Abstract
The protein kinase TBK1 is a central regulator of innate immune responses and autophagy, and ablation of either function has been linked to neuroinflammatory or degenerative diseases. Autophagy is an intracellular process that recycles old or damaged proteins and organelles. In recent years, the TBK1-dependent regulation of autophagy pathways has been characterized. However, the autophagy-dependent regulation of TBK1 activity awaits further clarification. Here, we observed that TBK1 is recruited to SQSTM1/p62-containing aggregates via the selective autophagy receptor TAX1BP1. In these aggregates, TBK1 phosphorylates SQSTM1/p62 at serine 403 and thus presumably regulates the efficient engulfment and clearance of these structures. We found that TBK1 activation is strongly increased if FIP200, a component of the autophagy-inducing ULK1 complex, is not present or cannot bind to TAX1BP1. Given our collective findings, we hypothesize that FIP200 ensures the inducible activation of TBK1 at SQSTM1/p62 condensates.
Similar content being viewed by others
Introduction
(Macro-)autophagy is an intracellular recycling process that maintains cellular homeostasis by degrading old or damaged proteins and organelles. Autophagy can be either nonselective or selective with regard to its cargo. Nonselective autophagy occurs at basal levels in most cell types but is also inducible under conditions of nutrient depletion. The process is initiated with the nucleation of a phagophore, which engulfs bulk cargo and expands into double-membraned vesicles called autophagosomes. Autophagosomes transport the cargo to lysosomes, in which the cargo is ultimately degraded. The initiation of autophagic processes is centrally regulated by two kinase complexes: (1) the UNC-51-like autophagy activating kinase 1 (ULK1) protein kinase complex containing the Ser/Thr kinase ULK1 and the adapter proteins autophagy-related (ATG) 13, ATG101 and focal adhesion kinase (FAK)-interacting protein of 200 kDa (FIP200); and (2) the class III phosphatidylinositol 3-kinase (PtdIns3K) lipid kinase complex containing the phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3/VPS34) and the associated proteins phosphoinositide 3 kinase regulatory subunit 4 (PIK3R4/VPS15), Beclin 1, ATG14, and nuclear receptor-binding factor 2 (NRBF2)1,2. Both kinase complexes are also involved in the regulation of selective autophagy, during which the cargo is specifically targeted by selective autophagy receptors (SARs)3,4. Selective autophagic processes are classified based on the type of degraded cargo, such as damaged mitochondria (mitophagy), protein aggregates (aggrephagy), or intracellular pathogens (xenophagy)3. The SARs sequestosome 1 (SQSTM1/p62), neighbor of BRCA1 gene 1 (NBR1), nuclear dot protein 52 kDa (NDP52), Tax1 binding protein 1 (TAX1BP1), and optineurin (OPTN) all belong to the SQSTM1/p62-like receptor (SLR) family and represent the best-studied family of SARs5. They simultaneously bind ubiquitin moieties on the cargo and phosphatidylethanolamine (PE)-conjugated ATG8 family proteins5, which are attached to the membrane of forming autophagosomes6. Recently, NDP52 was shown to attract the ULK1 complex to damaged mitochondria7 or cytosolic pathogens8 by binding to a C-terminal region of FIP200. Once recruited, ULK1 initiates the formation of the phagophore directly at the cargo9. The recruitment of the ULK1 complex is facilitated by TANK-binding kinase 1 (TBK1)7,8. The Ser/Thr kinase TBK1 is a central regulator of innate immune responses, but in recent years, its involvement in autophagy signaling pathways has been discovered. In addition to facilitating recruitment of the ULK1 complex to mitochondria or intracellular pathogens, TBK1 directly regulates other components of the autophagy signaling cascade, including AMP-activated protein kinase (AMPK)10, syntaxin 1711, and various SARs7,8,12,13,14,15,16,17. TBK1-dependent phosphorylation frequently modulates the binding affinities of SARs; for example, it has been reported that TBK1-catalyzed phosphorylation of SQSTM1/p62 at Ser403 enhances ubiquitin binding13,14,18.
The innate immune response is the first line of defense during viral infections. Pattern recognition receptors (PRRs) detect specific bacterial or viral components, ultimately inducing type I interferon (IFN) and proinflammatory cytokine production to prevent viral invasion and replication19. PRRs include Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and cytosolic DNA receptors such as cyclic GMP-AMP synthase (cGAS). Upon ligand binding, TLR3/4, RIG-I, and cGAS mediate the recruitment of TBK1 to the adaptor proteins TIR domain-containing adapter-inducing interferon-β (TRIF), mitochondrial antiviral signaling protein (MAVS), and stimulator of interferon genes (STING), respectively20. Local clustering leads to trans-autophosphorylation at Ser172 and thus activation of TBK121,22. Once activated, TBK1 phosphorylates interferon regulatory factor (IRF) 3 and IRF7, thereby inducing their dimerization and nuclear translocation. In the nucleus, IRF3 and IRF7 activate type I IFN gene expression23. TLR signaling, e.g., binding of the adaptor proteins TRIF and myeloid differentiation primary response 88 (MyD88) to Beclin 1, also induces autophagy24,25. On the other hand, autophagy can exert anti-inflammatory effects26, such as by targeting the RIG-I-MAVS axis27 or via TAX1BP1-mediated selective degradation of TRIF28. Furthermore, TAX1BP1 and the ubiquitin-editing enzyme A20 target TBK1 to inhibit the immune response by disrupting the TRAF3-TBK1 signaling complex29.
Mutations of the TBK1 gene are connected to several diseases, including childhood herpes simplex encephalitis (HSE), amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and normal-tension glaucoma (NTG) (reviewed in30). These diseases are caused either by dysregulated autophagy or by impaired IFN production30. Loss-of-function mutations in the TBK1 gene have been linked to ALS and FTD31,32. In turn, gain-of-function TBK1 mutations (i.e., gene duplication) have been reported in patients with NTG. Therefore, TBK1 must be tightly regulated to maintain cellular homeostasis. Although the involvement of TBK1 in the regulation of autophagy is known, the regulation of TBK1 itself during autophagic processes awaits further clarification.
In this study, we found that TBK1-TAX1BP1-SQSTM1/p62 aggregates develop in cells with autophagy impairment in general and FIP200 deficiency in particular. TBK1 is activated in these aggregates and phosphorylates SQSTM1/p62 at Ser403. These aggregates are likely caused by proteotoxic stress induced by inhibition of autophagy. The recruitment of TBK1 to these aggregates is mediated by TAX1BP1. The activation of TBK1 is clearly increased in cells if FIP200 is not present or cannot bind to TAX1BP1, indicating that FIP200—in addition to recruiting the downstream autophagy machinery—inhibits aberrant activation of TBK1. We propose that FIP200 controls the TBK1 activation threshold and ensures the inducibility of TBK1 activity.
Results
Loss of autophagy in general and loss of components of the ULK1 complex in particular lead to increased focal accumulation and activation of TBK1
To search for novel FIP200-interacting proteins, we performed mass spectrometry (MS) using GFP-FIP200 as bait. Among the identified binding partners were TBK1, TBKBP1/SINTBAD, and several autophagy receptors (TAX1BP1, SQSTM1/p62, and NBR1) (Table S1). The interaction between GFP-FIP200 and TBK1 was confirmed by immunopurification (Fig. 1). Recent reports have shown that TBK1 binds to the autophagy receptor NDP52 and facilitates ULK1 complex recruitment via FIP2007,8. In our immunopurification experiments, we observed that GFP-FIP200 bound to the autophagy receptors TAX1BP1 and NDP52 (Fig. 1). Accordingly, we assumed that FIP200 interacts with TBK1 via TAX1BP1 and/or NDP52. Since the cells used for MS and immunopurification were cultured in complete medium, it is likely that these interactions are constitutive.
FIP200 interacts with TBK1. Flp-In T-REx 293 cells inducibly expressing GFP or GFP-FIP200 were treated with 0.1 µg/ml doxycycline for 16 h to induce expression. Afterwards, the GFP proteins were purified overnight using GFP-trap beads. Cleared cellular lysates (input) and purified proteins were immunoblotted for the indicated proteins. The full-length blots are presented in Figure S5.
