Abstract
Chagas disease remains a major neglected disease in Colombia. We aimed to characterize Trypanosoma cruzi transmission networks in the Sierra Nevada de Santa Marta (SNSM) region, to shed light on disease ecology and help optimize control strategies. Triatomines were collected in rural communities and analyzed for blood feeding sources, parasite diversity and gut microbiota composition through a metagenomic and deep sequencing approach. Triatoma dimidiata predominated, followed by Rhodnius prolixus, Triatoma maculata, Rhodnius pallescens, Panstrongylus geniculatus and Eratyrus cuspidatus. Twenty-two species were identified as blood sources, resulting in an integrated transmission network with extensive connectivity among sylvatic and domestic host species. Only TcI parasites were detected, predominantly from TcIb but TcIa was also reported. The close relatedness of T. cruzi strains further supported the lack of separate transmission cycles according to habitats or triatomine species. Triatomine microbiota varied according to species, developmental stage and T. cruzi infection. Bacterial families correlated with the presence/absence of T. cruzi were identified. In conclusion, we identified a domestic transmission cycle encompassing multiple vector species and tightly connected with sylvatic hosts in the SNSM region, rather than an isolated domestic transmission cycle. Therefore, integrated interventions targeting all vector species and their contact with humans should be considered.
Similar content being viewed by others
Introduction
Chagas disease remains a major neglected disease in many American countries, including Colombia, despite intensive efforts at vector control to reduce transmission to humans. There are an estimated 6–8 million persons in the Americas with Chagas disease, causing a burden of 29,000,000 DALYs and a health care cost of $24.73 billion1,2,3. In Colombia, there are over 400,000 cases of T. cruzi infection, and over 5000 new cases per year due to vectorial transmission4.
In particular, the Sierra Nevada de Santa Marta (SNSM) region in northeastern Colombia is considered as a hyper-endemic region, with a human seroprevalence of 16–57% in some communities5,6,7,8. Such a high prevalence is associated with complex T. cruzi transmission cycles that involve several species of triatomines, including Triatoma dimidiata, Triatoma maculata, Rhodnius prolixus, Rhodnius pallescens, Rhodnius pictipes, Rhodnius neivai, Panstrongylus geniculatus, Panstrongylus rufotuberculatus, and Eratyrus cuspidatus and an extensive diversity of potential mammalian hosts5,6,9,10,11. These vector species show diverse levels of adaptation to the domestic environment, with R. prolixus and T. dimidiata able to colonize houses, and possibly R. pallescens, while T. maculata is mostly found in peridomiciles, often associated with bird nests, and E. cuspidatus is mostly intrusive and more rarely observed6,12,13. Nonetheless, several of these species have been found to frequently blood feed on humans5,14, and can contribute to the risk of human infection15.
Furthermore, additional layers of complexity are added to these T. cruzi transmission cycles when considering parasite diversity, and its potential interactions with triatomine gut microbiota, which may modulate parasite development16,17,18. Indeed, of T. cruzi as a species has been divided into seven discrete typing units (DTUs TcI to TcVI and Tcbat)19,20, which represent highly stable near-clade parasite populations21,22,23. This genetic structure is thought to be maintained by the mostly clonal propagation of T. cruzi21,24, although recent evidences indicate the occurrence of sexual reproduction and recombination associated with a high level of inbreeding, at least within TcI DTU25,26. In Northeastern Colombia, TcI largely predominates, although some non-TcI DTUs have also been reported5. Within TcI, TcIa, TcIb and TcId subgroups27,28 have been identified by PCR genotyping in patients, vectors and some reservoir hosts, and these initial studies suggested some overlap among sylvatic and domestic parasite transmission cycles6,29. A better understanding of these potential overlaps and the contribution of different triatomine species to human transmission is critical to optimize vector control strategies and ensure their efficacy30.
While some of these aspects have been investigated in an isolated manner in previous studies as detailed above, their integration to understand transmission cycles and Chagas disease ecology has been limited. Furthermore, new approaches based on deep sequencing of selected markers allow for an unprecedented depth of analysis to disentangle feeding and parasite transmission cycles from different vectors and the multiple interactions shaping T. cruzi parasite diversity31,32. In this study, we aimed to characterize T. cruzi transmission networks based on the identification of vertebrate feeding host diversity as well as parasite and microbial diversity in triatomines from the SNSM region in northwestern Colombia, to shed light on Chagas disease ecology in the region and help optimize current control strategies.
Results
Triatomine diversity
Our study included the analysis of a convenience sample of 42 bugs collected inside and around rural houses (Table 1) in Guajira and Magdalena departments in the SNSM in northeastern Colombia (Fig. 1). Analysis of ITS-2 and Cyt B sequences confirmed the morphological identification of species for adults and allowed the identification of the nymphs. The most abundant species was T. dimidiata, followed by R. prolixus, T. maculata, R. pallescens, P. geniculatus and E. cuspidatus (Table 1).
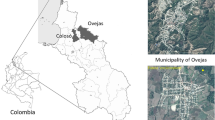
Map of the study area. Triatomine collections were performed in the indicated localities of the SNSM region. Insert: map of Colombia. This map was created in QGIS 3.4 (https://www.qgis.org/en/site/).
Triatomine feeding profiles
Analysis of blood meal sources based on 12 s RNA sequences indicated that bugs fed on a wide diversity of vertebrate host species, with a total of 22 species identified as blood sources, covering mammals, birds, reptiles, and amphibians. However, most blood meals focused on a more limited diversity of vertebrate species, including domestic (human, dog, pig) and sylvatic species (squirrel, opossum, porcupine, or anteater) (Fig. 2A). Notably, while the triatomine species collected in this study shared the same macrohabitat and were found in sympatry, their feeding profiles varied significantly (Fig. 2A,B). R. prolixus and E. cuspidatus presented the highest proportion of blood meals on humans (> 95%), followed by R. pallescens, T. dimidiata, and P. geniculatus. On the other hand, T. maculata was the species with the lowest proportion of blood meals on humans (30%). As noted before in other triatomine species, most bugs had fed on multiple host species, ranging from 2 to 11 species, and the number of feeding host was comparable among triatomine species (Fig. 2C). Overall, female bugs had fed on 3.5 ± 0.5 host species, males on 4.9 ± 0.6 host species and nymphs on 4.0 ± 0.5 host species (ANOVA F = 1.70, P = 0.19), indicating a similar host-switching behavior among these triatomines (Fig. 2C).
