Abstract
Worldwide the increase in multi-resistant bacteria due to misuse of traditional antibiotics is a growing threat for our health. Finding alternatives to traditional antibiotics is thus timely. Probiotic bacteria have numerous beneficial effects and could offer safer alternatives to traditional antibiotics. Here, we use the nematode Caenorhabditis elegans (C. elegans) to screen a library of different lactobacilli to identify potential probiotic bacteria and characterize their mechanisms of action. We show that pretreatment with the Lactobacillus spp. Lb21 increases lifespan of C. elegans and results in resistance towards pathogenic methicillin-resistant Staphylococcus aureus (MRSA). Using genetic analysis, we find that Lb21-mediated MRSA resistance is dependent on the DBL-1 ligand of the TGF-β signaling pathway in C. elegans. This response is evolutionarily conserved as we find that Lb21 also induces the TGF-β pathway in porcine epithelial cells. We further characterize the host responses in an unbiased proteome analysis and identify 474 proteins regulated in worms fed Lb21 compared to control food. These include fatty acid CoA synthetase ACS-22, aspartic protease ASP-6 and vitellogenin VIT-2 which are important for Lb21-mediated MRSA resistance. Thus, Lb21 exerts its probiotic effect on C. elegans in a multifactorial manner. In summary, our study establishes a mechanistic basis for the antimicrobial potential of lactobacilli.
Similar content being viewed by others
Introduction
Ever since the discovery of antibiotic substances, microorganisms have shown the ability to develop resistance towards antimicrobial compounds. With the extensive use of antibiotics in agriculture and healthcare systems globally, there is a rapidly growing collection of multidrug-resistant pathogenic bacteria1. MRSA is a major cause of nosocomial infections. MRSA is an opportunistic pathogen, often residing on the host without notice. However, infections may originate from preceding asymptomatic colonization or spread to compromised individuals from healthy carriers2. The bacteria colonize the surfaces of the host organism such as skin, nasal cavities and the intestine2,3. MRSA can result in a versatile spectrum of infections, ranging from superficial to life threatening. As MRSA is resistant to many antibiotics, treating infections has become challenging and will probably lead to increased severe infections and MRSA-related deaths in the future4. Therefore, there is an urgent need for alternative treatment strategies.
Probiotic bacteria potentially offer an easy and inexpensive solution as prophylactic treatment to enhance resistance towards MRSA. It is particularly attractive that unlike traditional antibiotics the entire microbiota is not affected by probiotics. The definition of probiotics are living microorganisms that are able to confer a beneficial effect to a host organism when supplemented adequately5. Some probiotic mechanisms of action are known: fighting pathogens by competition of anchoring sites6,7,8, signal interference9 or secreting antimicrobial agents10, maintaining the intestinal barrier integrity11,12, and modulation of host immune system12,13. It is becoming increasingly clear that the effects of probiotics are strain and species specific and highly diverse14. Therefore, to personalize treatment in the future further mechanistic deconvolution of the individual probiotic strains is essential for properly tailored supplementation of strains according to the state of the host organism15. The study of individual bacterial strains is often laborious and cost-inefficient in most model systems. However, the small nematode C. elegans offers a simple yet powerful way of investigating host-microbe interactions and host responses.
C. elegans is a non-parasitic nematode naturally found in rotting fruits and organic matter16. The ease of culturing this bacterivore, combined with its short lifecycle and lifespan, its fully annotated genome and available genetic tools have made C. elegans a valuable model organism for studying various complex biological processes such as ageing17 and innate immunity18,19. Being a bacterivore, C. elegans can be used to study probiotic effects on a host organism20. Lactic acid bacteria have been shown to augment pathogen tolerance in C. elegans21. Several studies have revealed that the innate immune system of C. elegans is often involved in the host response to probiotic bacteria22,23,24,25,26,27,28. The immune system of C. elegans comprises several conserved pathways, one being the evolutionarily conserved TGF-β pathway, which regulates both growth, development and immunity29. DBL-1 is one of the ligands of the TGF-β pathway and it regulates growth30 and innate immunity31,32,33. DBL-1 signaling controls transcription of several anti-bacterial agents, such as lysosomes and C-lectins31,34. Consequently, dbl-1 deficient mutants are generally more susceptible to a range of pathogens29. Recently, DBL-1 signaling was shown to be important for maintaining the natural abundance of members within the Enterobacter family in the natural microbiota of C. elegans. Interestingly, dbl-1 deficiency changes the role of these bacteria from commensal to pathogenic35.
Here we present the identification of a new probiotic Lactobacillus spp. Lb21, which both increases longevity and resistance towards MRSA in the C. elegans model. Testing factors within the host innate immune pathways revealed that the DBL-1 ligand of the TGF-β pathway was essential to obtain the Lb21-mediated tolerance to MRSA. To broaden our view of potentially important factors, we used mass spectrometry-based protein quantification to assess the changes in protein expression between worms fed Lb21 and worms fed control food. We found that ACS-22, a homologue of human Fatty Acid Transporter 1 (FATP1), VIT-2, a vitellogenin lipoprotein, and ASP-6, an aspartyl protease, were important for the nematodes to obtain full Lb21-mediated protection from MRSA. Taken together, our results suggest a multifaceted mechanism behind the increased pathogenic tolerance elicited by Lb21 preconditioning and a potential pretreatment strategy for MRSA infections.