It has been previously reported that the C terminus of FIP200 mediates binding to the autophagy receptor(s) NDP52 and/or SQSTM1/p627,8,33. The interaction between the C terminus of FIP200 (amino acids 1352–1441) and NDP52 has been shown to be mandatory for the recruitment of the ULK1 complex to damaged mitochondria or intracellular bacteria, respectively7,8, whereas the binding of the C-terminal FIP200 claw domain (amino acids 1494–1594) to SQSTM1/p62 promotes autophagosome formation at SQSTM1/p62-ubiquitin condensates33. To analyze the potential role of FIP200 and, in particular, its C terminus in TBK1 activation, we made use of fip200 knockout (KO) mouse embryonic fibroblasts (MEFs) that were transfected with an empty vector or cDNA encoding wild-type FIP200 or a variant lacking the C-terminal part of FIP200 (amino acids 1369–1594, ΔCT). Notably, we observed that loss of FIP200 led to aberrant accumulation of activated TBK1, as determined by immunofluorescence (IF) using antibodies specific for TBK1 phosphorylated at Ser172 (pS172) (Fig. 2A). This finding is in accordance with those of a study by Goodwin et al. investigating ferritinophagy34. Expression of wild-type FIP200 in fip200 KO MEFs blocked the accumulation of phospho-TBK1. In contrast, the C-terminally truncated version of FIP200 reduced the focal localization of phospho-TBK1 but was not able to completely prevent it (Fig. 2A). Since autophagy is blocked in FIP200-deficient MEFs35, we also investigated the influence of generally defective autophagy on TBK1 regulation. For this, we examined the phosphorylation status of TBK1 in MEFs deficient in ATG3 via immunoblotting and IF (Fig. 2A, Figure S1A). ATG3 mediates the E2-like conjugation of LC3 to PE, which is an essential step during autophagy6. Atg3 KO cells showed greater TBK1 activation than wild-type MEFs (Fig. 2A, Figure S1A). We observed that blockade of autophagy with the lysosomal V-ATPase inhibitor bafilomycin A1 (BafA1) increased TBK1 activation in wild-type cells but not in atg3 KO cells (Figure S1A). Next, we investigated whether depletion of ATG13, another component of the ULK1 complex, affects TBK1 activation. IF analysis of atg13 KO cells revealed that both TBK1 and FIP200 formed puncta (Fig. 2B). Transient expression of GST-TBK1 showed that the observed TBK1 and FIP200 puncta colocalized, and partially circular structures were detectable (Fig. 2C). To enable better comparison, the phospho-TBK1-positive puncta per cell were counted in all mentioned cell lines, and the size and intensity of each punctum were determined (Fig. 2D). Loss of FIP200 led to an increase in the number of puncta per cell. Moreover, the sizes and intensities of these phospho-TBK1-positive structures were increased. Removal of the FIP200 C-terminal domain increased the number of TBK1 aggregates by a factor of 3. The sizes and intensities of the resulting structures were also increased (by ~ 5 × per cell and ~ 9 × per cell, respectively) (Fig. 2D). In atg13 KO cells, the intensity of the phospho-TBK1 puncta was slightly increased, but the number and area were not as high as those in fip200 KO MEFs. Similarly, there were fewer phospho-TBK1 aggregates in atg3 KO cells than in fip200 KO MEFs, and the phospho-TBK1 aggregates in atg3 KO cells were smaller and less intense than those observed in fip200 KO MEFs (Fig. 2D).
Loss of components of the ULK1 complex leads to aberrant accumulation and increased activation of TBK1. (A) fip200 KO MEFs were transfected with an empty vector or cDNA encoding full-length FIP200 (FIP200 WT) or C-terminally truncated FIP200 (amino acids 1369–1594, FIP200 ΔCT). These transfectants, atg3 KO and Atg3 WT MEFs were fixed in 4% PFA and immunostained for TBK1 pS172. Representative sections are depicted. (B) atg13 KO MEFs transfected with an empty vector or cDNA encoding WT ATG13 were fixed in 4% PFA and immunostained for TBK1 pS172 and FIP200. Representative sections are depicted. (C) For costaining of TBK1 and FIP200, atg13 KO MEFs were transiently transfected with cDNA encoding GST-TBK1, fixed in 4% PFA, and immunostained for FIP200 and GST. Representative sections are depicted. For IF imaging, DAPI was used to stain nuclei. (D) The number, area, and intensity of TBK1 pS172-positive structures in at least 198 cells per cell line were quantified using ImageJ 1.53c and normalized to the values in FIP200 WT cells. The means + SDs of three independent experiments are shown. (E) Cleared cellular lysates of MEF ulk1/2 DKO, fip200 KO and atg13 KO MEFs transfected with an empty vector or the respective WT cDNA (encoding FIP200, ATG13, ULK1, or ULK2) were immunoblotted for the indicated proteins. The full-length blots are presented in Figure S6. The densities of the bands on immunoblots from three independent experiments were quantified and normalized to those of Vinculin. The normalized densities of all samples of each cell line were then normalized to that of the respective reconstituted cell line. Only the relative levels of TBK1 pS172 are shown. The boxes represent the highest and the lowest value, while the centerline shows the mean. Statistical analysis was performed using two-way ANOVA with uncorrected Fisher's LSD test. n.s. = not significant; *p < 0.05; ***p < 0.001.
Next, we investigated TBK1 activation in cells deficient for single components of the ULK1 complex by immunoblotting. Briefly, MEFs deficient in either ATG13 or ULK1/ULK2 were transfected with cDNA encoding wild-type ATG13, ULK1, or ULK2. As determined by immunoblotting, loss of ATG13 increased TBK1 activation, while loss of ULK1/2 led to only a slight increase in TBK1 activation. The absence of FIP200 had the strongest effect on TBK1 activation (Fig. 2E). Next, we analyzed whether FIP200 also influences TBK1 activation in human cell lines by knocking down FIP200 levels in HeLa cells or differentiated THP-1 cells with siRNA. Knockdown of FIP200 with three different siRNAs increased TBK1 activation in both cell lines, indicating the general validity of our observations (Figure S1B). The immunoblots for Flp-In T-REx 293 cells and MEFs (Figs. 1 and 2E) suggested that overexpression of FIP200 does not affect TBK1 protein levels, and this was confirmed by quantification (Figure S1C). Collectively, these data suggest that autophagy in general and the ULK1 complex in particular are important for the regulation of TBK1 activation. Within the ULK1 complex, FIP200 seems to be the main effector controlling TBK1 activation.
Phospho-TBK1 aggregates are positive for the autophagy receptors TAX1BP1 and SQSTM1/p62
We observed that loss of FIP200 or deletion of its C terminus resulted in aberrant TBK1 accumulation. Normally, protein aggregates are cleared by aggrephagy. Since autophagy receptors recognize the cargoes during selective autophagy and connect them to the autophagy machinery, we analyzed whether the receptors TAX1BP1, NDP52, OPTN and SQSTM1/p62 localize to TBK1 aggregates. As determined by IF, TAX1BP1 and SQSTM1/p62 colocalized with TBK1 aggregates caused by loss of FIP200, while NDP52 and OPTN did not (Fig. 3A). Similar observations were made for cells expressing the C-terminally truncated version of FIP200 (Fig. 3B). It has previously been reported that TBK1 phosphorylates SQSTM1/p62 at Ser403 and that this phosphorylation ensures efficient autophagosomal engulfment of ubiquitinated mitochondria13. Phospho-SQSTM1/p62 was highly abundant within the TBK1-TAX1BP1-SQSTM1/p62 aggregates (Fig. 3A,B). We also analyzed whether the C-terminally truncated variant of FIP200 was recruited to TBK1 aggregates. The expressed FIP200 ΔCT showed a cytosolic distribution and no colocalization with TBK1 aggregates (Fig. 3B). Additionally, some of the TBK1-positive aggregates that formed because of FIP200 loss were positive for the autophagy marker LC3, while others were not. Similar observations were made for cells expressing FIP200 ΔCT (Figure S2A). It has been previously reported that the Golgi apparatus acts as a platform for TBK1 activation36. Thus, we investigated whether TBK1 aggregates occur in specific organelles. Antibodies against ERGIC-53/p58, ERp72 and Golgin97 were used to stain the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC), the ER, and the Golgi complex, respectively. As determined by IF, TBK1 aggregates did not colocalize with any of these organelles (Figure S2B). Taken together, these data suggest that TBK1 accumulates together with TAX1BP1 and SQSTM1/p62 in large aggregates/condensates when binding to FIP200 is abolished.
TBK1 aggregates are positive for the autophagy receptors TAX1BP1 and SQSTM1/p62 pS403. (A + B) fip200 KO MEFs transfected with an empty vector (A) or cDNA encoding FIP200 ΔCT (B) were fixed in 100% MeOH and immunostained for TBK1 in combination with TBK1 pS172, FIP200, SQSTM1/p62 pS403, NDP52, or OPTN. Representative sections of three independent experiments are depicted. The bar graphs represent the pixel intensities of the areas indicated by the respective dashed red arrows shown in the insets.
TBK1 aggregation depends on TAX1BP1
Since we observed that TAX1BP1 colocalizes with TBK1 aggregates, we next investigated whether TBK1-SQSTM1/p62 aggregate formation itself depends on TAX1BP1. Specifically, we decreased TAX1BP1 expression via RNA interference (RNAi) in fip200 KO, FIP200 wild-type (WT) and FIP200 ΔCT cells and analyzed TBK1 activation and SQSTM1/p62 phosphorylation at Ser403 by immunoblotting. Although we clearly detected focal localization of TBK1 in FIP200 ΔCT cells (Fig. 2A,D), the pS172 signal detected by immunoblotting resembled that of FIP200 WT cells. Knockdown of TAX1BP1 decreased TBK1 activation and SQSTM1/p62 phosphorylation in all cell lines (Fig. 4A–C). Interestingly, we observed that SQSTM1/p62 migrated at a higher molecular weight in fip200 KO MEFs and in cells expressing the C-terminally truncated FIP200 variant than in WT cells, presumably reflecting increased phosphorylation of SQSTM1/p62, albeit not necessarily only at Ser403 (Fig. 4A). These results indicate that TBK1-SQSTM1/p62 aggregate formation and the phosphorylation of SQSTM1/p62 at Ser403 in the aggregates depend on the autophagy receptor TAX1BP1.