Triatomine feeding profiles. (A) Feeding profile of the indicated triatomine species. Vertebrate feeding hosts are color coded as indicated. Numbers in parenthesis on the right indicate sample size. Note the different feeding proportion on humans. (B) NMDS analysis of feeding profiles among triatomine species. PERMANOVA, P = 0.027 (R. pallescens, P. geniculatus and E. cuspidatus were excluded from the statistical analysis due to the limited sample size for these species). (C) Number of concomitant feeding hosts species detected in single bugs according to triatomine species (left panel) and according to sex/developmental stage (right panel). T. dim: T. dimidiata; R. pro: R. prolixus; P. gen: P. geniculatus; R. pal: R. pallescens; T. mac: T. maculata.
The co-occurrence of host species in multiple blood meals from individual bugs provides direct evidence of possible cross-species transmission pathways of T. cruzi parasite when occurring among mammalian species. This information together with the overall feeding profiles of bugs was used to build a parasite transmission network (Fig. 3A). As mentioned above, at least 22 vertebrate species were found to be involved in this network, emphasizing the ecological diversity of blood feeding hosts for triatomines in the region and the complexity of their interactions in T. cruzi transmission. Indeed, most host species in the network were well connected (as evidenced by a network density of 0.424), which provides extensive opportunities for cross-species transmission of T. cruzi. Importantly, there were extensive connections between domestic, synanthropic, and sylvatic mammalian species, suggesting that T. cruzi parasites infecting opossums, porcupines or anteaters could be transmitted to domestic animals and humans (and possibly vice-versa). A few hosts were somewhat isolated in the network, as indicated by a high network centralization statistic (0.633). Furthermore, multiple non-competent hosts including birds, amphibians and reptiles also served as blood sources and help maintain triatomine populations. Finally, an average network heterogeneity of 0.556 indicated the lack of hub nodes or dominant hosts in the network topology.
Triatomine feeding and parasite transmission networks. (A) Global triatomine feeding and T. cruzi transmission network in the SNSM region. This network integrates triatomine species diversity and their relative abundance, as well as the co-occurrence of host species in multiple blood meals from individual bugs, and triatomine blood feeding profiles. The size of vertebrate hosts nodes is proportional to the frequency of blood meals on each host species, and the width of the edges connecting the hosts is proportional to the frequency of their co-occurrence in single bug blood meals. Solid edges link mammalian hosts (green circles), which are competent hosts for T. cruzi, while dotted edges link non-competent hosts (amphibians: light blue triangles, birds: blue octagons, and reptiles: orange diamonds). (B–D) Feeding and parasite transmission networks for R. prolixus, T. dimidiata and T. maculata, respectively. For clarity, only the genus of hosts is indicated. Networks are based on data from 12, 17, and 6 bugs, respectively.
Nonetheless, analysis of triatomine species for which sample size was sufficient revealed that each was involved in very different networks. Indeed, R. prolixus network included the lowest diversity of feeding hosts, with only nine species, but these were much more connected as indicated by a high network density (0.750) and a low network centralization (0.321) (Fig. 3B). Humans were also a clear dominant host and network hub as indicated by a low network heterogeneity (0.248). On the other hand, T. dimidiata was involved in a more diverse network with 18 host species highlighting a more opportunistic feeding behavior (Fig. 3C). This resulted in hosts being somewhat less connected, as indicated by a lower network density (0.353) and a higher network centralization (0.662), and no clear network hubs or host dominance was detected (network heterogeneity of 0.678). Finally, T. maculata network topology was more similar to that of R. prolixus, although a high diversity of up to 11 host species was involved. These were well connected (network density 0.740 and network centralization 0.311) (Fig. 3D).
T. cruzi infection
Trypanosoma cruzi infection was detected in 18/42 bugs (42.8%), corresponding to 1/3 (33.3%) in P. geniculatus and R. pallescens, 0/12 (0%) in R. prolixus, 10/17 (58.8%) in T. dimidiata, and 5/6 (83.3%) in T. maculata. Infection rate tended to be higher in adult bugs compared to nymphs (14/26, 53.8% vs 3/11, 27.2%, respectively, χ2 = 2.27, df = 1, P = 0.13). T. rangeli was also detected in one of the R. pallescens bug.
Trypanosoma cruzi parasites infecting bugs were genotyped using the mini-exon marker, and sequences were obtained through deep sequencing. A total of 287 mini-exon sequences (of 453 bp in length) were obtained, corresponding to 5–38 haplotypes per bug, ranging in frequency from 0.5 to 52.3%. All sequences belonged to TcI DTU. Phylogenetic analysis of these sequences showed that unique clusters of sequences were detected in several bugs (Fig. 4). Such clusters of sequences were likely paralogous sequences derived from a single parasite strain/clone, given the multicopy nature of the mini-exon marker, and illustrated well the Russian-doll pattern previously described22,33,34. A few bugs presented sequences from more than one cluster, suggesting potential infection with more than one strain of T. cruzi. Several bugs also shared nearly identical or closely similar parasite sequence haplotypes, indicating that they were infected with closely similar parasite strains. Furthermore, no genetic clustering of parasite sequences according to vector species was observed, and similar parasite haplotypes could be found in multiple triatomine species. This observation strengthened the extensive overlap of T. cruzi transmission cycles among these vector species, as indicated in Fig. 3.
Phylogenetic analysis of T. cruzi mini-exon diversity. A total of 287 mini-exon sequences are shown that are color-coded for each individual bug analyzed. The corresponding triatomine species are indicated in the outer ring. Reference sequences from TcIa (Raccoon70 strain), TcIb (FcHcl1 strain) and TcId (SylvioX10 strain) are also included in the tree.
To better understand the evolutionary history of TcI strains present in this region, we performed a further phylogenetic analysis including mini-exon sequences from additional strains from a more diverse geographic origin. Nearly all sequences from this study clustered within a large monophyletic group that included TcIb sequences from FcHcl1 reference strain as well as some sequences from Ecuador (Fig. 5). Thus, most parasites circulating among these bugs belonged to TcIb. One notable exception was one T. maculata bug, which harbored sequences that clustered with TcIa sequences from Panama, Venezuela, Mexico, and the USA, forming another well-defined monophyletic group. It is noteworthy that some other T. cruzi strains from Colombia clustered within TcIa and TcId subgroups, highlighting the diversity of TcI strains in this country. TcIa and TcIb parasites would have diverged about 55,000 years ago (± 20,000 years), and were clearly distinct from TcId parasites mostly found in Argentina and Brazil, which may include more than one cluster of sequences as it is a more diverse group28. TcIe included some of the oldest TcI parasite strains, mostly from Bolivia, which may represent the geographic origin of ancestral TcI DTU in the continent. Mantel test confirmed a significant isolation by distance of these TcI strains across the American continent (R = 0.37, P = 0.0001, Fig. 5 insert).
Phylogenetic analysis of TcI strains from northeastern Colombia. The best tree is shown based on Akaike information criteria, and T. cruzi sequences are color-coded according to their country of origin as indicated. The main TcI subgroups corresponding to TcIa, TcIb, TcId and TcIe are highlighted in different colors. Uncertainty in time of divergence is indicated for significant nodes (> 50% support). Insert: Mantel test indicating significant isolation by distance (R = 0.37, P = 0.0001).