Results
Lb21 extends lifespan in C. elegans
To find novel probiotic bacteria, we obtained a library of 93 different lactobacilli from DuPont Nutrition Biosciences ApS, and used increased lifespan of C. elegans as a screening end-point to limit the number of probiotic candidates (Fig. 1a). To minimize developmental effects the worms were fed the standard food source Escherichia coli OP50 until adulthood and subsequently transferred to the individual lactobacilli. After 14 days the plates were scored for populations with increased survival compared to control populations fed OP50. This initial round of screening identified 15 strains with probiotic potential (Fig. 1a). Interestingly, the remaining lactobacilli had no or even detrimental effects on survival so these were not examined further. We decided to focus on one of the beneficial strains, Lb21. Since comparison between two bacterial strains of closer family is likely to confer more information of the underlying mechanisms than comparing more distant families, we included the Lactobacillus strain Lb23 for comparison, as it did not increase lifespan. To enable comparison between previously published studies, the standard C. elegans feeding strain E. coli OP50 was also included as a control strain.
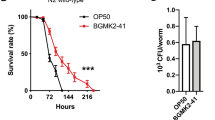
Identifying probiotic lactobacilli strains using C. elegans. (a) Outline of screening setup to detect bacteria that extend nematode lifespan compared to control bacteria, E. coli OP50 and strains isolated in the screen. (b) Lifespan of rrf-3(pk1426) mutants feeding on Lactobacillus spp. Lb21 compared to control bacteria, OP50. (c) Lifespan of rrf-3(pk1426) mutants feeding on Lactobacillus strain Lb23 compared to control bacteria, OP50. (d) CFU levels on day 5 of animals fed OP50 expressing GFP (OP50-GFP) until adulthood (day 3) and then shifted to Lb21, Lb23 or OP50 for 48 h (n = 10). (e) Localization and level of OP50-GFP after 5 days ± UV treatment of plates seeded with OP50-GFP. Scale bar = 100 μm. Graphs show a representative experiment from several independent experiments. See Table S1 for mean ± SD and log-rank test significance.
Longitudinal lifespan analyses of Lb21 revealed that mean survival was extended from 9–38% by Lb21 compared to OP50, whereas Lb23 had a 13% decreased mean lifespan compared to OP50 (Fig. 1b,c, Fig S1, and Table S1). Simply shifting worms from an E. coli diet to a Lactobacillus diet is not per se sufficient to increase lifespan indicating that probiotic effects are highly strain and species specific.
Because Lb21 and Lb23 affected lifespan in opposite directions, we first wanted to establish whether they were both able to colonize C. elegans and thereby establish a microbiota in the adult intestine. Using CFU, we evaluated the level of colonization in the intestine of 5-days old adults after 2 days of pretreatment with Lb21, Lb23 or OP50, respectively. In agreement with previous studies33,36 we found that OP50 colonized the intestine. Importantly, Lb21 and Lb23 were both able to attach to the intestinal tract and establish a gut microbiota in adult C. elegans (Fig. 1d). Hence, the differences in longevity were not a result of one Lactobacillus but not the other being unable to colonize the C. elegans intestine.
Establishment of a C. elegans microbiota happens during middle age (around day 4) due to declining pharyngeal grinder function37. In our pretreatment protocol, we have used OP50 expressing GFP (OP50-GFP) to verify that no OP50 colonization was formed prior to exposure to Lb21 and Lb23. Without UV irradiation of OP50-GFP, the intestinal tract was fully colonized by day 5, whereas no GFP was detected in animals fed UV-treated OP50-GFP (Fig. 1e). Hence, the UV treatment prevented OP50 from colonizing the intestine during development, and the effects of Lb21 and Lb23 are not complicated by intestinal colonization of residual OP50.
To discern the differences between Lb21 and Lb23 we sequenced their genomes (Table S2). This revealed that Lb21 in fact comprised two different Lactobacillus species, one Lactiplantibacillus plantarum strain (94.3%), referred to as Lb21.1, and one Levilactobacillus brevis strain (5.7%), herein referred to as Lb21.2. The Lb23 was confirmed to be a Levilactobacillus brevis. Interestingly, a lock-in lifespan analysis showed that Lb21.1 increased longevity compared to Lb21.2, but the mixture of them were superior compared to the individual strains (Fig S2). Therefore, we chose to focus on the Lb21 spp. (referred to as Lb21) instead of Lb21.1 and Lb21.2 individually. Some fluctuation in the ratios of the two individual species cannot be avoided, which may explain some of the variation seen in the lifespan analyses (Table S1).
Our screening strategy is based on the general relationship between resistance to different stresses and extended lifespan. Most long-lived animals are also resistant to various stressors38,39. Hence, we hypothesized that probiotic bacteria capable of increasing lifespan would likely confer resistance towards pathogenic bacteria such as MRSA. These would be prime candidates for alternatives to traditional antibiotics and thus help reduce the problem of bacterial resistance1.