Aberrant TBK1 accumulation depends on TAX1BP1. (A) fip200 KO, FIP200 WT, and FIP200 ΔCT MEFs were transfected with either 20 nM negative control siRNA or 20 nM Tax1bp1 siRNA for 72 h, and cleared cellular lysates were immunoblotted for the indicated proteins. The FIP200 cDNAs used for reconstitution of fip200 KO MEFs encoded an N-terminal FLAG peptide. The densities of the bands on immunoblots from three independent experiments were quantified and normalized to those of Actin. The full-length blots are presented in Figure S7. (B) To compare basal expression levels between the cell lines used, the normalized densities of the untransfected samples were normalized to the signal of FIP200 WT MEFs. (C) To analyze the influence of siRNA transfection, the normalized densities of all samples of each cell line were normalized to that of the corresponding untransfected sample. Only the relative levels of TAX1BP1, TBK1 pS172, and SQSTM1/p62 pS403 are shown. The boxes represent the highest and the lowest value, while the centerline shows the mean. Two-way ANOVA with uncorrected Fisher's LSD test was used to determine differences between treatments. n.s. = not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Impaired autophagy in fip200 KO MEFs contributes to aberrant TBK1 aggregation and activation
Since defective autophagy increased TBK1 activation in atg3 KO MEFs (Figure S1A), we next investigated whether nonfunctional autophagy was the only mechanism leading to increased TBK1 activation in fip200 KO MEFs by analyzing starvation-induced bulk autophagy. We performed an LC3 turnover assay in fip200 KO, FIP200 WT and FIP200 ΔCT-expressing cells. Upon autophagy induction, lipidated LC3-II is increasingly generated and then degraded in autolysosomes37. The V-ATPase inhibitor BafA1 blocks lysosomal degradation, leading to accumulation of LC3-II37. In fip200 KO MEFs, we did not observe LC3-II accumulation under either normal or starvation conditions, confirming that autophagy signaling was defective. However, expression of FIP200 WT and FIP200 ΔCT restored basal and starvation-induced autophagy (Fig. 5A,B). Similar to the case in Atg3 WT cells (Figure S1A), TBK1 activation was increased in FIP200 WT and FIP200 ΔCT MEFs after inhibition of autophagy with BafA1. Additionally, starvation led to reduced SQSTM1/p62 levels and decreased TBK1 phosphorylation. However, TBK1 and SQSTM1/p62 phosphorylation was increased in fip200 KO cells and was unaffected by all treatment conditions (Fig. 5A). FIP200 ΔCT cells also resembled FIP200 WT cells during bulk autophagy with regard to TBK1 phosphorylation (see also Fig. 4A), whereas FIP200 ΔCT cells represented an intermediate status between fip200 KO and FIP200 WT cells with regard to SQSTM1/p62 phosphorylation. Collectively, these findings indicate that the TBK1 accumulation in FIP200 ΔCT cells detected by IF (Fig. 2A) was not solely attributable to impaired autophagy signaling, since bulk autophagic flux appeared rather normal in these cells.
The C terminus of FIP200 is dispensable for starvation-induced autophagy. (A) fip200 KO, FIP200 WT, and FIP200 ΔCT MEFs were cultured in complete or starvation (EBSS) medium in either the absence or presence of 10 nM BafA1 for 2 h. Cleared cellular lysates were immunoblotted for the indicated proteins. The FIP200 cDNAs used for reconstitution of fip200 KO MEFs encoded an N-terminal FLAG peptide. The full-length blots are presented in Figure S8. (B) The densities of the bands on immunoblots from three independent experiments were quantified and normalized to those of Actin. The normalized densities of all samples of each cell line were then normalized to that of the respective untreated control sample. Only the relative levels of LC3-II are shown. The boxes represent the highest and the lowest value, while the centerline shows the mean. Two-way ANOVA with uncorrected Fisher's LSD test was used to determine differences between treatments. n.s. = not significant; *p < 0.05; ****p < 0.0001.
Loss of FIP200 further enhances TBK1 accumulation and activation caused by defective autophagy
Since we observed a clear difference between atg3 KO and fip200 KO MEFs with regard to phospho-TBK1 accumulation by IF (Fig. 2A) and a rather normal autophagic flux in FIP200 ΔCT cells (Fig. 5A), we speculated that inhibited autophagy could not be the sole cause for the aberrant TBK1 activation. Accordingly, we next investigated whether loss of FIP200 in autophagy-deficient cells can further increase TBK1 accumulation and activation. We performed FIP200 knockdown experiments in atg3 KO cells and analyzed TBK1 activation and SQSTM1/p62 phosphorylation by immunoblotting. SiRNA-mediated knockdown of FIP200 led to increased TBK1 activation and SQSTM1/p62 phosphorylation in atg3 KO MEFs (Fig. 6A). We also investigated TBK1 accumulation by IF. Upon transfection of nontargeting or Fip200-targeting siRNA, we observed punctate or circular TBK1-positive structures (Fig. 6B,C). Fip200 siRNA led to the removal of FIP200 from these aggregates and to an increased size of the ring-shaped structures. Similar to the findings in fip200 KO cells, the TBK1 structures also contained phosphorylated SQSTM1/p62 (Fig. 6C). Collectively, these data show that the combination of defective autophagy and loss of FIP200 leads to aberrant accumulation of TBK1-TAX1BP1-SQSTM1/p62 aggregates. It appears that the presence of FIP200 at these aggregates ensures the inducibility of TBK1 activation.
Loss of FIP200 further enhances TBK1 accumulation and activation caused by defective autophagy. (A) Atg3 WT and atg3 KO MEFs were transfected with either 20 nM negative control siRNA or 20 nM Fip200 siRNA for 72 h, and the cleared cellular lysates were immunoblotted for the indicated proteins. The asterisk indicates a nonspecific band. The full-length blots are presented in Figure S9. The densities of the bands on immunoblots from three independent experiments were quantified and normalized to those of Actin. For each protein, the normalized densities of atg3 KO samples were normalized to that of the respective untransfected sample. The boxes represent the highest and the lowest value, while the centerline shows the mean. Two-way ANOVA with Tukey's multiple comparisons test was used to determine differences between treatments. n.s. = not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. (B + C) atg3 KO MEFs were transfected with either 20 nM negative control siRNA or 20 nM Fip200 siRNA for 72 h and immunostained for TBK1 pS172 (B; fixed in 4% PFA) or TBK1 in combination with either FIP200 or SQSTM1/p62 pS403 (C; fixed in 100% MeOH). For (B), the number, area, and intensity of TBK1 pS172-positive structures of at least 232 cells per cell line were quantified using ImageJ 1.53c and normalized to those of untransfected atg3 KO cells. The boxes represent the highest and the lowest value, while the centerline shows the mean from four independent experiments. For IF imaging, DAPI was used to stain nuclei.
The IFN-β response upon TLR3/TLR4 stimulation is not significantly affected in fip200 KO or FIP200 ΔCT-expressing cells
Since TBK1 is involved not only in autophagy signaling but also in the regulation of innate immune responses, we examined whether loss of FIP200 influences TLR3/TLR4-induced IFN-β production through TBK1-TAX1BP1-SQSTM1/p62 aggregate formation. Fip200 KO, FIP200 WT and FIP200 ΔCT MEFs were either transfected with poly I:C or treated with lipopolysaccharide (LPS), and IFN-β gene expression was analyzed by reverse transcription (RT)-quantitative PCR (qPCR). In all three cell lines, poly I:C treatment resulted in clear upregulation of IFN-β mRNA expression, whereas the response to LPS was rather weak (Fig. 7A). Although TBK1 was highly activated in fip200 KO MEFs, the activation did not result in increased IFN-β production under basal or stimulated conditions compared to that in FIP200 WT cells (Fig. 7A). In contrast, IFN-β production appeared rather impaired in fip200 KO and FIP200 ΔCT MEFs after poly I:C treatment, but the differences were not statistically significant (Fig. 7A). Additionally, the receptors STING and TRIF, which recruit TBK1 during innate immune responses, did not colocalize with TBK1-TAX1BP1-SQSTM1/p62 aggregates (Figures S3A and S3B). In addition to IFN-β production, we also examined TBK1 and SQSTM1/p62 phosphorylation by immunoblotting. Neither poly I:C nor LPS induced major changes in the (increased) phosphorylation status of TBK1 and SQSTM1/p62 in fip200 KO MEFs (Fig. 7B,C). Expression of wild-type FIP200 slightly restored the inducibility of TBK1 phosphorylation upon LPS treatment. In FIP200 ΔCT MEFs, poly I:C and LPS treatment also resulted in slightly increased TBK1 phosphorylation accompanied by a simultaneous decrease in SQSTM1/p62 phosphorylation (Fig. 7B,C). Taken together, the results suggest that FIP200 deficiency desensitizes cells to TLR3-/TLR4-engaging stimuli with regard to TBK1 activation. However, this desensitization does not result in increased IFN-β production under basal or TLR3-/TLR4-stimulated conditions. If FIP200 is present but not capable of interacting with TAX1BP1 and/or SQSTM1/p62 (as in FIP200 ΔCT MEFs), the inducibility of TBK1 activation upon TLR3/TLR4 engagement is partially restored.
FIP200 deficiency desensitizes cells to TLR3-/TLR4-engaging stimuli. (A–C) fip200 KO, FIP200 WT, and FIP200 ΔCT MEFs were transfected with 2 µg/ml poly I:C or treated with 1 µg/ml LPS for the indicated times. dH2O was used for mock transfection. (A) RT-qPCR analyses were performed to determine the relative IFN-β expression. All values were normalized to those of control samples of FIP200 WT MEFs. The boxes represent the highest and the lowest value, while the centerline shows the mean from three independent experiments. (B) Lysates of poly I:C-transfected or LPS-treated cells were immunoblotted for the indicated proteins. Representative immunoblots of three independent experiments are depicted. The full-length blots are presented in Figure S10. (C) The densities of the bands on immunoblots from three independent experiments (except for 3 h poly I:C transfection, which was performed only once) were quantified and normalized to those of Actin. The normalized densities of all samples were then normalized to that of the respective untreated sample of FIP200 WT cells. Only the relative levels of TBK1 pS172 and SQSTM1/p62 pS403 are shown. The boxes represent the highest and the lowest value, while the centerline shows the mean.