Bacterial microbiota
Analysis of the gut microbiota of these triatomines revealed an extensive diversity of bacteria as up to 77 families were identified, with proportions ranging from 0.5% up to over 85% (Fig. 6A). Eratyrus cuspidatus was excluded from further analysis due to its sample size. Comparison of alpha diversity within species indicated that the level of diversity was comparable among these triatomines, as indicated by Shannon and Chao1 diversity indices, although R. pallescens tended to present a somewhat higher diversity (Fig. 6B). However, beta diversity was significantly different among species (PERMANOVA, F = 1.73, P = 0.006), indicating species-specific differences in composition of the microbiota (Fig. 6C).
Composition of triatomine gut microbiota. (A) Proportion of bacterial families according to triatomine species. Taxonomic groups are color-coded as indicated and numbers in parenthesis on the right indicate sample size for each species. (B) Comparison of Chao1 (top panel) and Shannon (bottom panel) diversity indices among species. There were no significant differences among species (F = 0.82, P = 0.52 and F = 1.47, P = 0.23, respectively). (C) NMDS analysis of beta diversity among species. Microbiota composition was significantly different among species (PERMANOVA, F = 1.73, P = 0.006).
Thus, the abundance of some bacterial families varied among triatomine species. For example, Pseudomonadaceae and Alcaligenaceae were more abundant in P. geniculatus and R. pallescens, Mycobacteriaceae in R. prolixus, Paenibacilaceae in R. prolixus and T. maculata, Clostridiaceae in R. pallescens and T. dimidiata, among others (Supplementary Fig. S1). Noteworthy, Wolbachia was not identified in any of the bug analyzed.
We next focused on T. dimidiata microbiota, since we had a sufficient sample size to assess its diversity according to bug developmental stage/sex. Alpha diversity was significantly higher in nymphs compared to male and female adult bugs, as indicated by Chao1 index (F = 5.82, P = 0.015), but this did not reached statistical significance for Shannon index (F = 2.64, P = 0.11) (Fig. 7A). Multiple bacterial families such as Mycobacteriaceae, Salinisphaeraceae, Methylobacteriaceae, or Nocardiospsaceae were only detected in nymphs, and others such as Pseudonocardiaceae, Streptomycetaceae, or Sphingomonadaceae were more abundant in nymphs than in adult bugs. Beta diversity was also significantly different between adults and nymphs (PERMANOVA F = 1.84, P = 0.031), confirming differences in microbiota composition (Fig. 7B). These data indicated that microbiota composition changed during the development of the bugs, and that the emergence of adults was accompanied by a reduction in bacterial diversity in T. dimidiata.
Microbiota diversity in T. dimidiata. (A) Alpha diversity assessed by Chao1 (top panel) and Shannon (Bottom panel) indices. Nymphs presented a significantly higher diversity based on Chao1 index (F = 5.82, P = 0.015), but not according to Shannon index (F = 2.64, P = 0.11). (B) NMDS analysis of beta diversity between adults and nymphs, indicating a significant difference in microbiota composition (PERMANOVA F = 1.84, P = 0.031).
Finally, we evaluated potential associations between T. cruzi parasites and the bacterial microbiota since they share the gut environment and may interact. We assessed pairwise correlations of T. cruzi with bacterial families, and found that T. cruzi infection was positively correlated with the presence of Rhizobiaceae (R = 0.39), and negatively correlated with the presence of Burkhoderiaceae (R = − 0.35), Porphyromonadaceae (R = − 0.39), and Enterobacteriaceae (R = − 0.51) (Supplementary Fig. S2). Further analysis focusing on T. dimidiata indicated that T. cruzi infection was positively correlated with the presence of Kineosporiaceae (R = 0.56), and negatively correlated with the presence of Brevibacteriaceae (R = − 0.52), Dermabacteriaceae (R = − 0.62) and Enterobacteriaceae (R = − 0.57) (Fig. 8). These data suggest potential interactions between T. cruzi and the bacterial community in triatomine guts, which may favor or limit T. cruzi development, and thus vectorial capacity.
Correlation among bacterial families and T. cruzi in T. dimidiata. The network illustrates the diversity of bacterial families in T. dimidiata, indicated as nodes which size is proportional to the abundance of each family (except for T. cruzi). Green nodes indicate bacterial families found in uninfected bugs, and orange nodes indicate those found in T. cruzi infected bugs. Edges link nodes/families that are significantly correlated in the bugs (R > 0.35 and P < 0.05). T. cruzi infection was positively correlated (red edge) with the presence of Kineosporiaceae (R = 0.56), and negatively correlated (blue edges) with the presence of Brevibacteriaceae (R = − 0.52), Dermabacteriaceae (R = − 0.62) and Enterobacteriaceae (R = − 0.57).
Discussion
Due to the complexity of T. cruzi transmission in the SNSM region in northeastern Colombia, we aimed to characterize parasite transmission networks based on the identification of vertebrate feeding host, parasite and microbial diversity in triatomine vectors. The diversity of vector species collected in our study is in general agreement with previous studies15. Thus, T. dimidiata and R. prolixus remain vectors of major concern in this region, but other species also need to be considered. Indeed, analysis of blood-feeding sources confirmed that all species collected fed on humans, as noted before5,14, although the frequency varied according to triatomine species. This highlights that all five species are epidemiologically relevant and need to be considered for vector control in the region.
Nonetheless, each triatomine species presented unique feeding profiles, which resulted in T. dimidiata, T. maculata, and R. prolixus being involved in very different feeding and parasite transmission networks. In addition, the predominance of adult bugs may indicate recent invasions rather than well-established colonies, except for R. prolixus which colonization is well documented. Thus, although these vectors species can be considered sympatric in the region, there are likely differences in their microhabitat leading to these differences in feeding profiles. For example, T. dimidata appeared as the most opportunistic vector, feeding on the broadest range of hosts that include a large diversity of domestic and sylvatic species, as seen before in southern Mexico31. These data suggest a high dispersal capacity of this vector between habitats, as also observed in the Andean region of Colombia35. On the other hand, R. prolixus had a more limited range of hosts, suggesting a more restricted microhabitat, in agreement with its frequent association with palm trees in multiple regions15,36,37,38. The high proportion of blood meals on humans is also in agreement with its role as a major vector species associated with frequent intrusions into human dwellings as well as its ability to colonize them36,39. Finally, T. maculata in the region appeared as a rather peridomestic vector feeding frequently on domestic hosts species, and further studies should help clarify the feeding profiles of P. geniculatus and R. pallescens. These data also illustrate well the sensitivity and usefulness of the metabarcoding and deep sequencing approach used here, which allowed the identification of a rather large diversity of feeding hosts from a limited number of bugs, as highlighted before40,41.