Lb21 increases MRSA tolerance but not E. coli ETEC resistance in vivo
Consistent with our hypothesis, we found that pretreatment with Lb21 for 48 h prior to MRSA challenge significantly enhanced survival of C. elegans in an MRSA killing assay compared to control worms. In independent experiments, survival was increased by approximately 40% for two different clinical MRSA isolates, MRSA 43484 and MRSA 64, respectively, compared to OP50-pretreated animals (Fig. 2a,b, Table S1). Despite being a Lactobacillus Lb23 provided no or only modestly increased tolerance to MRSA compared to OP50 preconditioning (Fig. 2a,b, and Table S1). Thus, consistent with the longevity result simply shifting the diet from E.coli to a Lactobacillus does not offer probiotic effects. It needs to be the right Lactobacillus strain. In agreement with the observed effect on longevity, the Lb21 spp. provided more resistance to MRSA than the two individual subspecies (Fig S3).
Lb21 but not Lb23 increases MRSA tolerance in C. elegans. (a) Survival of rrf-3(pk1426) mutants pretreated with Lb21, Lb23 and OP50 for 48 h before being shifted to MRSA strain 43484. (b) Survival of rrf-3(pk1426) mutants pretreated with Lb21, Lb23 and OP50 for 48 h before being shifted to MRSA strain 64. (c) Survival of rrf-3(pk1426) mutants after pretreatment with Lb21, Lb23 and OP50 for 48 h before being challenged with the pathogen ETEC F18. Graphs show a representative experiment from several independent experiments. See Table S1 for mean ± SD and log-rank test significance. (d) CFU level on day 6 of Lb21, Lb23 or OP50 and MRSA 43484 in rrf-3(pk1426) mutants pretreated for 48 h with Lb21, LB23 or OP50, respectively (3 technical replicates, n = 10 animals pr. group).
The protective effect of Lb21 on MRSA resistance is remarkable and next we wanted to test the generality of the elicited protection. Many bacteria are pathogenic to C. elegans including enterotoxigenic E. coli (ETEC). To this end, we pretreated animals with Lb21, Lb23 and OP50 and subsequently exposed them to the gram-negative ETEC F1840. Neither Lb21 nor Lb23 were able to protect the nematodes from ETEC F18-induced death (Fig. 2c).
Since the Lb21-mediated protective effect was restricted to MRSA, our next step was to evaluate if Lb21 extended C. elegans survival by displacement of MRSA in the intestinal tract. We measured CFU levels in animals exposed to MRSA 43484 for 24 h. Prior to MRSA exposure worms were pretreated with Lb21, Lb23 or OP50 for 48 h. MRSA was equally able to colonize the intestine of C. elegans both after Lb21 and Lb23 pretreatment, although residual Lb21 and Lb23 were present at comparable levels (Fig. 2d). Since MRSA can displace the initial colonization by both Lb21 and Lb23 the protecting effect of Lb21 is not a result of merely outcompeting MRSA attachment in the intestinal tract.
Observing a small amount of Lb21 left after MRSA recolonization, we assessed that the positive effect on pathogen resistance could result from bacteria-bacteria interaction rather than from a host-response to Lb21. Therefore, we investigated the growth of MRSA in the presence of Lb21 and Lb23 in vitro (Fig S4). Neither Lb21 nor Lb23 affected the growth of MRSA or vice versa, implying that the decrease in mortality mediated by Lb21 is triggered by a host response and not directly via inter-bacterial interactions. This is consistent with our observation that MRSA colonization is not inhibited by Lb21 pretreatment.
Since Lb21 did not directly impair MRSA colonization the protective effect could be mediated by C. elegans responding to a substance secreted by the probiotic bacteria. To test this, we examined the effect of pretreatment with Lb21 bacterial supernatant but it did not confer protection against MRSA (Fig S5 and Table S1). Hence, Lb21 does not secrete an active compound mediating the increased MRSA tolerance. Rather presence of Lb21 cells is needed for at proper host response. Interestingly, UV irradiation of Lb21 did not abolish the protective effect (Fig S5 and Table S1) suggesting that dividing bacteria are not necessary for increased MRSA resistance.
Lb21 mediates its effect via the DBL-1 ligand of the TGF-β pathway
To better understand the Lb21-mediated host response, we turned to C. elegans mutants previously shown to be involved in the responses to probiotic bacteria. We performed MRSA killing assays with mutants of four different anti-aging and immune pathways.
The forkhead box O (FOXO) transcription factor homologue DAF-16 is negatively regulated by the insulin-like pathway (IIS), and is responsible for transcription of genes involved in extended longevity, innate immunity and stress resistance41. We observed that pretreating daf-16(mu86) mutants with Lb21 increased their tolerance to MRSA compared to OP50-preconditioned control animals (Fig. 3a and Table S1). Thus, Lb21-mediated MRSA tolerance is independent of the DAF-2/DAF-16 IIS. Similarly, neither the p38 MAPK immune pathway nor the toll-like receptor of C. elegans, TOL-1, were necessary for the Lb21-mediated response, as Lb21 pretreatment resulted in increased pathogen tolerance in both pmk-1(km25) and tol-1(nr2033) mutants (Fig. 3b,c).