Discussion
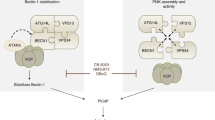
As a component of the autophagy-inducing ULK1 complex, FIP200 is centrally involved in bulk and selective autophagy. However, FIP200 also participates in several additional cellular signaling pathways38. TBK1 is a central regulator of both autophagic and inflammatory signaling39. Here, we observed that TBK1, TAX1BP1 and SQSTM1/p62 form protein aggregates in MEFs lacking FIP200. Within these structures, TBK1 is activated by trans-autophosphorylation and in turn phosphorylates SQSTM1/p62 at Ser403, thus further promoting the formation of SQSTM1/p62-ubiquitin condensates. TBK1 activation and SQSTM1/p62 phosphorylation depend on TAX1BP1, indicating that this autophagy receptor mediates the recruitment of TBK1 to SQSTM1/p62 condensates. We observed that TBK1 activation is strongly enhanced in cells lacking FIP200. On the one hand, this might be caused by the inhibition of autophagy and thus the generally inhibited/reduced clearance of SQSTM1/p62 condensates. On the other hand, our data suggest that the constitutive binding of FIP200 to TBK1 via TAX1BP1 keeps TBK1 in an inducible state. Accordingly, FIP200 controls the TBK1 activation threshold at SQSTM1/p62-positive protein aggregates. Given this collective evidence, we hypothesize that the TAX1BP1-TBK1-phospho-SQSTM1/p62 axis engages in positive feedforward regulation of aggrephagy that is controlled by the recruitment of FIP200 (scheme depicted in Figure S4).
In 2008, it was reported that FIP200 binds to ULK1/2 and is required for autophagosome formation35. Since then, several reports have confirmed the central role of FIP200 in autophagy signaling, including in selective autophagic processes. Studies using conditional fip200 KO mice have shown that loss of FIP200 leads to accumulation of SQSTM1/p62-positive protein aggregates in osteoblasts40, retinal pigment epithelial cells41, and mammary tumor cells42 due to impairment of autophagy. In addition, recent reports have indicated that recruitment of the ULK1 complex to damaged mitochondria or invading bacteria is mediated by the binding of FIP200 to the autophagy receptor NDP527,8. Furthermore, it has been found that the C-terminal claw domain of FIP200 binds to the autophagy receptor SQSTM1/p62 and thus promotes autophagosome formation at SQSTM1/p62-ubiquitin condensates33. We observed that FIP200 also associates with the autophagy receptor TAX1BP1, similarly resulting in the association of FIP200 with SQSTM1/p62-positive condensates. Furthermore, it appears that NDP52 and/or OPTN is not recruited to these structures. Generally, our observations are in line with the findings of a recent study indicating that the clearance of protein aggregates is mediated by TAX1BP143. Apparently, both SQSTM1/p62 and TAX1BP1 are present in protein aggregates and can recruit FIP200. The binding of FIP200 to these two autophagy receptors is not necessarily mutually exclusive, since binding of TAX1BP1 (and NDP52) has been reported to occur outside the claw domain8. However, TAX1BP1 is clearly responsible for TBK1 activation and SQSTM1/p62 phosphorylation within these structures.
Several types of crosstalk exist between TBK1 and autophagy. On the one hand, TBK1 regulates several components of the autophagy signaling cascade, including AMPK10, syntaxin 1711, and various autophagy receptors7,8,12,13,14,15,16,17. We observed that the TBK1-dependent phosphorylation of SQSTM1/p62 at Ser403 was increased in fip200 KO MEFs. Such phosphorylation has been implicated in the SQSTM1/p62-dependent phase separation/clustering of polyubiquitinated proteins and the selective autophagic clearance of these protein aggregates18,44,45. Furthermore, it has been confirmed that TBK1 can catalyze this phosphorylation13,14. We propose that the TAX1BP1-dependent recruitment of TBK1 to protein aggregates and the subsequent phosphorylation of SQSTM1/p62 at Ser403 represent a feedforward loop that ensures efficient phase separation, engulfment, and clearance of the cargo. Apparently, this function needs to be controlled by FIP200, since TBK1 might increase the size and potentially the insolubility of the protein aggregates via uncontrolled SQSTM1/p62 phosphorylation. This conclusion is in line with the observations of Turco et al., who reported that knockdown of FIP200 increases both the number and volume of SQSTM1/p62 puncta33. As mentioned above, SQSTM1/p62-positive protein aggregates have already been reported for various cell lines obtained from conditional fip200 KO mice40,41,42. Additionally, TBK1 inhibition has been found to prevent the generation of insoluble protein aggregates in hepatocytes46. Interestingly, we observed that the phosphorylated-Ser403 (pS403) variant of SQSTM1/p62 shifted to an increased molecular weight. Ser403 phosphorylation itself might have contributed to this phenomenon; however, we cannot exclude the possibility that additional phosphorylation events were involved. Turco et al. reported that phosphorylation of the FIP200-interacting region in SQSTM1/p62 (i.e., Ser349/Thr350, Ser365, Ser366, Ser370/Thr375) increases the affinity of SQSTM1/p62 for the FIP200 claw domain47. It is possible that phosphorylation of this region increases as a compensatory mechanism to counteract FIP200 deficiency and thus contributes to the alteration in molecular weight.
On the other hand, TBK1 activation is controlled by autophagy, as confirmed by our observations. We found that inhibition of autophagy via targeting of various autophagy-related genes (e.g., ATG3, FIP200, ATG13, ULK1) or via treatment with BafA1 increased TBK1 activation. Furthermore, starvation-induced autophagy decreased TBK1 activation, but starvation did not affect TBK1 activation in autophagy-incompetent cells. Yang et al. also observed that starvation-induced autophagy represses TBK1 activation, while inhibition of autophagy increases TBK1 activation48. The authors suggested that NDP52 and SQSTM1/p62 promote autophagy of phospho-TBK1 complexes48. It has also been reported that TBK1 is directly targeted by ULK110. Goodwin et al. analyzed TBK1 activation during lysosomal ferritin flux in cells deficient in either of the ULK1 complex subunits. Similar to our results, their findings revealed that among the alterations, FIP200 depletion had the greatest impact on TBK1 activation; the authors also observed TAX1BP1-TBK1-positive structures34. In that case, the authors suggested that a compensatory relationship exists between the ULK1/2 complex and TBK1 activation, at least for lysosomal ferritin flux34. We propose that FIP200 exerts an additional specific function during aggrephagy/selective autophagy that cannot be compensated for by the other subunits of the ULK1 complex. Furthermore, we propose that this additional function of FIP200 is independent of its autophagy-regulating function. These propositions are supported by two observations. First, we detected increased phospho-TBK1 levels in cells deficient in ULK1/2, ATG13, or ATG3. However, the levels in these cells were still lower than those in fip200 KO MEFs. We believe that the number and/or size of protein aggregates was increased in all these cell lines with autophagy deficiency/impairment and that this effect certainly contributed to TBK1 activation. However, these data simultaneously suggest that the recruitment of the ULK1 complex is not the sole function of FIP200 during aggrephagy. Of note, Turco et al. reported that ULK1 is still recruited to SQSTM1/p62 condensates in cells lacking FIP200 but that ATG16L1 recruitment is abolished33. This observation was recently substantiated in a preprint from the same group reporting that FIP200 is dispensable for the recruitment of the upstream autophagy machinery to the condensates but necessary for phosphatidylinositol 3-phosphate formation, WIPI2 recruitment, and ULK1 kinase activation49. Second, we observed increased phospho-TBK1 levels in cells expressing the ΔCT variant of FIP200. However, in these cells, normal starvation-induced autophagy occurred. It has been reported that the ULK1 complex is organized by a C-shaped FIP200 N-terminal domain dimer50. Accordingly, it is possible that the C-terminal domain of FIP200 exerts some additional functions that are independent of its canonical autophagy function. Notably, Jun-Lin Guan’s group recently reported that FIP200 can limit AZI2/NAP1-TBK1-IRF signaling independent of its canonical autophagy function51,52. They proposed that—in fip200 KO cells—phospho-TBK1-containing phase condensates accumulate, resulting in TBK1 hyperactivation and sustained proinflammatory signaling51,52. This mechanism is in line with a report describing that cellular stress leads to incorporation of the TBK1 adaptors TBKBP1/SINTBAD and AZI2/NAP1 into membraneless organelles that control the threshold of TBK1 activation53. Whether the TBKBP1/SINTBAD-AZI2/NAP1-membraneless organelles and the SQSTM1/p62 aggregates represent different or overlapping phase condensates remains to be investigated. We detected TBKBP1/SINTBAD in our MS analyses of anti-FIP200 immunopurifications (see Table S1), but we did not observe differences in TBKBP1/SINTBAD puncta formation in FIP200 WT, FIP200 ΔCT or fip200 KO cells (data not shown). It is tempting to speculate that different compositions of phase condensates can regulate different TBK1 signaling outcomes in different cellular systems. Here, we observed an effect not on IFN-β production but rather on SQSTM1/p62 phosphorylation. Generally, the relative contributions of the different protein–protein interactions for the heterotrimeric control of TBK1 activation at phase condensates, i.e., the interactions between FIP200 and TAX1BP1 (see this report and8), the interactions between TAX1BP1 and TBKBP1/SINTBAD or AZI2/NAP116, and the interactions between FIP200 and TBKBP1/SINTBAD or AZI2/NAP1, await further clarification8,51. Furthermore, it has been suggested that TAX1BP1 can directly associate with TBK112, adding another level of complexity. Currently, we speculate that the binding of FIP200 to TAX1BP1 and/or TBKBP1/SINTBAD-AZI2/NAP1 is sufficient for control of TBK1 activation either via mediation of the selective removal of TBK1 or via steric hindrance of TBK1 trans-autophosphorylation.