Despite the feeding specificities of each triatomine species, they also shared many host species, which resulted in an integrated transmission network showing extensive connectivity among sylvatic and domestic host species, including humans. This is a strong evidence of the presence of a single parasite transmission cycle encompassing all hosts and vector species in the region, rather than separate sylvatic and domestic cycles. Furthermore, no single mammalian species emerged as a key host/reservoir to maintain T. cruzi circulation, and parasite maintenance would be rather evenly spread over an important diversity of domestic and sylvatic mammals. In this context, manipulating host community structure to reduce parasite transmission to humans as proposed in other settings42 is unlikely to be very effective.
Analysis of T. cruzi sequences confirmed previous studies showing the presence of TcIa and TcIb in Northern Colombia5,6,15 and highlighted the predominance of TcIb in these vectors. TcIb was also found to predominate on nearby Margarita island as assessed by PCR15. The close relatedness of the T. cruzi strains, which clustered in a large TcIb monophyletic group, further support the conclusion that there are no separate transmission cycles according to domestic or sylvatic habitats, nor according to triatomine species. Interestingly, on a continental scale, we detected a significant genetic structuring of TcI parasites, as proposed before22,43, and the subgroups proposed before again formed clear clusters, except possibly TcId27,28,44. Also, the frequency of the respective TcI subgroups seems to vary according to the geographic region, with TcIa predominant in North America, and TcId and TcIe predominant in South America. In Colombia, a mixture of TcIa, TcIb and TcId subgroups is observed, and TcIb may be predominant, at least in this region. Remarkably, TcIe from Bolivia appeared as the most ancestral TcI, and may represent the source of TcI in the Americas with a more recent expansion and diversification in Central and North America. Further studies with additional strains and markers should help clarify how TcI subgroups have dispersed across the continent45.
Triatomines were also found to host species-specific microbiota, in general agreement with preliminary studies in R. pallescens and T. maculata46. Nonetheless, the microbiota of R. pallescens observed here was rather different from that of other populations in Panama, which also presented important differences among sites, raising questions on the stability of microbiota composition within species47. On the other hand, T. dimidiata microbiota included predominantly Bacillales, Enterobacteriales and Clostridales orders in Northern Colombia and Southern Mexico, although some minor differences could be observed31. Similarly, in T. sanguisuga, no differences in microbiota were observed among sites32. Importantly, Wolbachia was not detected in any of the species studied here, in agreement with multiple observations in triatomines31,32,46,48, but contrasting with its previous detection in R. pallescens from Panama47. As reported before for R. prolixus48, we also found evidence of ontogenic changes in microbiota in T. dimidiata, with an important reduction in diversity in adults compared to nymphs. This is in agreement with the key role of some endosymbionts in triatomine development49, but further studies are needed to identify the taxonomic groups involved and the nature of their interactions with the bugs. Finally, we confirmed important interactions of triatomine microbiota with T. cruzi infection, as observed in other triatomine species32,50. Thus, Kineosporiaceae, Brevibacteriaceae, Dermabacteriaceae and Enterobacteriaceae are emerging as taxonomic groups that may interact with T. cruzi in T. dimidiata. Importantly, Enterobacteriaceae have also been found to be negatively associated with T. cruzi infection in Triatoma sanguisuga18, suggesting that comparable interactions may occur across triatomine species. These observations raise the possibility of identifying bacterial species making triatomine bugs resistant to T. cruzi infection for vector control. Such approaches would be similar to Wolbachia and dengue virus in Aedes, or Microsporidia and Plasmodium in Anopheles51,52. A better understanding of these interactions in multiple triatomine species would be key to further evaluate the feasibility of such strategies.
Our study presents nonetheless some limitations, the main one due to the limited sample size and convenience sampling of bugs. Thus, further studies of additional bugs and sites would help strengthen our observations. Also, while the connection of the bugs collected here with sylvatic habitat is clearly evidenced by blood meals on sylvatic host species, further analyses of bugs from sylvatic habitats would be important to refine our understanding of parasite transmission cycles in this habitat.
In conclusion, we identified here an integrated T. cruzi transmission network based on vector feeding profiles, which evidenced extensive connectivity among sylvatic and domestic feeding hosts through multiple triatomine species. Limited parasite diversity lent further support to a single parasite transmission cycle encompassing all hosts and vector species in the region, rather than an isolated domestic transmission cycle. As a consequence, vector control focusing on a single species is unlikely to be effective, and no single reservoir host may be targeted. In this situation, integrated interventions targeting all vector species and their contact with humans should be considered, as discussed previously30. Finally, the limited parasite diversity further suggests important constraints, which may reflect a necessary adaptation of parasites having to be able to survive in a wide diversity of host species and interact with different bacterial communities in several vector species. Expanding such studies is needed to assess how host, vector, and parasite diversity are interconnected.
Methods
Study area
The SNSM is located in the extreme northwest of South America, north of Colombia, between latitude 10°–11° north and longitude 72°–74° west. It is made up of a mountainous group with a pyramidal shape and a triangular base of about 120 km on a side, extending from the Caribbean plain, at sea level, to a height of 5775 m on the Bolívar and Colón peaks. The study sites covered the northern slopes and associated lowlands within SNSM.
Ethical statement
The study was approved by the Institutional Bioethics Committee of the Cooperative University of Colombia (UCC). Permission was obtained from homeowners to search their premises for bugs. All methods were carried out in accordance with relevant guidelines and regulations.
Triatomine collection and DNA extraction
Triatomines were collected between January 2017 and August 2019 by a combination of community participation, manual searches and chicken-baited traps in several localities in the Guajira and Magdalena departments in the SNSM (Fig. 1). The sampling was done in the communities of Gaira (urban, located in the city of Santa Marta), Cacahualito and Calabazo in the rural area of Santa Marta, near the natural national Tayrona park, and Limonal-Carrizal (rural area) in the Guajira department. Two environments were covered: intradomicile and peridomicile. The dwellings included in the study were a convenience sample from a pre-existing list of houses with triatomine infestation from the Santa Marta's health office.
Three triatomine collection methods were used: active search, passive search and community search. The active search of adults and nymphs was carried out inside the dwellings (intra-domicile) in dark places, under furniture, beds, mats, and mattresses, inside the cracks of the walls, spaces between ceiling and ceiling, behind the doors and pictures, and in the peridomicile in the vegetation, corrals of domestic animals, caves, among others. The passive search for triatomines was done with traps that use a chicken live bait53. The traps were placed in peridomestic palm trees within 20 m of the houses, for an approximate exposure period of 12 h between 18:00 and 6:00 h. The inhabitants were instructed on recognizing the triatomine insects, and on the collection method. A plastic container with paper folded inside and covered with tulle was used in order to store the insects.