DBL-1 is essential for Lb21-mediated MRSA tolerance. (a) Double mutant daf-16(mu86);rrf-3(pk1426) survival compared to survival of rrf-3(pk1426) pretreated with either OP50 or Lb21 for 48 h before being shifted to MRSA 43484. (b) Double mutant rrf-3(pk1426);pmk-1(km25) survival compared to survival of rrf-3(pk1426) pretreated with either OP50 or Lb21 before being shifted to MRSA 43484. (c) Double mutant tol-1(nr2033);rrf-3(pk1426) survival compared to survival of rrf-3(pk1426) pretreated with either OP50 or Lb21 before being shifted to MRSA 43484. (d) Double mutant rrf-3(pk1426);dbl-1(nk3) survival compared to survival of rrf-3(pk1426) pretreated with either OP50 or Lb21 before being shifted to MRSA 43484. Graphs show a representative experiment from several independent experiments. See Table S1 for mean ± SD and log-rank test significance.
Another key immune pathway of C. elegans is the TGF-β pathway29,31. DBL-1 is one ligand of this pathway and is involved in signaling of both growth and immune responses29. Intriguingly, we did not observe an Lb21-induced extension of survival in dbl-1(nk3) mutants compared to controls (Fig. 3d). Thus, Lb21-mediated MRSA resistance is dependent on DBL-1.
Because probiotic bacteria may exert multifactorial effects, we wondered whether there were other potential modes of actions of Lb21. To identify such mechanisms we decided to examine changes in the proteome following Lb21 pretreatment.
Unbiased proteome-based discovery of regulated proteins
We used an unbiased approach to investigate the proteomes of Lb21-fed, Lb23-fed and OP50-fed animals by using Mass Spectrometry-based protein identification and relative quantification.
Based on the DDA raw data files and using the ProteinPilot Software (Sciex) we identified 2173 unique C. elegans protein groups. These were used to generate a spectral library for protein identifications and relative quantifications across the three sample groups using SWATH data and the Spectronaut (Biognosys) default analysis pipeline. Across all samples, 1980 C. elegans proteins (groups) were quantifiable. A heat map representing the hierarchical clustering of the replicates based on log(2) transformed intensities of all proteins illustrates that the samples cluster according to the three groups (Fig. 4a).
C. elegans proteome response to Lb21, Lb23 and OP50. (a) Heat map showing hierarchical clustering of the C. elegans proteome response to Lb21, Lb23 and OP50, based on log(2) transformed intensities of all quantifiable proteins. (b) Venn diagram showing the number of proteins with regulated abundance when two of the three food sources are compared pairwise. A total of 474 proteins showed regulated abundance in either of the comparisons. (c) A subset of regulated proteins between Lb21 and Lb23. (d) A subset of regulated proteins between Lb21 and OP50. See Table S3 for list of all regulated proteins.
In total, 474 proteins showed significant regulation between either of the groups. 308 of the regulated proteins were altered in abundance in Lb21 fed worms versus OP50 fed worms, while only 63 proteins were altered in Lb21 versus Lb23 fed worms (Fig. 4b). Of these, 27 proteins were regulated in both Lb23 and OP50 versus Lb21 (all regulated proteins are listed in Table S3). All regulated proteins were GO annotated and GO enrichment analysis of the 21 proteins upregulated in response to Lb21 compared to Lb23 revealed mainly a biological process enrichment in organic acid metabolic processes and lipid transport and localization. Enrichment analysis of the 42 proteins that were down regulated in Lb21-fed compared to Lb23-fed nematodes, was merely centered on macromolecular catabolic processes and defense response towards other organisms and stimuli (Table 1). Full GO enrichment analysis of all groups in Lb21 compared to OP50 and P-values are listed in Table S4.
ACS-22, VIT-2 and ASP-6 are involved in Lb21 mediated MRSA resistance
To test the in vivo relevance for MRSA resistance of significantly regulated proteins identified in the proteome analysis, we chose to test the effect of Lb21 pretreatment of mutants for several proteins found to be up or downregulated.
The C-type lectin CLEC-65 and the serpin SRP-7 were found to be down- and upregulated, respectively, in the Lb21/Lb23 proteomic data and the mutants clec-65(ok2337) and srp-7(ok1090) were tested for their ability to confer an Lb21 rescuing effect. However, they did not have an effect on Lb21-mediated MRSA tolerance (Table S1).