The involvement of LC3 in this process (removal of SQSTM1/p62 condensates and/or regulation of TBK1 activation) needs to be clarified. In the abovementioned study on TBK1 hyperactivation at phase condensates, the authors used a FIP200 variant that cannot bind ATG1351. In cells expressing this mutant, LC3 lipidation is blocked54. The authors speculated that FIP200 can—through its ability to bind cargo receptors—bypass the need for LC3 and initiate phagophore formation at the cargo during selective autophagy52. However, alternative recruitment mechanisms might also play roles. It has been reported that deletion of the FIP200 claw domain does not completely prevent the targeting of SQSTM1/p62-positive cargo to lysosomes33 and that there is residual LC3 lipidation in cells expressing a FIP200 claw domain mutant that cannot bind to SQSTM1/p6249. We clearly observed partial colocalization of LC3 and TBK1 in both fip200 KO cells and cells expressing our ΔCT variant lacking both the interaction site with TAX1BP1 and the claw domain. At present, we cannot differentiate whether this colocalization was caused by alternative recruitment of the LC3 lipidation machinery or perhaps by lipidation-independent recruitment of LC3 molecules.
Protein aggregation is a hallmark of neurodegenerative diseases. Additionally, several mutations in TBK1 have been identified that underlie neuroinflammatory diseases, and dysregulation of TBK1 is strongly connected to these diseases (reviewed in30). For example, treatment with the TBK1 inhibitor BX795 abrogates the aberrant insolubility of an optineurin mutant (E50K) that can be found in patients suffering from NTG55. Additionally, ALS-associated TBK1 mutations affect the phosphorylation of different autophagy receptors56. A role of FIP200 in neuronal homeostasis has also been recognized. Liang et al. reported that neural-specific loss of FIP200 resulted in cerebellar degeneration accompanied by progressive neuronal loss, spongiosis, and neurite degeneration57. The authors also reported accumulation of SQSTM1/p62 and ubiquitinated protein aggregates57. Interestingly, the observed phenotypes developed earlier and were somewhat more severe in neural-specific FIP200 conditional KO mice than in Atg5 or Atg7 conditional KO mice57. We speculate that the FIP200-dependent regulation of TBK1 activity at SQSTM1/p62 condensates might contribute to these observations, establishing enforced recruitment of FIP200 to these condensates—next to pharmacological regulation of TBK1 activity—as a promising therapeutic approach.
Materials and methods
Antibodies and reagents
Antibodies against β-Actin (western blot [WB]: 1:20,000, clone AC-74, Sigma-Aldrich, #A5316), ATG13 (1:1000, Sigma-Aldrich, #SAB4200100), ATG3 (WB: 1:1000, Cell Signaling Technology, #3415), ERGIC-53/p58 (IF: 1:25, Sigma-Aldrich, #E1031), ERp72 (IF: 1:100, clone D70D12, Cell Signaling Technology, #5033), FIP200 ([1] IF: 1:500, Proteintech, #17250-1-AP, [2] WB: 1:1000, Bethyl Laboratories, #A301-536A or [3] WB: 1:1000, Bethyl Laboratories, #A301-574A, used for THP-1 and HeLa cells), FLAG (WB: 1:1000, clone M2, Sigma-Aldrich, #F1804), GAPDH (WB: 1:5000, clone 6C5, Abcam, #ab8245), GFP (WB: 1:1000, clone 3H9, ChromoTek, #3H9), Golgin97 (IF: 1:100, clone D8P2K, Cell Signaling Technology, #13192), GST (IF: 1:800, clone 26H1, Cell Signaling Technology, #2624), HA (WB: 1:2000, BioLegend, #901501), LC3B (WB: 1:1000, IF: 1:200, Cell Signaling Technology, #2775), NDP52 (IF: 1:50, Proteintech, #12229-1-AP), OPTN (IF: 1:50, Proteintech, #10837-1-AP), SQSTM1/p62 (WB: 1:1000, PROGEN, #GP62-C), SQSTM1/p62 pS403 (WB: 1:1000, IF: 1:400, clone D8D6T, Cell Signaling Technology, #39786), STING (IF: 1:100, Proteintech, #19851-1-AP), TAX1BP1 ([1] IF: 1:50, Proteintech, #14424-1-AP or [2] WB: 1:1000, Bethyl Laboratories, #A303-792A), TBK1 ([1] IF: 1:50, WB: 1:1000, clone A-6, Santa Cruz Biotechnology, #sc-398366 or [2] WB: 1:1000, clone D1B4, Cell Signaling Technology, #3504, used for THP-1 and HeLa cells), TBK1 pS172 (IF: 1:50, WB: 1:1000, clone D52C2, Cell Signaling Technology, #5483), TRIF (IF: 1:100, clone E-7, Santa Cruz Biotechnology, #sc-514384), ULK1 (1:1000, clone D8H5, Cell Signaling Technology, #8054), and Vinculin (WB: 1:2000, clone hVIN-1, Sigma-Aldrich, #V9131) were used. For immunoblot analyses, IRDye 800- or IRDye 680-conjugated secondary antibodies were used (LI-COR Biosciences, #926-68070 [680RD goat anti-mouse IgG], #926-68071 [680RD goat anti-rabbit IgG], #926-32211 [800CW goat anti-rabbit IgG], #926-32219 [800CW goat anti-rat IgG], and #926-68077 [680RD donkey anti-guinea pig IgG]). The secondary antibodies for IF analyses were purchased from Jackson ImmunoResearch (Alexa Fluor 488-AffiniPure goat anti-rabbit IgG, 1:500, #111-545-003 and Alexa Fluor 647-AffiniPure goat anti-mouse IgG, 1:500, #115-605-003). Other reagents used were BafA1 (Sigma-Aldrich, #B1793), LPS from Escherichia coli O55:B5 (Sigma-Aldrich, #L4524), PMA (Sigma-Aldrich, #P1585), polyinosinic:polycytidylic acid (poly I:C; Sigma-Aldrich, #P9582), and puromycin (InvivoGen, #ant-pr). Dimethyl sulfoxide (DMSO; AppliChem, #A3672) was used to dissolve BafA1.
Constructs and siRNAs
Human cDNA encoding FLAG-tagged full-length FIP200 (isoform 1) was amplified from p3xFLAG-CMV10-hFIP200 (kindly provided by Noboru Mizushima and previously described in35; Addgene plasmid #24300; http://n2t.net/addgene:24300; RRID: Addgene_24300) and cloned into the multiple cloning site of the oncoretroviral vector S91I2Pco. This vector expressed FIP200 cDNA and an internal ribosomal entry site (IRES)-puromycin resistance gene (paca) cassette optimized for human codon usage using the GeneArt online tool (Thermo Fisher Scientific) under the control of the SFFV promoter in the 5'LTR. The codons encoding the C terminus of FIP200 (amino acids 1369–1594, CT) were removed by site-directed mutagenesis using Q5 High-Fidelity 2X Master Mix (New England Biolabs, #M0492). The sequences of oligonucleotides used for mutagenesis PCR were TAAAATTCTGCAGTCGAC (FIP200 Δ1369-1594 fwd) and TATCAAATCTTTATCCCGTTC (FIP200 Δ1369-1594 rev). The FIP200-encoding S91I2Pco vectors (both full-length and ΔCT) harbored cDNAs encoding the amino acid exchange D818Y. Murine Ulk1 and Ulk2 cDNAs were kindly provided by Do-Hyung Kim (Addgene plasmid #31960; http://n2t.net/addgene:31960; RRID: Addgene_31960) and Masaaki Muramatsu (Addgene plasmid #22898; http://n2t.net/addgene:22898; RRID: Addgene_22898), respectively. pMSCVblast-MYC-mULK1 was generated by amplifying MYC-Ulk1 using the oligonucleotides 5´ gattaactcgagATGGAGCAAAAGCTCATTTCTGAGG 3´ (forward) and 5´ ctagttgttaacTCAGGCATAGACACCACTCAGC 3´ (reverse), digestion with XhoI and HpaI (Thermo Scientific #FD0694 and #FD1034) and cloning into pMSCVblast digested with the same enzymes. Subsequently, pMSCVblast-3xFLAG-mULK1 was generated by replacing the codons encoding the MYC tag in the pMSCVblast-MYC-mULK1 vector with codons encoding 3xFLAG by site-directed mutagenesis using the oligonucleotides 5´ gattataaagatcatgacatcgattacaaggatgacgatgacaagGGATTCCTGGAGCCGGGCCGCGGCGGCGTCGAG 3´ (forward) and 5´ accgtcatggtctttgtagtccatCGTTAACCTCGAGAGATCTAATTCCGGCGCCTAGAGAAGGAGTGAGGG 3´ (reverse). After amplification, the PCR product was treated with KDL enzyme mix (NEB # M0554S) and transformed into competent cells. pMSCVzeo-3xHA-mULK2 was generated by amplification of the 3xHA-mULK2 fragment with the oligonucleotides 5´gcgccggaattagatctctcgaggttaacgATGGCATACCCTTATGACGTAC 3´ (forward) and 5´ ggaaaagcgcctcccctacccggtagaattTCACACAGTTGCAGTGCTACAG 3´ (reverse) and insertion into the pMSCVzeo plasmid (a gift from David Mu; Addgene plasmid #75088; http://n2t.net/addgene:75088; RRID: Addgene_75088). For this, pMSCVzeo was linearized with EcoRI (Thermo Fisher Scientific #FD0274), and both the 3xHA-mULK2 PCR product and the linearized pMSCVzeo were treated with 10 U of Klenow fragment (Thermo Fisher Scientifc #EP0054), purified and annealed in the presence of ligase buffer (Thermo Fisher Scientific #B69). The reaction was incubated for 30 min at 37 °C and used to transform competent cells. Mouse Fip200 (ON-TARGETplus siRNA SMARTpool, #L-041191-01-0005), mouse Tax1bp1 (ON-TARGETplus siRNA SMARTpool, #L-055360-01-0005) and negative control (ON-TARGETplus nontargeting pool, #D-001810-10-20) siRNAs were obtained from Dharmacon (Horizon Discovery Group). Human FIP200 (#HSS114818 [#1], #HSS114819 [#2] and #HSS190643 [#3]), human GAPDH (#12935-140, RNAi Positive Control) and negative control (#12935-300, Medium GC Duplex) Stealth siRNAs were obtained from Thermo Fisher Scientific. Lipofectamine RNAiMAX (Thermo Fisher Scientific, #13778-150) was used as a transfection reagent to knock down FIP200 and TAX1BP1 in MEFs, and Viromer Blue (Biozym Scientific, #230005) was used to knock down FIP200 and GAPDH in HeLa and THP-1 cells. Seventy-two hours after transfection, cells were harvested for immunoblotting or fixed and stained for IF.