A total of 42 bugs were analyzed, including adults and nymphs. For adults, species were identified using morphological keys54. Triatomines were kept at 4 °C in individual vials until DNA extraction, handled with gloves, and processed on a bench cleaned with bleach (~ 0.6% sodium hypochlorite) and ethanol 70%. Each insect was dissected in a sterile disposable petri dish using a new scalpel blade each time to prevent cross-contamination.
DNA was extracted from the distal portion of the triatomines’ abdomen using Qiagen DNeasy Blood & Tissue kit following the manufacturer’s instructions, in a dedicated laboratory. DNA samples were stored at − 20 °C until used.
PCR amplification of selected markers
We amplified specific markers for the simultaneous identification of triatomine species, vertebrate blood meals, T. cruzi parasites and gut microbiota as described before31,32. Briefly, we amplified a fragment of triatomine Internal Transcribed Spacer (ITS)-2, using primers ITS2TdF (5′-TGGAAATTTTCTGTTGTCCACA-3′) and ITS2Tria (5′-CAGACAATGCCTAGATGCGA3′) or primers ITS2_200F (5′-TCGYATCTAGGCATTGTCTG-3′) and ITS2_200R (5′-CTCGCAGCTACTAAGGGAATCC-3′) for species identification18,40. Because these primers fail to consistently amplify ITS2 sequences from Rhodnius species, we also used primers CYTB7432F (5′-GGACG(AT)GG(AT)ATTTATTATGGATC-3′) and CYTB7433R (5′-GC(AT)CCAATTCA(AG) GTTA(AG)TAA-3′)55 to amplify a fragment of Cytochrome B for these samples. To identify triatomine blood meal sources, we used two sets of primers targeting the 12S rRNA gene of vertebrates. Similarly, two sets of primers were used to amplify a 140 bp and a 400 bp fragment of the bacterial 16S rRNA gene, respectively, as described previously56,57, to characterize the composition of the microbiota. Finally, samples were tested for the presence of T. cruzi parasites through amplification of satellite DNA with primers TcZ1/TcZ2. Positive samples were genotyped by PCR amplification of the mini-exon sequence using a multiplex PCR as described by Souto et al., which gives PCR products of different sizes according to the DTU58, and newly designed primers that amplify a larger fragment of 500 bp of this marker59. PCR reactions were carried out in a C1000 Touch™ Thermal Cycler (Hercules, CA, USA), and PCR products were separated in 2% agarose gels, stained with ethidium bromide and analyzed with Bio-Rad Gel Doc XR+ with Image Lab™ software (Hercules, CA, USA). PCR reaction preparation, PCR apmplification and gel electrophoresis were performed in separate laboratory rooms, to prevent contaminations. In addition, all PCR reactions included positive and negative controls.
Next generation sequencing
PCR products for blood meal source, bacterial microbiota, T. cruzi genotypes and triatomine ITS2 were pooled for each bug and purified for sequencing on a MiSeq (Illumina) platform (paired sequencing). For this, PCR products were end-repaired and indexed/tagged during library preparation to allow for multiplexing of individual bugs for the sequencing. Each bug/library was used to generate 500,000–1,000,000 reads, corresponding to 1000–100,000 reads per marker. Raw Fastq sequences files were imported in Geneious 11.0.2 software for analysis and mapped to each specific DNA marker using reference sequences. Sequences were filtered for quality and length and trimmed of PCR primer sequences and of any other added sequences from the indexing. Raw sequence data are available in NCBI SRA database under BioProject PRJNA701552 (BioSample accession number SAMN17885581–SAMN17885622).
Sequence and data analysis
Potential sequence chimera were filtered using UCHIME60. Sequence variants were then identified using FreeBayes SNP/variant tool61, to distinguish between sequencing errors/artifacts and significant sequence variants. For triatomine ITS-2 and CytB sequence analysis, haplotypes were aligned with reference sequences of the species for a precise taxonomic identification of the triatomines. Novel ITS-2 and CytB sequences were deposited in GenBank under accession number MW262950–262967 and MW267836–MW267846.
For blood meal sources, 12S sequences were analyzed by MEGABLAST against the entire “nr” GenBank database. A sequence match with > 97% identity was used for species/genus identification. Sequences were further filtered to remove low abundance sequences (< 0.5%) and normalized using the Total Sum Scaling (TSS) method, to account for variability in sequencing depth among samples. Rarefaction curves were elaborated in Microbiome Analyst62, at the individual and group level, to estimate species richness of our sampling of feeding host diversity. Beta diversity of feeding sources among triatomine species was analyzed by Non-Metric Dimensional Scaling (NMDS), based on Bray–Curtis Index distance measures, and statistical significance of differences in feeding hosts among groups was evaluated by permutational MANOVA (PERMANOVA). We also compared the number of host species detected in single triatomines. Data on the proportion of blood-feeding sources and multiple blood meal sources in single bugs were used to construct feeding networks and parasite transmission pathways in Cytoscape 3.831,32. Separate networks were elaborated for R. prolixus, T. dimidiata and T. maculata. Network density, network centralization and network heterogeneity descriptive statistics were calculated as implemented in Cytoscape to summarize network topography and compare networks.
Parasite sequences from the mini-exon gene were first aligned with sequences from reference strains covering all DTUs (TcI: SylvioX10, FcHcl1, Raccoon70; TcII: Tu18, TcIII: M6241; TcIV: CanIII, TcV: SC43; TcVI: CLBrener; TcBat: TCC949cl3), and phylogenetic trees were constructed using the Approximately-Maximum Likelihood method as implemented in FastTree, wich uses the general time-reversible (GTR) model of nucleotide evolution63. All mini-exon sequences were deposited in GenBank database under accession numbers MW256825–MW257111. Mini-exon sequences from multiple TcI strains from across the Americas were then used for further comparison (Supplementary Table S1). TcI sequences were analyzed in Beast 2.6.264 and the best model based on Akaike information criteria was based on a HKY substitution model for a constant coalescent population, and the molecular clock was set based on substitution frequency of 7.1 × 10–8 as previously established for T. cruzi65. The model was run for 10 million iterations to obtain the maximum credibility tree. Isolation by distance was tested using Mantel test as implemented in Past3 software66 and pair-wise geographic distances among strains were based on their country of origin.
