assisted selection.
Data availability
The phenotypic and genotypic data analyzed in this study were obtained from North Florida Holsteins (Bell, FL), and Council on Dairy Cattle Breeding (Bowie, MD). These datasets were used under agreement, and hence, are not publicly available. However, data are available upon request to FP and with permission of North Florida Holsteins, and Cooperative Dairy DNA Repository.
References
Norman, H. D., Wright, J. R., Hubbard, S. M., Miller, R. H. & Hutchison, J. L. Reproductive status of Holstein and Jersey cows in the United States. J. Dairy Sci. 92, 3517–3528. https://doi.org/10.3168/jds.2008-1768 (2009).
Santos, J. E., Thatcher, W. W., Chebel, R. C., Cerri, R. L. & Galvao, K. N. The effect of embryonic death rates in cattle on the efficacy of estrus synchronization programs. Anim. Reprod. Sci. 82–83, 513–535. https://doi.org/10.1016/j.anireprosci.2004.04.015 (2004).
Diskin, M. G., Parr, M. H. & Morris, D. G. Embryo death in cattle: An update. Reprod. Fertil. Dev. 24, 244–251. https://doi.org/10.1071/RD11914 (2011).
Wiltbank, M. C. et al. Pivotal periods for pregnancy loss during the first trimester of gestation in lactating dairy cows. Theriogenology 86, 239–253. https://doi.org/10.1016/j.theriogenology.2016.04.037 (2016).
Cabrera, V. E. Economics of fertility in high-yielding dairy cows on confined TMR systems. Animal 8(Suppl 1), 211–221. https://doi.org/10.1017/S1751731114000512 (2014).
De Vries, A. Economic value of pregnancy in dairy cattle. J. Dairy Sci. 89, 3876–3885. https://doi.org/10.3168/jds.S0022-0302(06)72430-4 (2006).
Diskin, M. G. & Morris, D. G. Embryonic and early foetal losses in cattle and other ruminants. Reprod. Domest. Anim. 43(Suppl 2), 260–267. https://doi.org/10.1111/j.1439-0531.2008.01171.x (2008).
Bamber, R. L., Shook, G. E., Wiltbank, M. C., Santos, J. E. & Fricke, P. M. Genetic parameters for anovulation and pregnancy loss in dairy cattle. J. Dairy Sci. 92, 5739–5753. https://doi.org/10.3168/jds.2009-2226 (2009).
Carthy, T. R., Ryan, D. P., Fitzgerald, A. M., Evans, R. D. & Berry, D. P. Genetic parameters of ovarian and uterine reproductive traits in dairy cows. J. Dairy Sci. 98, 4095–4106. https://doi.org/10.3168/jds.2014-8924 (2015).
Gershoni, M., Ezra, E. & Weller, J. I. Genetic and genomic analysis of long insemination interval in Israeli dairy cattle as an indicator of early abortions. J. Dairy Sci. 103, 4495–4509. https://doi.org/10.3168/jds.2019-17482 (2020).
VanRaden, P. M., Olson, K. M., Null, D. J. & Hutchison, J. L. Harmful recessive effects on fertility detected by absence of homozygous haplotypes. J. Dairy Sci. 94, 6153–6161. https://doi.org/10.3168/jds.2011-4624 (2011).
Tsuruta, S. & Misztal, I. THRGIBBS1F90 for estimation of variance components with threshold-linear models. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, vol. 89, 27–31 (2006).
Wang, H., Misztal, I., Aguilar, I., Legarra, A. & Muir, W. M. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 94, 73–83. https://doi.org/10.1017/s0016672312000274 (2012).
Aguilar, I., Misztal, I., Tsuruta, S., Legarra, A. & Wang, H. PREGSF90–POSTGSF90: Computational Tools for the Implementation of Single-step Genomic Selection and Genome-wide Association with Ungenotyped Individuals in BLUPF90 Programs (2014).
Durinck, S. et al. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. https://doi.org/10.1093/bioinformatics/bti525 (2005).
Peñagaricano, F., Weigel, K. A., Rosa, G. J. & Khatib, H. Inferring quantitative trait pathways associated with bull fertility from a genome-wide association study. Front. Genet. 3, 307. https://doi.org/10.3389/fgene.2012.00307 (2012).