Cell lines
Fip200 KO MEFs were kindly provided by Jun-Lin Guan (Department of Cancer Biology, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA) and have been described previously58. Ulk1/2 double knockout (DKO) MEFs were kindly provided by Tullia Lindsten (Memorial Sloan Kettering Cancer Center, New York City, New York, USA) and have been described previously59. Plat-E cells were kindly provided by Toshio Kitamura (Institute of Medical Science, University of Tokyo, Japan) and have been described previously60. The Plat-E cells were transfected with 1.9 µg of the S91I2Pco- or pMSCV-based retroviral vectors using FuGENE 6 Transfection Reagent (Promega, #E2692) to produce recombinant retroviruses. After 48 h, retroviral supernatant was collected and added together with 3 µg/ml polybrene (Sigma, #9268) to fip200 KO or ulk1/2 DKO MEFs. After 72 h, cells were selected in medium containing puromycin (2.5 µg/ml; InvivoGen #ant-pr-1), blasticidin (35 µg/ml; InvivoGen #ant-bl-05), or zeocin (Invitrogen #46-0509). Atg13 KO MEFs were kindly provided by Noboru Mizushima (Department of Biochemistry and Molecular Biology, Graduate School and Faculty of Medicine, University of Tokyo, Japan), and their reconstitution has been described previously61. Wild-type and atg3 KO MEFs were kindly provided by Masaaki Komatsu (Department of Physiology, Juntendo University Graduate School of Medicine, Tokyo, Japan) and have been described previously62. Wild-type HeLa cells were kindly provided by Richard J. Youle (National Institute of Neurological Disorders and Stroke, Bethesda, Maryland, USA). THP-1 cells were obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (#ACC 16). The procedure for generating Flp-In T-REx 293 cells inducibly expressing a protein of interest has been previously described63. Briefly, full-length human FIP200 cDNA was amplified and cloned into the vector pcDNA5/FRT/TO-GFP. The expression constructs (GFP or GFP-FIP200) were then cotransfected with pOG44 into Flp-In T-REx 293 cells (Invitrogen, R780-07). Stable transfectants were selected with 200 μg/ml hygromycin B (Invitrogen, 10687-010) and 5 μg/ml blasticidin (Invitrogen, A11139-02). MEFs, HeLa cells and Flp-In T-REx 293 cells were cultured in high-D-glucose Dulbecco’s modified Eagle’s medium (DMEM, Thermo Fisher Scientific, #41965-039) supplemented with 10% FCS (GE Healthcare, #A15-101 or Thermo Fisher Scientific, #10270-106), 100 U/ml penicillin and 100 μg/ml streptomycin at 37 °C and 5% CO2. THP-1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific, #61870-010) supplemented with 10% FCS, 100 U/ml penicillin, 100 μg/ml streptomycin and 10 mM HEPES solution (Sigma-Aldrich, #H0887). For amino acid starvation, cells were washed once with PBS and starved in EBSS (Thermo Fisher Scientific, #24010043) for 2 h. For induction of GFP or GFP-FIP200 expression, Flp-In T-REx 293 cells were incubated in complete medium containing 0.1 doxycycline (Clontech Laboratories, #631311) for 16 h.
IF
For IF microscopy, MEFs were seeded on glass coverslips (Marienfeld). For staining of total TBK1, cells were fixed in cold methanol for 15 min at 4 °C. Whenever total TBK1 was not stained, cells were fixed in 4% paraformaldehyde (PFA) for 15 min at 4 °C, quenched with 50 mM NH4Cl for 15 min and permeabilized with 50 µg/ml digitonin (Sigma-Aldrich, #D141) for 5 min. The fixed samples were blocked with 3% BSA (Roth, #8076) for 30 min and incubated with primary antibodies diluted in 3% BSA for 1–2 h. Afterwards, the samples were washed three times with PBS, incubated with secondary antibodies diluted in 3% BSA for 30 min, and washed once with 0.2% Tween-20 (Sigma-Aldrich, #P1379) and three times with PBS. Finally, the cells were embedded in ProLong Glass Antifade Mountant (Thermo Fisher Scientific, #P36980) containing 1 µg/ml DAPI (Roth, #6335.1). Representative images were collected with an Axio Observer 7 fluorescence microscope (Carl Zeiss Microscopy) using a 40x/1,4 Oil DIC M27 Plan-Apochromat objective (Carl Zeiss Microscopy) and an ApoTome 2 (Carl Zeiss Microscopy) using the 488 nm and 647 nm channels. A lateral shift in channel 647 was corrected using ZEN 2.3 (Carl Zeiss Microscopy) as follows: x = −0.82 px (63.5 nm) and y = 0.52 px (40.3 nm). The average displacement values were estimated from five images of multispectral beads (FocalCheck fluorescence microscope test slide #1 A5, Invitrogen, # F36909), and the analysis was based on the channel registration tool of the NanoJ toolbox for ImageJ using a single block to obtain linear displacement values64. The pixel intensities of the areas indicated by dashed red arrows were measured with ZEN 2.3 lite (Carl Zeiss Microscopy) and are depicted in bar graphs.
Immunoblotting and immunopurification
For immunoblotting, cells were harvested following the indicated treatment, pelletized, flash-frozen and lysed in lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10 μM Na2MoO4, 1 mM Na3VO4, 50 mM NaF, 5 mM Na4P2O7, 1% [v/v] Triton X-100 [Carl Roth, #3051.2], and protease inhibitor cocktail [Sigma-Aldrich, #P2714]) for 30 min on ice. The lysates were clarified by centrifugation at 18 000 rcf and 4 °C for 15 min. The protein concentrations were determined by the Bradford method, and the samples were adjusted to equal concentrations. The samples were then prepared via addition of sample buffer (125 mM Tris–HCl [pH 6.8], 17.2% [v/v] glycerol, 4.1% [w/v] SDS [AppliChem GmbH, #A7249], 200 μg/ml bromophenol blue, 2% [v/v] β-mercaptoethanol), heated to 95 °C for 5 min and subjected to SDS-PAGE. Afterwards, the proteins were transferred to PVDF membranes (Merck Millipore, #IPFL00010) and analyzed using the indicated primary antibodies and appropriate secondary antibodies (LI-COR Biosciences). The signals were detected using an Odyssey Infrared Imaging system (LI-COR Biosciences) and quantified with Image Studio 5.25 (LI-COR Biosciences). For affinity purification of GFP-tagged proteins, cells were flash-frozen and lysed in lysis buffer containing 0.3% CHAPS (Roth, #1479.2) as detergent. The lysates were clarified and incubated with GFP-trap beads (ChromoTek, #gta-200) overnight with rotation. The purified proteins were washed three times with lysis buffer and analyzed by immunoblotting.
RT-qPCR
Total RNA was isolated from approximately 1 × 106 cells using a NucleoSpin RNA II Kit (Macherey–Nagel, #740955.250) according to the manufacturer’s instructions. The isolated RNA was either directly used for reverse transcription or stored at − 80 °C. First-strand cDNA was generated using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, #4368814) according to the manufacturer’s instructions with 1 μg of isolated total RNA plus additional RNase inhibitor (Thermo Fisher Scientific, #10777019) and standard cycling conditions (10 min at 25 °C, 120 min at 37 °C, 5 min at 85 °C). The generated cDNA was directly used for qPCR or stored at − 20 °C. qPCR analysis was performed using an Applied Biosystems 7300 Real Time PCR System (Thermo Fisher Scientific) and GoTaq qPCR Master Mix (Promega, #A6001). The sequences of the oligonucleotides used for RT-qPCR were TGACAGGATGCAGAAGGAGA (β-Actin left), CGCTCAGGAGGAGCAATG (β-Actin right), CAGGCAACCTTTAAGCATCAG (IFN-β left), and CCTTTGACCTTTCAAATGCAG (IFN-β right). For each gene, technical triplicates were prepared, each of which contained 25 ng of total cDNA and a primer concentration of 500 nM. Amplification was performed with a standard temperature profile (2 min at 50 °C, 10 min at 95 °C, and 40 cycles of 15 s at 95 °C and 1 min 60 °C) followed by a dissociation run (15 s at 95 °C, 1 min at 60 °C, gradual increase of 1 °C/min up to 95 °C). The cycle threshold (CT) values were computed automatically. Relative gene expression was calculated by the 2-∆∆CT method, and β-Actin was used as a reference gene. Additionally, the values were normalized to those of control samples. The experiments were performed in triplicate.