For microbiota composition, bacterial 16S sequences were analyzed using a Bayesian classifier from the Ribosomal Database Project67. Taxonomic identification of bacteria was made at a threshold of > 97% sequence identity at the level of bacterial families, Rarefaction curves were elaborated in Microbiome Analyst62. Alpha diversity of microbiota communities (within bugs) was assessed by calculating Chao1 and Shannon indexes, which were compared by Student t-test or ANOVA, depending on the number of groups compared. Beta diversity (among triatomine species or sub-populations) was compared by Non-Metric Dimensional Scaling (NMDS), based on Bray–Curtis Index distance measures, and statistical significance of differences among groups was evaluated by permutational MANOVA (PERMANOVA). We assessed potential differences among triatomine species, and within species among sex/developmental stage. Finally, correlation networks were elaborated to evaluate potential associations among bacterial families of the microbiota and T. cruzi infection, based on Spearman rank correlation. A global network was elaborated, as well as a network for T. dimidiata only. All analyses were performed as implemented in Microbiome Analyst software62.
References
Hotez, P. J. et al. An unfolding tragedy of chagas disease in North America. PLoS Negl. Trop. Dis. 7(10), e2300. https://doi.org/10.1371/journal.pntd.0002300 (2013) (PMID: 24205411).
Hotez, P. J., Bottazzi, M. E., Franco-Paredes, C., Ault, S. K. & Periago, M. R. The neglected tropical diseases of Latin America and the Caribbean: A review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl. Trop. Dis. 2(9), e300. https://doi.org/10.1371/journal.pntd.0000300 (2008) (PMID: 18820747).
Lee, B. Y., Bacon, K. M., Bottazzi, M. E. & Hotez, P. J. Global economic burden of Chagas disease: A computational simulation model. Lancet Infect. Dis. 13(4), 342–348. https://doi.org/10.1016/S1473-3099(13)70002-1 (2013) (PMID: 23395248).
WHO. Chagas disease in Latin America: An epidemiological update based on 2010 estimates. Wkly. Epidemiol. Rec. 90(6), 33–43 (2015) (PMID: 25671846).
Pena-Garcia, V. H., Gomez-Palacio, A. M., Triana-Chavez, O. & Mejia-Jaramillo, A. M. Eco-epidemiology of Chagas disease in an endemic area of Colombia: Risk factor estimation, Trypanosoma cruzi characterization and identification of blood-meal sources in bugs. Am. J. Trop. Med. Hyg. 91(6), 1116–1124. https://doi.org/10.4269/ajtmh.14-0112 (2014) (PMID: 25331808).
Mejia-Jaramillo, A. M. et al. Genotyping of Trypanosoma cruzi in a hyper-endemic area of Colombia reveals an overlap among domestic and sylvatic cycles of Chagas disease. Parasit. Vectors. 7, 108. https://doi.org/10.1186/1756-3305-7-108 (2014) (PMID: 24656115).
Dib, J. C., Agudelo, L. A. & Velez, I. D. Prevalencia de patologías tropicales y factores de riesgo en la comunidad indígena de Bunkwimake, Sierra Nevada de Santa Marta. DUAZARY. 3(1), 38–44 (2006).
Parra-Henao, G. et al. In search of congenital Chagas disease in the Sierra Nevada de Santa Marta, Colombia. Am. J. Trop. Med. Hyg. 101(3), 482–483. https://doi.org/10.4269/ajtmh.19-0110 (2019) (PMID: 31264558).
Guhl, F., Aguilera, G., Pinto, N. & Vergara, D. Actualización de la distribución geográfica y ecoepidemiología de la fauna de triatominos (Reduviidae: Triatominae) en Colombia. Biomedica. 27(Suppl 1), 143–162 (2007) (PMID: 18154255).
Parra-Henao, G., Suarez-Escudero, L. C. & Gonzalez-Caro, S. Potential distribution of Chagas disease vectors (Hemiptera, Reduviidae, Triatominae) in Colombia, based on Ecological Niche Modeling. J. Trop. Med. 2016, 1439090. https://doi.org/10.1155/2016/1439090 (2016) (PMID: 28115946).
Rodriguez-Mongui, E., Cantillo-Barraza, O., Prieto-Alvarado, F. E. & Cucunuba, Z. M. Heterogeneity of Trypanosoma cruzi infection rates in vectors and animal reservoirs in Colombia: A systematic review and meta-analysis. Parasit. Vectors. 12(1), 308. https://doi.org/10.1186/s13071-019-3541-5 (2019) (PMID: 31221188).
Dib, J., Barnabe, C., Tibayrenc, M. & Triana, O. Incrimination of Eratyrus cuspidatus (Stal) in the transmission of Chagas’ disease by molecular epidemiology analysis of Trypanosoma cruzi isolates from a geographically restricted area in the north of Colombia. Acta Trop. 111(3), 237–242. https://doi.org/10.1016/j.actatropica.2009.05.004 (2009) (PMID: 19442641).
Parra Henao, G., Angulo, V., Jaramillo, N. & Restrepo, M. Triatominos (Hemiptera: Reduviidae) de ka Sierra Nevada de Santa Marta, Colombia. Aspectos epidemiológicos, entomológicos y de distribución. Rev. CES Med. 23(1), 17–26 (2009).
Hernandez, C. et al. Untangling the transmission dynamics of primary and secondary vectors of Trypanosoma cruzi in Colombia: Parasite infection, feeding sources and discrete typing units. Parasit. Vectors. 9(1), 620. https://doi.org/10.1186/s13071-016-1907-5 (2016) (PMID: 27903288).
Cantillo-Barraza, O., Chaverra, D., Marcet, P., Arboleda-Sanchez, S. & Triana-Chavez, O. Trypanosoma cruzi transmission in a Colombian Caribbean region suggests that secondary vectors play an important epidemiological role. Parasit. Vectors. 7, 381. https://doi.org/10.1186/1756-3305-7-381 (2014) (PMID: 25141852).
Weiss, B. & Aksoy, S. Microbiome influences on insect host vector competence. Trends Parasitol. 27(11), 514–522. https://doi.org/10.1016/j.pt.2011.05.001 (2011) (PMID: 21697014).
Azambuja, P., Garcia, E. S. & Ratcliffe, N. A. Gut microbiota and parasite transmission by insect vectors. Trends Parasitol. 21(12), 568–572 (2005) (PMID: 16226491).
Dumonteil, E. et al. Interactions among Triatoma sanguisuga blood feeding sources, gut microbiota and Trypanosoma cruzi diversity in southern Louisiana. Mol Ecol. 29(19), 3747–3761 (2020).
Zingales, B. et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: Second revision meeting recommends TcI to TcVI. Mem. Inst. Oswaldo Cruz. 104(7), 1051–1054 (2009) (PMID: 20027478).
Zingales, B. et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 12(2), 240–253. https://doi.org/10.1016/j.meegid.2011.12.009 (2012) (PMID: 22226704).
Tibayrenc, M. & Ayala, F. J. The population genetics of Trypanosoma cruzi revisited in the light of the predominant clonal evolution model. Acta Trop. 151, 156–165. https://doi.org/10.1016/j.actatropica.2015.05.006 (2015) (PMID: 26188332).