Wang, P., Pereira, F. A., Beasley, D. & Zheng, H. Presenilins are required for the formation of comma- and S-shaped bodies during nephrogenesis. Development 130, 5019–5029. https://doi.org/10.1242/dev.00682 (2003).
Mirnics, K. et al. Molecular signatures of neurodegeneration in the cortex of PS1/PS2 double knockout mice. Mol. Neurodegener. 3, 14. https://doi.org/10.1186/1750-1326-3-14 (2008).
Uchiyama, Y. et al. Kif26b, a kinesin family gene, regulates adhesion of the embryonic kidney mesenchyme. Proc. Natl. Acad. Sci. USA 107, 9240–9245. https://doi.org/10.1073/pnas.0913748107 (2010).
Sun, J. & Ren, D. IER3IP1 deficiency leads to increased beta-cell death and decreased beta-cell proliferation. Oncotarget 8, 56768–56779. https://doi.org/10.18632/oncotarget.18179 (2017).
Sferruzzi-Perri, A. N., Sandovici, I., Constancia, M. & Fowden, A. L. Placental phenotype and the insulin-like growth factors: Resource allocation to fetal growth. J. Physiol. 595, 5057–5093. https://doi.org/10.1113/JP273330 (2017).
Bergman, D., Halje, M., Nordin, M. & Engstrom, W. Insulin-like growth factor 2 in development and disease: A mini-review. Gerontology 59, 240–249. https://doi.org/10.1159/000343995 (2013).
Janssen, A. B., Tunster, S. J., Heazell, A. E. & John, R. M. Placental PHLDA2 expression is increased in cases of fetal growth restriction following reduced fetal movements. BMC Med. Genet. 17, 17. https://doi.org/10.1186/s12881-016-0279-1 (2016).
Tunster, S. J., Van De Pette, M. & John, R. M. Isolating the role of elevated Phlda2 in asymmetric late fetal growth restriction in mice. Dis. Model Mech. 7, 1185–1191. https://doi.org/10.1242/dmm.017079 (2014).
Amirchaghmaghi, E. et al. The role of toll like receptors in pregnancy. Int. J. Fertil. Steril. 7, 147–154 (2013).
Purnell, E. T. et al. Influence of the preimplantation embryo development (Ped) gene on embryonic platelet-activating factor (PAF) levels. J. Assist. Reprod. Genet. 23, 269–273. https://doi.org/10.1007/s10815-006-9039-z (2006).
Roudebush, W. E. et al. Embryonic platelet-activating factor: An indicator of embryo viability. Hum. Reprod. 17, 1306–1310. https://doi.org/10.1093/humrep/17.5.1306 (2002).
Padmakumar, V. et al. Detection of differential fetal and adult expression of chloride intracellular channel 4 (CLIC4) protein by analysis of a green fluorescent protein knock-in mouse line. BMC Dev. Biol. 14, 24. https://doi.org/10.1186/1471-213X-14-24 (2014).
Zhang, Z. et al. Wnt/beta-catenin signaling pathway in trophoblasts and abnormal activation in preeclampsia (review). Mol. Med. Rep. 16, 1007–1013. https://doi.org/10.3892/mmr.2017.6718 (2017).
Aouache, R., Biquard, L., Vaiman, D. & Miralles, F. Oxidative stress in preeclampsia and placental diseases. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19051496 (2018).
Nomura, Y., Hirata, Y., Kiuchi, K. & Oh-Hashi, K. Translational and post-translational regulation of mouse cation transport regulator homolog 1. Sci. Rep. 6, 28016. https://doi.org/10.1038/srep28016 (2016).
Yoshizawa, T. et al. Vascular abnormalities in the placenta of Chst14−/− fetuses: Implications in the pathophysiology of perinatal lethality of the murine model and vascular lesions in human CHST14/D4ST1 deficiency. Glycobiology 28, 80–89. https://doi.org/10.1093/glycob/cwx099 (2018).
Steinhauser, C. B. et al. Fructose synthesis and transport at the uterine-placental interface of pigs: Cell-specific localization of SLC2A5, SLC2A8, and components of the polyol pathway. Biol. Reprod. 95, 108. https://doi.org/10.1095/biolreprod.116.142174 (2016).