In-gel digestion and MS
For MS analysis, gel pieces were reduced, alkylated and digested with trypsin as described elsewhere65. MS was performed as previously described66: Peptides were extracted with 0.1% trifluoroacetic acid and subjected to liquid chromatography (LC). For peptide separation over a 130 min LC gradient, an Ultimate 3000 Rapid Separation LC system (Dionex/Thermo Fisher Scientific, Idstein, Germany) equipped with an Acclaim PepMap 100 C18 column (75 μm inner diameter, 25 cm length, 2 µm particle size from Thermo Fisher Scientific, Bremen, Germany) was used. MS analysis was carried out on an Orbitrap Elite mass spectrometer (Thermo Scientific, Bremen, Germany) operating in positive mode and equipped with a nano-electrospray ionization source. The capillary temperature was set to 275 °C, and the source voltage was set to 1.4 kV. Survey scans were carried out in the Orbitrap mass analyzer over a mass range from 350 to 1700 m/z at a resolution of 60 000 (at 400 m/z). The target value for the automatic gain control was 1 000 000, and the maximum fill time was 200 ms. The 20 most intense peptide ions (minimum signal intensity 500, excluding singly charged ions and ions with a charge state ≥ 4) were isolated, transferred to the linear ion trap (linear trap quadrupole, LTQ) part of the instrument and fragmented using collision-induced dissociation. The peptide fragments were analyzed using a maximum fill time of 200 ms and an automatic gain control target value of 100 000 with a mass range set depending on the parent mass using normal scan mode. Already fragmented ions were excluded from fragmentation for 45 s.
Computational MS data analysis
The computational analysis of the MS data was based on previously described protocols67. Peptide and protein identification and quantification were performed using MaxQuant (version 1.5.0.3, MPI for Biochemistry, Planegg, Germany) applying standard parameters. Searches were carried out based on 20183 Homo sapiens protein entries downloaded from UniProtKB on 26th November 2014. Methionine oxidation and acetylation at the protein N termini were set as variable modifications, and carbamidomethylation at cysteines was considered as fixed modification. Peptides and proteins were accepted with a false discovery rate set to 1%. Unique and razor peptides were used for label-free quantification. The minimal ratio count was set to two, and the “match between runs” option was enabled. The normalized intensities provided by MaxQuant were analyzed using the Perseus framework (version 1.5.0.15, MPI for Biochemistry, Planegg, Germany). Only proteins with a minimum of 3 valid values in total were considered for protein quantification. Proteins that were identified only by site or were marked as contaminants (from the MaxQuant contaminant list) were excluded from the analysis. For calculation of enriched proteins in the two groups, Student’s t-test was applied. The significance analysis was applied to log2-transformed values after replacement of missing values from a normal distribution (width 0.3, downshift 1.5) using an S0 constant = 0 and a 5% false discovery rate-based cutoff68. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD022563.
Statistical analysis
For immunoblotting, the density of each protein band was divided by the average density of all bands from the same protein on the membrane. The resulting ratios for the proteins of interest were normalized to the ratios of the corresponding loading controls, and the fold changes were calculated by dividing each normalized density ratio by the average density ratio of the indicated control lane (control lane: fold change = 1.00, n ≥ 3). For IF analyses, puncta and nuclei were quantified and analyzed using ImageJ 1.53c. A punctum-to-nucleus ratio was calculated to determine the number of puncta per cell. The macros used for quantifications are provided in Table S2. At least 50 cells per experiment were analyzed in at least three biological replicates. For comparisons between different groups, two-way ANOVA was performed using GraphPad Prism 7 (GraphPad Software).
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author. The plasmids generated in this study are available from the corresponding author without restriction upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium69 via the PRIDE70 partner repository with the dataset identifier PXD022563.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- ATG:
-
Autophagy-related
- AZI2/NAP1:
-
5-Azacytidine-induced 2
- cGAS:
-
Cyclic GMP-AMP synthase
- FIP200:
-
Focal adhesion kinase (FAK)-interacting protein of 200 kDa
- FTD:
-
Frontotemporal dementia
- HSE:
-
Herpes simplex encephalitis
- IFN:
-
Interferon
- IRF3/7:
-
Interferon regulatory factor 3/7
- (MAP1) LC3:
-
(Microtubule-associated protein 1A/1B) light chain 3
- MAVS:
-
Mitochondrial antiviral signaling protein
- MyD88:
-
Myeloid differentiation primary response 88
- NBR1:
-
Neighbor of BRCA1 gene 1
- NDP52:
-
Nuclear dot protein 52 kDa
- NRBF2:
-
Nuclear receptor-binding factor 2
- NTG:
-
Normal-tension glaucoma
- OPTN:
-
Optineurin
- PIK3C3/VPS34:
-
Phosphatidylinositol 3-kinase catalytic subunit type 3
- PIK3R4/VPS15:
-
Phosphoinositide 3-kinase regulatory subunit 4
- PRR:
-
Pattern recognition receptor
- RIG-I:
-
Retinoic acid-inducible gene I
- RLR:
-
RIG-I-like receptor
- SAR:
-
Selective autophagy receptor
- SQSTM1/p62:
-
Sequestosome 1
- STING:
-
Stimulator of interferon genes
- TAX1BP1:
-
Tax1-binding protein 1
- TBK1:
-
TANK-binding kinase 1
- TBKBP1/SINTBAD:
-
TBK1-binding protein 1
- TLR3/4:
-
Toll-like receptor 3/4
- TRIF:
-
TIR-domain-containing adapter-inducing interferon-β
- ULK1/2:
-
UNC-51-like autophagy-activating kinase 1/2
References
Wesselborg, S. & Stork, B. Autophagy signal transduction by ATG proteins: From hierarchies to networks. Cell. Mol. Life Sci. https://doi.org/10.1007/s00018-015-2034-8 (2015).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell. Dev. Biol. 27, 107–132. https://doi.org/10.1146/annurev-cellbio-092910-154005 (2011).
Rogov, V., Dotsch, V., Johansen, T. & Kirkin, V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167–178. https://doi.org/10.1016/j.molcel.2013.12.014 (2014).
Johansen, T. & Lamark, T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432, 80–103. https://doi.org/10.1016/j.jmb.2019.07.016 (2020).
Kirkin, V. & Rogov, V. V. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell 76, 268–285. https://doi.org/10.1016/j.molcel.2019.09.005 (2019).
Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 63, 1–10. https://doi.org/10.1016/j.ceb.2019.12.001 (2020).
Vargas, J. N. S. et al. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell 74, 347-362.e346. https://doi.org/10.1016/j.molcel.2019.02.010 (2019).
Ravenhill, B. J. et al. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol. Cell 74, 320-329.e326. https://doi.org/10.1016/j.molcel.2019.01.041 (2019).
Walker, S. A. & Ktistakis, N. T. Autophagosome biogenesis machinery. J. Mol. Biol. https://doi.org/10.1016/j.jmb.2019.10.027 (2019).
Zhao, P. et al. TBK1 at the crossroads of inflammation and energy homeostasis in adipose tissue. Cell 172, 731–743. https://doi.org/10.1016/j.cell.2018.01.007 (2018).
Kumar, S. et al. Phosphorylation of syntaxin 17 by TBK1 controls autophagy initiation. Dev. Cell 49, 130-144.e136. https://doi.org/10.1016/j.devcel.2019.01.027 (2019).
Fu, T. et al. Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1. Proc. Natl. Acad. Sci. USA 115, E11651–E11660. https://doi.org/10.1073/pnas.1811421115 (2018).
Matsumoto, G., Shimogori, T., Hattori, N. & Nukina, N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum. Mol. Genet. 24, 4429–4442. https://doi.org/10.1093/hmg/ddv179 (2015).
Pilli, M. et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37, 223–234. https://doi.org/10.1016/j.immuni.2012.04.015 (2012).
Richter, B. et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. https://doi.org/10.1073/pnas.1523926113 (2016).
Thurston, T. L., Ryzhakov, G., Bloor, S., von Muhlinen, N. & Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221. https://doi.org/10.1038/ni.1800 (2009).
Wild, P. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233. https://doi.org/10.1126/science.1205405 (2011).
Matsumoto, G., Wada, K., Okuno, M., Kurosawa, M. & Nukina, N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289. https://doi.org/10.1016/j.molcel.2011.07.039 (2011).
Koyama, S., Ishii, K. J., Coban, C. & Akira, S. Innate immune response to viral infection. Cytokine 43, 336–341. https://doi.org/10.1016/j.cyto.2008.07.009 (2008).
Liu, S. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630. https://doi.org/10.1126/science.aaa2630 (2015).
Helgason, E., Phung, Q. T. & Dueber, E. C. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: The emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 587, 1230–1237. https://doi.org/10.1016/j.febslet.2013.01.059 (2013).
Ma, X. et al. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl. Acad. Sci. 109, 9378–9383. https://doi.org/10.1073/pnas.1121552109 (2012).
Tamura, T., Yanai, H., Savitsky, D. & Taniguchi, T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26, 535–584. https://doi.org/10.1146/annurev.immunol.26.021607.090400 (2008).
Shi, C. S. & Kehrl, J. H. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283, 33175–33182. https://doi.org/10.1074/jbc.M804478200 (2008).
Pattingre, S. et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939. https://doi.org/10.1016/j.cell.2005.07.002 (2005).
Galluzzi, L. & Green, D. R. Autophagy-independent functions of the autophagy machinery. Cell 177, 1682–1699. https://doi.org/10.1016/j.cell.2019.05.026 (2019).
Jounai, N. et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA 104, 14050–14055. https://doi.org/10.1073/pnas.0704014104 (2007).
Yang, Q. et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 13, 1–21. https://doi.org/10.1371/journal.ppat.1006600 (2017).
Parvatiyar, K., Barber, G. N. & Harhaj, E. W. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. J. Biol. Chem. 285, 14999–15009. https://doi.org/10.1074/jbc.M110.109819 (2010).
Ahmad, L., Zhang, S. Y., Casanova, J. L. & Sancho-Shimizu, V. Human TBK1: A gatekeeper of neuroinflammation. Trends Mol. Med. 22, 511–527. https://doi.org/10.1016/j.molmed.2016.04.006 (2016).