Majeau, A., Murphy, L., Herrera, C. & Dumonteil, E. Assessing Trypanosoma cruzi parasite diversity through comparative genomics: Implications for disease epidemiology and diagnostics. Pathogens. 10, 212. https://doi.org/10.3390/pathogens10020212 (2021).
Flores-Ferrer, A., Marcou, O., Waleckx, E., Dumonteil, E. & Gourbière, S. Evolutionary ecology of Chagas disease; what do we know and what do we need?. Evol. Appl. 11(4), 470–487. https://doi.org/10.1111/eva.12582 (2017).
Tibayrenc, M., Kjellberg, F. & Ayala, F. J. A clonal theory of parasitic protozoa: The population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc. Natl. Acad. Sci. USA 87, 2414–2418 (1990).
Berry, A. S. F. et al. Sexual reproduction in a natural Trypanosoma cruzi population. PLoS Negl. Trop. Dis. 13(5), e0007392. https://doi.org/10.1371/journal.pntd.0007392 (2019) (PMID: 31107905).
Schwabl, P. et al. Meiotic sex in Chagas disease parasite Trypanosoma cruzi. Nat. Commun. 10(1), 3972. https://doi.org/10.1038/s41467-019-11771-z (2019) (PMID: 31481692).
Falla, A. et al. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors and humans. Acta Trop. 110(1), 15–21 (2009) (PMID: 19135020).
Cura, C. I. et al. Trypanosoma cruzi I genotypes in different geographical regions and transmission cycles based on a microsatellite motif of the intergenic spacer of spliced-leader genes. Int. J. Parasitol. 40(14), 1599–1607. https://doi.org/10.1016/j.ijpara.2010.06.006 (2010) (PMID: 20670628).
Rodriguez, I. B. et al. Transmission dynamics of Trypanosoma cruzi determined by low-stringency single primer polymerase chain reaction and southern blot analyses in four indigenous communities of the Sierra Nevada de Santa Marta, Colombia. Am. J. Trop. Med. Hyg. 81(3), 396–403 (2009) (PMID: 19706903).
Waleckx, E., Gourbière, S. & Dumonteil, E. Intrusive triatomines and the challenge of adapting vector control practices. Mem. Inst. Oswaldo Cruz. 110(3), 324–338 (2015).
Dumonteil, E. et al. Detailed ecological associations of triatomines revealed by metabarcoding and next-generation sequencing: Implications for triatomine behavior and Trypanosoma cruzi transmission cycles. Sci. Rep. 8(1), 4140. https://doi.org/10.1038/s41598-018-22455-x (2018) (PMID: 29515202).
Dumonteil, E. et al. Interactions among Triatoma sanguisuga blood feeding sources, gut microbiota and Trypanosoma cruzi diversity in southern Louisiana. Mol. Ecol. https://doi.org/10.1111/mec.15582 (2020) (PMID: 32749727).
O’Connor, O., Bosseno, M. F., Barnabe, C., Douzery, E. J. & Breniere, S. F. Genetic clustering of Trypanosoma cruzi I lineage evidenced by intergenic miniexon gene sequencing. Infect. Genet. Evol. 7(5), 587–593. https://doi.org/10.1016/j.meegid.2007.05.003 (2007) (PMID: 17553755).
Villanueva-Lizama, L., Teh-Poot, C., Majeau, A., Herrera, C. & Dumonteil, E. Molecular genotyping of Trypanosoma cruzi by next-generation sequencing of the mini-exon gene reveals infections with multiple parasite DTUs in Chagasic patients from Yucatan, Mexico. J. Inf. Dis. 219(12), 1980–1988 (2019).
Parra-Henao, G., Angulo, V. M., Osorio, L. & Jaramillo, O. N. Geographic distribution and ecology of Triatoma dimidiata (Hemiptera: Reduviidae) in Colombia. J. Med. Entomol. 53(1), 122–129. https://doi.org/10.1093/jme/tjv163 (2016) (PMID: 26487247).
Angulo, V. M., Esteban, L. & Luna, K. P. Attalea butyracea proximas a las viviendas como posible fuente de infestacion domiciliaria por Rhodnius prolixus (Hemiptera: Reduviidae) en los Llanos Orientales de Colombia. Biomedica. 32(2), 277–285. https://doi.org/10.1590/S0120-41572012000300016 (2012) (PMID: 23242302).
Feliciangeli, M. D., Sanchez-Martin, M., Marrero, R., Davies, C. & Dujardin, J. P. Morphometric evidence for a possible role of Rhodnius prolixus from palm trees in house re-infestation in the State of Barinas (Venezuela). Acta Trop. 101(2), 169–177. https://doi.org/10.1016/j.actatropica.2006.12.010 (2007) (PMID: 17306204).
Fitzpatrick, S., Feliciangeli, M. D., Sanchez-Martin, M. J., Monteiro, F. A. & Miles, M. A. Molecular genetics reveal that silvatic Rhodnius prolixus do colonise rural houses. PLoS Negl. Trop. Dis. 2(4), e210. https://doi.org/10.1371/journal.pntd.0000210 (2008) (PMID: 18382605).
Lopez, G. & Moreno, J. Genetic variability and differentiation between populations of Rhodnius prolixus and R. pallescens, vectors of Chagas’ disease in Colombia. Mem. Inst. Oswaldo Cruz. 90, 353–357 (1995).
Dumonteil, E. et al. Detailed ecological associations of triatomines revealed by metabarcoding based on next-generation sequencing: linking triatomine behavioral ecology and Trypanosoma cruzi transmission cycles. Sci. Rep. 8(1), 4140. https://doi.org/10.1038/s41598-018-22455-x (2018).
Hernández-Andrade, A., Moo-Millan, J., Cigarroa-Toledo, N., Ramos-Ligonio, A., Herrera, C., Bucheton, B., et al. Metabarcoding: A powerful yet still underestimated approach for the comprehensive study of vector-borne pathogen transmission cycles and their dynamics. in Vector-Borne Diseases: Recent Developments in Epidemiology and Control (ed. Claborn, D.) 1–6. (Intechopen, 2020). https://doi.org/10.5772/intechopen.83110
Flores-Ferrer, A., Waleckx, E., Rascalou, G., Dumonteil, E. & Gourbière, S. Trypanosoma cruzi transmission dynamics in a synanthropic and domesticated host community. PLoS Negl. Trop. Dis. 13(12), e0007902. https://doi.org/10.1371/journal.pntd.0007902 (2019).
Llewellyn, M. S. et al. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 5(5), e1000410. https://doi.org/10.1371/journal.ppat.1000410 (2009) (PMID: 19412340).