Hellstrom, A. et al. Role of insulinlike growth factor 1 in fetal development and in the early postnatal life of premature infants. Am. J. Perinatol. 33, 1067–1071. https://doi.org/10.1055/s-0036-1586109 (2016).
Hopkins, K. M. et al. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell Biol. 24, 7235–7248. https://doi.org/10.1128/MCB.24.16.7235-7248.2004 (2004).
Suryanarayana, V., Deenadayal, M. & Singh, L. Association of CYP1A1 gene polymorphism with recurrent pregnancy loss in the South Indian population. Hum. Reprod. 19, 2648–2652. https://doi.org/10.1093/humrep/deh463 (2004).
Wang, R. N. et al. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 1, 87–105. https://doi.org/10.1016/j.gendis.2014.07.005 (2014).
Krishnan, M. L. et al. Integrative genomics of microglia implicates DLG4 (PSD95) in the white matter development of preterm infants. Nat. Commun. 8, 428. https://doi.org/10.1038/s41467-017-00422-w (2017).
Kee, Y. & Bronner-Fraser, M. To proliferate or to die: Role of Id3 in cell cycle progression and survival of neural crest progenitors. Genes Dev. 19, 744–755. https://doi.org/10.1101/gad.1257405 (2005).
Yang, F. et al. Inositol 1,4,5-trisphosphate receptors are essential for fetal-maternal connection and embryo viability. PLoS Genet. 16, e1008739. https://doi.org/10.1371/journal.pgen.1008739 (2020).
Klein, J. Functions and pathophysiological roles of phospholipase D in the brain. J. Neurochem. 94, 1473–1487. https://doi.org/10.1111/j.1471-4159.2005.03315.x (2005).
Cai, K., Lucki, N. C. & Sewer, M. B. Silencing diacylglycerol kinase-theta expression reduces steroid hormone biosynthesis and cholesterol metabolism in human adrenocortical cells. Biochim. Biophys. Acta 552–562, 2014. https://doi.org/10.1016/j.bbalip.2013.12.005 (1841).
Lu, B. et al. Expression and regulation of GPAT isoforms in cultured human keratinocytes and rodent epidermis. J. Lipid Res. 51, 3207–3216. https://doi.org/10.1194/jlr.M007054 (2010).
Wang, S. et al. The appropriate frequency and function of decidual Tim-3+CTLA-4+CD8+ T cells are important in maintaining normal pregnancy. Cell Death Dis. 10, 407. https://doi.org/10.1038/s41419-019-1642-x (2019).
Pasquin, S. et al. Cardiotrophin-like cytokine factor 1 exhibits a myeloid-biased hematopoietic-stimulating function. Front. Immunol. 10, 2133–2133. https://doi.org/10.3389/fimmu.2019.02133 (2019).
Singh, H. & Aplin, J. D. Adhesion molecules in endometrial epithelium: Tissue integrity and embryo implantation. J. Anat. 215, 3–13. https://doi.org/10.1111/j.1469-7580.2008.01034.x (2009).
Huppertz, B. & Herrler, A. Regulation of proliferation and apoptosis during development of the preimplantation embryo and the placenta. Birth Defects Res. C Embryo Today 75, 249–261. https://doi.org/10.1002/bdrc.20056 (2005).
Tosti, E., Boni, R. & Gallo, A. Ion currents in embryo development. Birth Defects Res. C Embryo Today 108, 6–18. https://doi.org/10.1002/bdrc.21125 (2016).
Brainard, A. M., Korovkina, V. P. & England, S. K. Potassium channels and uterine function. Semin. Cell Dev. Biol. 18, 332–339. https://doi.org/10.1016/j.semcdb.2007.05.008 (2007).
Sanz-Ezquerro, J. J., Münsterberg, A. E. & Stricker, S. Editorial: Signaling pathways in embryonic development. Front. Cell Dev. Biol. https://doi.org/10.3389/fcell.2017.00076 (2017).
Komiya, Y. & Habas, R. Wnt signal transduction pathways. Organogenesis 4, 68–75. https://doi.org/10.4161/org.4.2.5851 (2008).
Sonderegger, S., Pollheimer, J. & Knöfler, M. Wnt signalling in implantation, decidualisation and placental differentiation—review. Placenta 31, 839–847. https://doi.org/10.1016/j.placenta.2010.07.011 (2010).