Freischmidt, A. et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 18, 631–636. https://doi.org/10.1038/nn.4000 (2015).
Cirulli, E. T. et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–1441. https://doi.org/10.1126/science.aaa3650 (2015).
Turco, E. et al. FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol. Cell 74, 330-346.e311. https://doi.org/10.1016/j.molcel.2019.01.035 (2019).
Goodwin, J. M. et al. Autophagy-independent lysosomal targeting regulated by ULK1/2-FIP200 and ATG9. Cell Rep. 20, 2341–2356. https://doi.org/10.1016/j.celrep.2017.08.034 (2017).
Hara, T. et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510. https://doi.org/10.1083/jcb.200712064 (2008).
Pourcelot, M. et al. The Golgi apparatus acts as a platform for TBK1 activation after viral RNA sensing. BMC Biol. 14, 69. https://doi.org/10.1186/s12915-016-0292-z (2016).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. https://doi.org/10.1080/15548627.2015.1100356 (2016).
Gan, B. & Guan, J. L. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell. Signal. 20, 787–794. https://doi.org/10.1016/j.cellsig.2007.10.021 (2008).
Louis, C., Burns, C. & Wicks, I. TANK-binding kinase 1-dependent responses in health and autoimmunity. Front. Immunol. 9, 434. https://doi.org/10.3389/fimmu.2018.00434 (2018).
Liu, F. et al. Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J. Bone Miner. Res. 28, 2414–2430. https://doi.org/10.1002/jbmr.1971 (2013).
Yao, J. et al. Deletion of autophagy inducer RB1CC1 results in degeneration of the retinal pigment epithelium. Autophagy 11, 939–953. https://doi.org/10.1080/15548627.2015.1041699 (2015).
Wei, H. et al. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. https://doi.org/10.1101/gad.2051011 (2011).
Sarraf, S. A. et al. Loss of TAX1BP1-directed autophagy results in protein aggregate accumulation in the brain. Mol. Cell https://doi.org/10.1016/j.molcel.2020.10.041 (2020).
Sun, D., Wu, R., Zheng, J., Li, P. & Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415. https://doi.org/10.1038/s41422-018-0017-7 (2018).
Zaffagnini, G. et al. P62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37, 1–21. https://doi.org/10.15252/embj.201798308 (2018).
Cho, C. S. et al. Lipotoxicity induces hepatic protein inclusions through TANK binding kinase 1-mediated p62/sequestosome 1 phosphorylation. Hepatology 68, 1331–1346. https://doi.org/10.1002/hep.29742 (2018).
Turco, E., Fracchiolla, D. & Martens, S. Recruitment and activation of the ULK1/Atg1 kinase complex in selective autophagy. J. Mol. Biol. 432, 123–134. https://doi.org/10.1016/j.jmb.2019.07.027 (2020).
Yang, S. et al. Autophagy inhibition dysregulates TBK1 signaling and promotes pancreatic inflammation. Cancer Immunol. Res. 4, 520–530. https://doi.org/10.1158/2326-6066.CIR-15-0235 (2016).
Turco, E., Fischer, I. & Martens, S. FIP200 organizes the autophagy machinery at p62-ubiquitin condensates beyond activation of the ULK1 kinase. bioRxiv, 2020.2007.2007.191189, https://doi.org/10.1101/2020.07.07.191189 (2020).
Shi, X. et al. ULK complex organization in autophagy by a C-shaped FIP200 N-terminal domain dimer. J. Cell Biol. 219, 4. https://doi.org/10.1083/jcb.201911047 (2020).
Okamoto, T. et al. FIP200 suppresses immune checkpoint therapy responses in breast cancers by limiting AZI2/TBK1/IRF signaling independent of its canonical autophagy function. Can. Res. 80, 3580–3592. https://doi.org/10.1158/0008-5472.CAN-20-0519 (2020).
Yeo, S. K., Wang, C. & Guan, J. L. Role of FIP200 in inflammatory processes beyond its canonical autophagy function. Biochem. Soc. Trans. 48, 1599–1607. https://doi.org/10.1042/BST20191156 (2020).
Saul, V. V. et al. ULK1/2 restricts the formation of inducible SINT-speckles, membraneless organelles controlling the threshold of TBK1 activation. iScience 19, 527–544. https://doi.org/10.1016/j.isci.2019.08.001 (2019).
Chen, S. et al. Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev. 30, 856–869. https://doi.org/10.1101/gad.276428.115 (2016).
Minegishi, Y. et al. Enhanced optineurin E50K-TBK1 interaction evokes protein insolubility and initiates familial primary open-angle glaucoma. Hum. Mol. Genet. 22, 3559–3567. https://doi.org/10.1093/hmg/ddt210 (2013).
Ye, J. et al. Effects of ALS-associated TANK binding kinase 1 mutations on protein-protein interactions and kinase activity. Proc. Natl. Acad. Sci. USA 116, 24517–24526. https://doi.org/10.1073/pnas.1915732116 (2019).
Liang, C. C., Wang, C., Peng, X., Gan, B. & Guan, J. L. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 285, 3499–3509. https://doi.org/10.1074/jbc.M109.072389 (2010).
Gan, B. et al. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 175, 121–133. https://doi.org/10.1083/jcb.200604129 (2006).
Cheong, H., Lindsten, T., Wu, J., Lu, C. & Thompson, C. B. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl. Acad. Sci. USA 108, 11121–11126. https://doi.org/10.1073/pnas.1107969108 (2011).
Morita, S., Kojima, T. & Kitamura, T. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066. https://doi.org/10.1038/sj.gt.3301206 (2000).
Wallot-Hieke, N. et al. Systematic analysis of ATG13 domain requirements for autophagy induction. Autophagy 14, 743–763. https://doi.org/10.1080/15548627.2017.1387342 (2018).
Sou, Y. S. et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell 19, 4762–4775. https://doi.org/10.1091/mbc.E08-03-0309 (2008).
Löffler, A. S. et al. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 7, 696–706. https://doi.org/10.4161/auto.7.7.15451 (2011).
Laine, R. F. et al. NanoJ: A high-performance open-source super-resolution microscopy toolbox. J. Phys. D Appl. Phys. 52, 163001. https://doi.org/10.1088/1361-6463/ab0261 (2019).
Poschmann, G. et al. High-fat diet induced isoform changes of the Parkinson’s disease protein DJ-1. J. Proteome Res. 13, 2339–2351. https://doi.org/10.1021/pr401157k (2014).
Nehrenheim, L. et al. Native aortic valve derived extracellular matrix hydrogel for three dimensional culture analyses with improved biomimetic properties. Biomed. Mater. 14, 035014. https://doi.org/10.1088/1748-605X/ab0791 (2019).
Kuhnert, F. et al. Rapid single-step affinity purification of HA-tagged plant mitochondria. Plant Physiol. 182, 692–706. https://doi.org/10.1104/pp.19.00732 (2020).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995).
Deutsch, E. W. et al. The ProteomeXchange consortium in 2020: Enabling “big data” approaches in proteomics. Nucleic Acids Res. 48, D1145–D1152. https://doi.org/10.1093/nar/gkz984 (2020).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450. https://doi.org/10.1093/nar/gky1106 (2019).
Acknowledgements
We thank Jun-Lin Guan for providing the fip200 KO MEFs, Tullia Lindsten for providing the ulk1/2 DKO MEFs, Masaaki Komatsu for providing the atg3 KO MEFs, Noboru Mizushima for providing the atg13 KO MEFs, Toshio Kitamura for providing the Plat-E cells, and Richard J. Youle for providing the WT HeLa cells. We thank Antje Löffler, Maria Deak and Dario Alessi for generating the Flp-In T-REx 293 cells inducibly expressing GFP or GFP-FIP200. We thank Stefanie Scheu and Shafaqat Ali for providing the IFN-β primer sequences.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Deutsche Forschungsgemeinschaft STO 864/4-3, STO 864/5-1, STO 864/6-1 and GRK 2158 (to BS), and the Düsseldorf School of Oncology (to BS; funded by the Comprehensive Cancer Center Düsseldorf/Deutsche Krebshilfe and the Medical Faculty of the Heinrich-Heine-University Düsseldorf).
Author information
Authors and Affiliations
Contributions
D.S. designed the experiments, performed mutagenesis, and generated cell lines (FIP200 and FIP200 ΔCT). D.S. performed immunofluorescence and immunoblot analyses and supported RT-qPCR analyses. N.B. performed RT-qPCR analyses. M.J.M. performed cloning of cDNA constructs and generated cell lines (3xFLAG-ULK1 and 3xHA-ULK2). N.W.H. performed cloning of cDNA constructs and generated cell lines (ATG13). J.D. provided expertise on statistics. C.W. and H.H. supported retroviral infections. O.F., A.S. and K.S. performed mass spectrometric analyses and initial validation experiments. W.W., F.S., Y.S., L.B., AF, and C.P. gave technical support. D.S. and B.S. analyzed and interpreted the data and wrote the manuscript. B.S. supervised the project. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schlütermann, D., Berleth, N., Deitersen, J. et al. FIP200 controls the TBK1 activation threshold at SQSTM1/p62-positive condensates. Sci Rep 11, 13863 (2021). https://doi.org/10.1038/s41598-021-92408-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-92408-4
This article is cited by
-
A metabolite sensor subunit of the Atg1/ULK complex regulates selective autophagy
Nature Cell Biology (2024)
-
Autophagy and autophagy-related pathways in cancer
Nature Reviews Molecular Cell Biology (2023)
-
Non-canonical function of FIP200 is required for neural stem cell maintenance and differentiation by limiting TBK1 activation and p62 aggregate formation
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.