Herrera, C. et al. Genetic variability and phylogenetic relationships within Trypanosoma cruzi I isolated in Colombia based on Miniexon Gene Sequences. J. Parasitol. Res. https://doi.org/10.1155/2009/897364 (2009) (PMID: 20798881).
Zumaya-Estrada, F. A. et al. North American import? Charting the origins of an enigmatic Trypanosoma cruzi domestic genotype. Parasit. Vectors. 5, 226. https://doi.org/10.1186/1756-3305-5-226 (2012) (PMID: 23050833).
Montoya-Porras, L. M., Omar, T. C., Alzate, J. F., Moreno-Herrera, C. X. & Cadavid-Restrepo, G. E. 16S rRNA gene amplicon sequencing reveals dominance of Actinobacteria in Rhodnius pallescens compared to Triatoma maculata midgut microbiota in natural populations of vector insects from Colombia. Acta Trop. 178, 327–332. https://doi.org/10.1016/j.actatropica.2017.11.004 (2018) (PMID: 29154947).
Kieran, T. J. et al. Regional biogeography of microbiota composition in the Chagas disease vector Rhodnius pallescens. Parasit. Vectors. 12(1), 504. https://doi.org/10.1186/s13071-019-3761-8 (2019) (PMID: 31665056).
Rodriguez-Ruano, S. M. et al. Microbiomes of North American Triatominae: The grounds for Chagas Disease epidemiology. Front. Microbiol. 9, 1167. https://doi.org/10.3389/fmicb.2018.01167 (2018) (PMID: 29951039).
Eichler, S. & Schaub, G. A. Development of symbionts in triatomine bugs and the effects of infections with trypanosomatids. Exp. Parasitol. 100(1), 17–27 (2002).
Waltmann, A. et al. Hindgut microbiota in laboratory-reared and wild Triatoma infestans. PLoS Negl. Trop. Dis. 13(5), e0007383. https://doi.org/10.1371/journal.pntd.0007383 (2019) (PMID: 31059501).
Herren, J. K. et al. A microsporidian impairs Plasmodium falciparum transmission in Anopheles arabiensis mosquitoes. Nat. Commun. 11(1), 2187. https://doi.org/10.1038/s41467-020-16121-y (2020) (PMID: 32366903).
Moreira, L. A. et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139(7), 1268–1278. https://doi.org/10.1016/j.cell.2009.11.042 (2009) (PMID: 20064373).
Angulo, V. M. & Esteban, L. Nueva trampa para la captura de triatominos en habitats silvestres y peridomesticos. Biomedica. 31(2), 264–268. https://doi.org/10.1590/S0120-41572011000200015 (2011) (PMID: 22159544).
Lent, H. & Wygodzinsky, P. Revision of Triatominae (Hemiptera: Reduviidae), and their significance as vectors of Chagas’ disease. Bull. Am. Mus. Nat. His. 163, 123–520 (1979).
Monteiro, F. A. et al. Molecular phylogeography of the Amazonian Chagas disease vectors Rhodnius prolixus and R. robustus. Mol. Ecol. 12(4), 997–1006. https://doi.org/10.1046/j.1365-294x.2003.01802.x (2003) (PMID: 12753218).
Baker, G. C., Smith, J. J. & Cowan, D. A. Review and reanalysis of domain-specific 16s primers. J. Microbiol. Meth. 55, 541–555 (2003).
Heuer, H., Krsek, M., Baker, P., Smalla, K. & Wellington, E. M. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63(8), 3233–3241 (1997).
Souto, R. P., Fernandes, O., Macedo, A. M., Campbell, D. A. & Zingales, B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 83(2), 141–152 (1996) (PMID: 9027747).
Majeau, A., Herrera, C. & Dumonteil, E. An improved approach to Trypanosoma cruzi molecular genotyping by next-generation sequencing of the mini-exon gene. Methods Mol. Biol. 1955, 47–60 (2019).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16), 2194–2200. https://doi.org/10.1093/bioinformatics/btr381 (2011) (PMID: 21700674).
Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv preprint. (arXiv:1207.3907 [q-bio.GN]), 1–9. https://arxiv.org/abs/1207.3907v2 (2012).
Dhariwal, A. et al. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45(W1), W180–W188. https://doi.org/10.1093/nar/gkx295 (2017) (PMID: 28449106).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 5(3), e9490. https://doi.org/10.1371/journal.pone.0009490 (2010) (PMID: 20224823).
Bouckaert, R. et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 15(4), e1006650. https://doi.org/10.1371/journal.pcbi.1006650 (2019) (PMID: 30958812).
Torres-Silva, C. F. et al. Assessment of genetic mutation frequency induced by oxidative stress in Trypanosoma cruzi. Genet Mol Biol. 41(2), 466–474. https://doi.org/10.1590/1678-4685-GMB-2017-0281 (2018) (PMID: 30088612).
Hammer, Ø., Harper, D. A. T. & Ryan, P. D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4(1), 9 (2001).
Cole, J. R. et al. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42(Database issue), D633–D642. https://doi.org/10.1093/nar/gkt1244 (2014) (PMID: 24288368).
Acknowledgements
We thank the communities of the Sierra Nevada of Santa Martha for their assistance in the collection of triatomines. This work was funded in part by the Louisiana Board of Regents through the Board of Regents Support Fund [# LESASF (2018-21)-RD-A-19], a Carol Lavin Bernick Faculty Grant and Grant #632083 from Tulane University School of Public Health and Tropical Medicine to ED. JLD was a recipient of a Ph.D. Fellowship from CONACyT, Mexico (No. 457722) and a PROMUV Fellowship from Universidad Veracruzana, México (PROMUV-2019, S16017347). Funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E.D., C.H. and G.P.H. designed research; G.P.H., A.R.G., J.U.C., E.P. and C.M.S. performed field work; C.M.S. and J.L.D. conducted laboratory experiments; C.M.S., R.G. and J.L.D. provided analytic tools; C.M.S., R.G. and E.D. analyzed data; E.D., C.H. and C.M.S. drafted the manuscript; all authors contributed editing and revisions of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Murillo-Solano, C., López-Domínguez, J., Gongora, R. et al. Diversity and interactions among triatomine bugs, their blood feeding sources, gut microbiota and Trypanosoma cruzi in the Sierra Nevada de Santa Marta in Colombia. Sci Rep 11, 12306 (2021). https://doi.org/10.1038/s41598-021-91783-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-91783-2
This article is cited by
-
Humans as blood-feeding sources in sylvatic triatomines of Chile unveiled by next-generation sequencing
Parasites & Vectors (2023)
-
Exposure to Trypanosoma parasites induces changes in the microbiome of the Chagas disease vector Rhodnius prolixus
Microbiome (2022)
-
Towards environmental detection of Chagas disease vectors and pathogen
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.