The fatty acid CoA synthetase ACS-22 was upregulated in Lb21-fed animals compared to Lb23-fed animals. Therefore, we tested the MRSA resistance of acs-22(tm3236) mutants pretreated with Lb21, Lb23 and OP50. Intriguingly, Lb21-pretreated rrf-3;acs-22 double mutants did not gain a beneficial effect compared to Lb21-pretreated rrf-3 controls, rather a decrease in MRSA tolerance ranging from 16–32% was observed in the absence of ACS-22 (Fig. 5a, and Table S1). MRSA tolerance obtained from Lb21 is therefore dependent on ACS-22.
acs-22, vit-2 and asp-6 are important for the host response to Lb21. (a) Double mutant rrf-3(pk1426); acs-22(tm3236) pretreated with Lb21 before MRSA challenge compared to rrf-3(pk1426) pretreated with Lb21 before MRSA challenge. (b) Double mutant rrf-3(pk1426);vit-2(ok3211) pretreated with Lb21 before MRSA challenge compared to rrf-3(pk1426) pretreated with Lb21 before MRSA challenge. (c) Double mutant rrf-3(pk1426);asp-6(tm2213) pretreated with Lb21 before MRSA challenge compared to rrf-3(pk1426) pretreated with Lb21 before MRSA challenge. See Table S1 for additional data.
The proteome analysis showed that many vitellogenins were downregulated between Lb21/OP50 and upregulated between Lb21/Lb23 with VIT-2 upregulated between Lb21/Lb23 (Fig. 4c,d, Table S3). Therefore, we evaluated how vit-2(ok3211) mutants responded to probiotic pretreatment. We found that Lb21 pretreatment of rrf-3;vit-2 mutants did not result in increased tolerance to MRSA with the mean survival decreased by 35% compared to Lb21 pretreated rrf-3 controls (Fig. 5b, Table S1). Thus, VIT-2 is part of the Lb21-mediated response, as vit-2 mutants did not acquire MRSA tolerance.
The aspartic protease ASP-6 was found to be downregulated between both groups in the proteomic analysis (Lb21/OP50 and Lb21/Lb23, Fig. 4c,d). Therefore, we assessed the level of MRSA-induced deaths in Lb21/Lb23/OP50-pretreated asp-6 (tm2213) mutants. Interestingly, rrf-3;asp-6 mutants did not obtain full Lb21-mediated MRSA resistance compared to Lb21-pretreated rrf-3 worms, with the asp-6 mutants exhibiting a reduction in MRSA tolerance ranging from 12–45% (Fig. 5c, Table S1). This revealed that ASP-6 is required for a part of the Lb21-induced MRSA resistance.
TGF-β production upon Lb21 administration in an epithelial cell line
There is increasing evidence in mammals suggesting that intestinal epithelial cell-derived TGF-β is regulated by the microbiota42. To test if the Lb21-mediated TGF-β response is evolutionarily conserved we examined the response in a cell line to Lb21 administration. As there is a growing problem in the pig production industry of antimicrobial resistance, hereunder MRSA, we chose to use a pig epithelial cell line (IPEC-J2).
Using RT-PCR we tested the effect of the bacteria supernatant from Lb21 and Lb23 on IPEC-J2 cells, under different concentrations. The probiotic bacteria were grown in TSB media and initially we verified that this did not increase the expression of TGF-β mRNA (Fig. 6a). At the concentration of 1 × 107 CFU/mL, supernatant of Lb21 but not Lb23 induced a significant higher amount of TGF-β mRNA (P < 0.05, Fig. 6a). Interestingly, the supernatant of Lb21 upregulated TGF-β mRNA expression in a dose dependent manner whereas the supernatant of Lb23 did not increase TGF-β mRNA expression (Fig. 6b). At the concentration of 2 × 107 CFU/mL, the supernatant of Lb21 induced a significant higher quantity of TGF-β than that of Lb23 (P < 0.01, Fig. 6b).
Lb21 but not Lb23 enhanced TGF-β expression of pig epithelial cells (IPEC-J2). Pig epithelial cells (IPEC-J2) were cultured with bacterial supernatant (Lb21 or Lb23, respectively) for 6 h. Cells were then collected and TGF-β mRNA was measured by RT-PCR. The experiments were performed twice with a total of 6 replicates. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01; n.s. not statistically significant. (a) The TSB media did not increase TGF-β mRNA. However, the Lb21 supernatant at 107 CFU/mL significantly increased TGF-β expression in IPEC-J2 cells. (b) TGF-β induction in IPEC-J2 cells by the Lb21 supernatant is dose-dependent.
Discussion
Probiotics are increasingly attracting attention as dietary health supplements in food and feed but our understanding of their mechanisms of action is incomplete. However, it is clear that among other mechanisms probiotic bacteria can compete with pathogenic bacteria for attachment sites and modulate immune and metabolic responses in the host. It is also evident, that probiotic effects often are strain and species specific, and probiotic bacteria might function as alternatives to traditional antibiotics to combat the increase in multi-resistant bacteria. It is thus relevant to identify novel probiotic bacteria. Here we report identification of a potential probiotic Lactobacillus spp. Lb21 as well insight to the mechanisms of action resulting in increased lifespan and MRSA tolerance in C. elegans.
Combining genetic analysis and unbiased proteomic profiling we find that Lb21-mediated MRSA tolerance is dependent on multiple host factors including DBL-1, the ligand of the TGF-β pathway, ASP-6, VIT-2 and ACS-22. We propose that the Lb21-mediated MRSA resistance is the result of multiple biological processes (Fig. 7).