Woods, L., Perez-Garcia, V. & Hemberger, M. Regulation of placental development and its impact on fetal growth-new insights from mouse models. Front. Endocrinol. 9, 570–570. https://doi.org/10.3389/fendo.2018.00570 (2018).
Gibb, S., Maroto, M. & Dale, J. K. The segmentation clock mechanism moves up a notch. Trends Cell Biol. 20, 593–600. https://doi.org/10.1016/j.tcb.2010.07.001 (2010).
Tang, C. et al. Glioma-associated oncogene 2 is essential for trophoblastic fusion by forming a transcriptional complex with glial cell missing-a. J. Biol. Chem. 291, 5611–5622. https://doi.org/10.1074/jbc.M115.700336 (2016).
Takai, H. et al. Placental sonic hedgehog pathway regulates fetal growth via the IGF axis in preeclampsia. J. Clin. Endocrinol. Metab. 104, 4239–4252. https://doi.org/10.1210/jc.2019-00335 (2019).
Boopathy, G. T. K. & Hong, W. Role of hippo pathway-YAP/TAZ signaling in angiogenesis. Front. Cell Dev. Biol. 7, 49–49. https://doi.org/10.3389/fcell.2019.00049 (2019).
Radi, Z. A., Marusak, R. A. & Morris, D. L. Species comparison of the role of p38 MAP kinase in the female reproductive system. J. Toxicol. Pathol. 22, 109–124. https://doi.org/10.1293/tox.22.109 (2009).
Mudgett, J. S. et al. Essential role for p38alpha mitogen-activated protein kinase in placental angiogenesis. Proc. Natl. Acad. Sci. USA. 97, 10454–10459. https://doi.org/10.1073/pnas.180316397 (2000).
Ni, N. & Li, Q. TGFβ superfamily signaling and uterine decidualization. Reprod. Biol. Endocrinol. 15, 84–84. https://doi.org/10.1186/s12958-017-0303-0 (2017).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293. https://doi.org/10.1016/j.cell.2012.03.017 (2012).
Martin-Bermudo, M. D. & Brown, N. H. Uncoupling integrin adhesion and signaling: The betaPS cytoplasmic domain is sufficient to regulate gene expression in the Drosophila embryo. Genes Dev. 13, 729–739. https://doi.org/10.1101/gad.13.6.729 (1999).
Docheva, D., Popov, C., Alberton, P. & Aszodi, A. Integrin signaling in skeletal development and function. Birth Defects Res. C Embryo Today 102, 13–36. https://doi.org/10.1002/bdrc.21059 (2014).
Machaty, Z. Signal transduction in mammalian oocytes during fertilization. Cell Tissue Res. 363, 169–183. https://doi.org/10.1007/s00441-015-2291-8 (2016).
Woollett, L. A. Maternal cholesterol in fetal development: Transport of cholesterol from the maternal to the fetal circulation. Am. J. Clin. Nutr. 82, 1155–1161. https://doi.org/10.1093/ajcn/82.6.1155 (2005).
Chrzanowska-Wodnicka, M., White, G. C. II., Quilliam, L. A. & Whitehead, K. J. Small GTPase Rap1 is essential for mouse development and formation of functional vasculature. PLoS ONE 10, e0145689. https://doi.org/10.1371/journal.pone.0145689 (2016).
Acknowledgements
The authors thank Donald Bennink for providing the data and the Cooperative Dairy DNA Repository and the Council on Dairy Cattle Breeding for facilitating the access to the genotypes.
Author information
Authors and Affiliations
Contributions
F.P., A.S., and R.S.B. conceived and designed the study. A.S. and R.S.B. collected the data. A.S. analyzed the data. F.P. and R.S.B. contributed to interpretation of results. A.S. wrote the manuscript. All authors read, edited, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sigdel, A., Bisinotto, R.S. & Peñagaricano, F. Genes and pathways associated with pregnancy loss in dairy cattle. Sci Rep 11, 13329 (2021). https://doi.org/10.1038/s41598-021-92525-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-92525-0
This article is cited by
-
Genetic improvement of economic traits in Murrah buffalo using significant SNPs from genome-wide association study
Tropical Animal Health and Production (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.