Lb21 affects C. elegans in a multifaceted manner. The protection against MRSA mediated by Lb21 is dependent on DBL-1 and the effect could likely be mediated by TGF-β controlled immune genes. MRSA tolerance is also dependent on ACS-22 and the underlying mechanism could be both improved intestinal barrier function and altered fat metabolism. VIT-2 and ASP-6 are also required for the protective effect of Lb21 towards MRSA infection. Black lines indicate interactions shown in this study and grey lines indicate possible mechanisms of action based on published studies.
The involvement of the TGF-β ligand DBL-1 in the probiotic host response is interesting. The TGF-β pathway is a part of the innate immune system and signaling regulates the expression of different immune effectors, such as lectins and lysozymes29,31. Our data supports previous studies which have reported that the innate immune system is required for probiotics to have a positive effect43. Though DBL-1 is mainly expressed in neurons30, the receptors and co-Smads of the TGF-β signaling pathway are highly expressed in the pharynx, epidermis and intestine29,44, indicating a role for TGF-β signaling in these tissues. Since the intestinal cells act as the an important line of defense in C. elegans45, it is possible that Lb21 enhances DBL-1 mediated activation of downstream factors in the intestinal cells and thereby primes the C. elegans immune system to resist a subsequent infection. Furthermore, since body size is regulated by DBL-1 TGF-β signaling in the hypodermis46, and the fact that DBL-1 signaling regulates anti-microbial peptide expression in the hypodermis47, Lb21-modulated DBL-1 activity in the hypodermis could potentially be important for the increased MRSA tolerance possibly through regulation of cuticle collagens48. This is also supported by the fact that bacterial colonization of C. elegans is regulated via TGF-β signaling in the hypodermis in addition to the pharynx35. Additionally, AVA interneurons produce DBL-1 shown to be important for the sensing of bacteria in C. elegans and this olfactory learning happens through SMA-6 receptor in the hypodermis49. Taken together, DBL-1 signaling could play a role in the recognition of Lb21, if dbl-1 mutants are unable to sense the bacteria and thus mount a beneficial response.
Involvement of TGF-β signaling in the Lb21 probiotic response was further established when the mRNA expression of TGF-β was only significantly increased in a pig epithelial cell line upon Lb21 supernatant administration and not induced by Lb23. Conservation of probiotic mechanisms between C. elegans and pig has previously been reported. A study comparing C. elegans and a porcine intestinal epithelial cell line as screening platforms for selecting probiotic bacteria concluded that they were largely able to identify the same probiotic bacteria50. Furthermore, similar host responses were induced in both models in agreement with our observations.
In addition to DBL-1, our study identified several other proteins involved in the probiotic response. The C. elegans genome comprises six vitellogenin lipoproteins, used in transportation of lipids from the intestine to oocytes to produce yolk51. Our proteome analysis identified the vitellogenins VIT-2, -3 and -6 as upregulated between Lb21 and Lb23, whereas VIT-1, -5 and -4 were downregulated between Lb21 and OP50. We confirmed that vit-2 was necessary to gain full protective effect of Lb21. VIT-2 is thought to be the ancestor of human apolipoprotein B, which is the major component of low density lipoprotein, which also facilitates transport of lipids52. Interestingly, there is a possible link between DBL-1 and VIT-2 as TGF-β signaling induces vit-2 transcription53. Consistent with our findings, VIT-2 has also previously been associated with pathogen resistance as vit-2 deficiency decreases resistance towards the pathogenic bacteria Photorhabdus luminescens54. It is therefore conceivable that VIT-2 via TGF-β signaling is important in a general immune response. Additionally, morbidity during C. elegans ageing is associated with the conversion of gut biomass conversion to yolk55. The vitellogenins are generally upregulated between Lb21 and Lb23 but downregulated between Lb21 and OP50, which suggests that moderate level of VITs are more beneficial than high levels of vitellogenins (OP50) or very low levels (Lb23). This is supported by the fact that inhibiting vitellogenesis reduces intestinal atrophy leading to increased lifespan55. Furthermore, RNAi inactivation of vit-2 and vit-5 increases lifespan56. The moderate level of VITs in Lb21-fed nematodes is perhaps advantageous by lowering the intestinal biomass conversion to yolk (intestinal atrophy), while sustaining an immune response via TGF-β.
The lysosome-located proteolytic asp-6 is primarily expressed in the intestine57, and it is upregulated during infections with Enterococcus faecalis, Erwinia carotovora, P. luminescens and Serratia marcescens58. We observed that ASP-2, -3, -5 and -6 were downregulated between Lb21/Lb23 and Lb21/OP50 suggesting that Lb21 is not sensed as an infection. Interestingly, asp-6 deficiency resulted in a negative effect on MRSA tolerance produced by Lb21 compared to control animals likely because APS-6 is needed during the normal response to MRSA infection.
Since ACS-22 was highly upregulated between Lb21 and Lb23, we hypothesized that lack of acs-22 would abolish the Lb21-mediated MRSA tolerance. Indeed, acs-22 mutants did not benefit from Lb21 pretreatment. Interestingly, there are several ways that ACS-22 could be required for Lb21-mediated MRSA resistance. ACS-22 is orthologous to the human Fatty Acid Transporter-1/-4 (FATP-1/-4) with predicted long-chain lipid transporter and CoA ligase activity59. acs-22 is primarily expressed in the intestine but also in the pharynx and specific head neurons59. Some studies have indicated that ACS-22 has a role in intestinal barrier maintenance60,61. Interestingly, a connection between probiotics, intestinal barrier function and acs-22 was recently reported61. Nematodes feeding on the lactic acid bacteria Lactobacillus bulgaricus exhibit normal acs-22 expression in the presence of toxic graphene oxide, whereas control animals have reduced levels of acs-22. The authors suggest that the beneficial effect of L. bulgaricus is dependent on maintaining a proper intestinal barrier through regulation of ACS-2261. Thus, it is plausible that Lb21 exerts its probiotic effects by increasing the integrity of the gut epithelial barrier by upregulating ACS-22. Additionally, ACS-22 has a predicted function in lipid metabolism59,61,62 which could also contribute to the probiotic effects of Lb21. The GO enrichment analysis (Table 1) supports this as it showed that processes within lipid transport, metabolism and localization were upregulated in Lb21-fed animals compared to Lb23-fed animals. We also note that several genes within fat metabolism are regulated by DBL-163 and the TGF-β pathway is important for lipid regulation in C. elegans as well as in humans and mammals64,65. This suggests that there could be a conserved link between ACS-22, VIT-2 and DBL-1 in Lb21-mediated MRSA resistance but the details of such interaction requires further investigation.
With probiotic and microbiota modulation of host health becoming increasingly clear, thorough knowledge and understanding of how the microbes affect their host organism are central for exploitation of probiotics in a correct and meaningful way. Several studies have examined the effect of probiotics alone and in combinations. Some report a detrimental effect of the combinations66, whereas others find a synergistic effect of probiotic mixtures67. Our sequencing data revealed that Lb21 comprises two different lactobacilli strains from distinct species: 94.3% L. plantarum (Lb21.1) and 5.7% L. brevis (Lb21.2). Interestingly, the combination of these strains augmented the effect of the individual strains regarding both lifespan extension and MRSA tolerance. We also observed that the genetic background of the host is important for the beneficial effect. This suggest that personalized medicine based approaches in the future could make probiotics even more effective.
Materials and methods
Bacterial strains
The lactobacilli strains were provided by DuPont Nutrition Biosciences ApS (Brabrand, Denmark), collected between 1980 and 2014 from various sources, mainly fermented milk, in different countries. The strains used in this study were Lactobacillus Lb21 and Lb23. Overnight (ON) cultures of lactobacilli were made by inoculating 75 μL aliquot culture in 10 mL De Man, Rogosa and Sharpe (MRS) broth (SIGMA) at aerobic conditions for 18–24 h depending on the strain under continuous shaking at 250 rpm. Aliquots were made from ON culture prepared from the provided DuPont stocks and stored at -80 °C in 25% glycerol. Nematode growth media (NGM) plates (17 g/L agar (SIGMA), 3 g/L NaCl, 2.5 g/L peptone (BD Difco) with 25 mM KPO4 (pH 6.0), 1 mM CaCl2, 1 mM MgSO4, and 5 μg/mL cholesterol (SIGMA) added after autoclaving) were seeded with 2:1 concentrated ON culture. The E. coli OP50 strain was inoculated in 10 mL Luria–Bertani (LB) broth at 37 °C for 18 h under continuous shaking and seeded without concentration. For all bacteria, seeded plates were left to dry for 48 h at room temperature. For supernatant experiments, ON cultures were centrifuged at 3095 rcf for 5 min and the supernatant was pipetted onto dry OP50 plates and left to dry for 48 h at room temperature. Two community-acquired MRSA isolates, MRSA 64 and MRSA 43484 (provided by K. Fuursted, Statens Serum Institut, Copenhagen, Denmark) were spread out from stock aliquot on Tryptic Soy (TS, SIGMA) agar plates and grown ON at 37 °C. Single colonies were inoculated in 10 mL TS broth for 18 h at 37 °C while shaking. The F18 fimbriated enterotoxigenic E. coli (ETEC) strain was inoculated in 10 mL LB broth ON at 37 °C while shaking.
C. elegans strains and maintenance
Animals were cultured at 20 °C and maintained on OP50 seeded NGM plates. C. elegans strains were purchased from the Caenorhabditis Genetic Center (CGC, Minnesota, USA), from the Tokyo Women’s Medical College (Tokyo, Japan) or generated by genetic crossing in our laboratory. C. elegans strains used in this study: NL2099 rrf-3(pk1426), SS104 glp-4(bn2), OLS77 daf-16(mu86);rrf-3(pk1426), OLS78 rrf-3(pk1426);pmk-1(km25), OLS84 tol-1(nr2033);rrf-3(pk1426), OLS86 rrf-3(pk1426);dbl-1(nk3), OLS114 rrf-3(pk1426);vit-2(ok3211), OLS88 rrf-3(pk1426);asp-6(tm2213), OLS89 rrf-3(pk1426);acs-22(tm3236), OLS117 rrf-3(pk1426);srp-7(ok1090), OLS118 glp-4(bn2);clec-65(ok2337). rrf-3 and glp-4 mutants are sterile at 25 °C and were used to preventing bagging and cross-contamination with offspring. glp-4 mutants were used instead of rrf-3 mutants when studying genes on chromosome II and to rule out allele specific effects.
Genotyping of the C. elegans mutants
Prior to experiments, C. elegans deletion mutants were verified by PCR. DNA templates were prepared from 1–10 worms incubated for 20 min at − 80 °C in 5 μL of a 100:1 solution of lysis buffer (10 mM Tris pH 8.3, 50 mL KCl, 0.45% NP-40, 0.45% Tween-20, 0.01% gelatin, 2.5 mM MgCl2) and proteinase K (10 mg/mL). Worms were lysed in an S1000™ Thermal Cycler (BIORAD) at 60 °C for 45 min and 15 min at 95 °C. DreamTaq MasterMix (FERMENTAS) was used for the PCR. Primers used in this study are shown in Table S5.
Longitudinal and lock-in lifespan analysis
To synchronize C. elegans populations, eggs were harvested from gravid hermaphrodites by means of a hypochlorite solution (0.25 mM KOH, 11.2 mM Sodium Hypochlorite (Sigma)). Worms were washed off NGM plates with S-basal solution (10 M NaCl, 0.05 M KPO4 in H2O) and centrifuged at 3095 rcf for 1 min, after which the supernatant was removed and 6 mL hypochlorite solution was added, and tubes were shaken vigorously. Eggs were washed in S-basal solution three times before pipetted onto UV-treated OP50 plates.
Synchronized eggs were allowed to develop at 25 °C on UV-treated OP50 for 3 days until adulthood. Hereafter, animals were shifted to the bacteria of interest. For longitudinal lifespan analysis, death events were scored every other day by touch provoked movement and surviving animals were transferred to fresh plates twice a week. For lock-in lifespan analysis, the total number of surviving and dead animals were monitored only once after 14 days.
All lifespan analyses were performed at 25 °C. GraphPad PRISM was used to plot data and perform log-rank test (Gehan-Breslow-Wilcoxon test), and P-values < 0.05 were set as significant. Lost worms were censored.
Pathogen killing assays
Three-days-old animals were preconditioned with Lb21, Lb23, or OP50 for 48 h before being shifted to MRSA 64, MRSA 43484 or F18 ETEC plates and death events scored every other day. All killing assay were performed at 25 °C. GraphPad PRISM was used to plot data and calculate log-rank test and the Gehan-Breslow-Wilcoxon test, and P-values < 0.05 were set as significant.
Colony forming units (CFU)
Eggs from hypochlorite treatment of rrf-3 animals developed into adults on UV-treated GFP expressing OP50 at 25 °C. Three-days-old animals were transferred to OP50, Lb21 and Lb23 for one hour and subsequently moved to fresh plates and left there for 2 days. A subset of the 5-days-old worms were used for colonization measurement and the rest moved to MRSA for 1 day. To see if colonization on MRSA was inhibited by probiotic pretreatment the bacterial load was also determined for 6-day-old worms on MRSA.
To determine colonization levels, ten worms were pooled in an Eppendorf tube with 500 µL S-basal with 0.1% triton X-100, which help disrupt the cuticle of the worms. Samples were made as technical replicates, 10 worms from each of 3 individual plates, collected at once. Worms were allowed to crawl on unspotted NGM plates for 45 min and then washed three times to minimize bacteria associated with the cuticle. A motor pestle was used to release the intestinal bacteria from the worm pellet. The resulting lysates were diluted in S-basal and plated onto selective media according to resistance and incubated overnight at 37 °C. Colonies were quantified and CFU/worm was calculated.
Proteome analysis
A detailed description can be found in the supporting information. Briefly, 3-days-old rrf-3 worms, developed on UV-treated OP50, were washed in S-basal and transferred to either Lb21, Lb23 or OP50 seeded NGM plates. On day 5, approximately 1000 worms per replicate were washed in S-basal and gently pelleted. Supernatants were aspirated and worm pellets were snap-frozen in liquid nitrogen and stored at − 80 °C for subsequent protein extraction and proteome analysis. Frozen worm pellets were thawed on ice, lysed, homogenized by beat-beating, and finally soluble protein fractions were recovered. The protein concentration of each sample was adjusted to 1 µg/µL, and enzymatic digestion of the proteins was performed by the addition of trypsin (Sigma-Aldrich/Merck). Proteins were identified and quantified using LC–MS/MS. For each sample, 5 µg peptide material was injected into an Eksigent NanoLC 415 system (AB/Sciex, MA, United States) coupled to TripleTOF 6600 mass spectrometer (AB/Sciex) and a DuoSpray Ion Source (AB/Sciex) controlled by Analyst TF 1.71 software (AB/Sciex).
Replicate analysis (n = 4 for Lb21 and Lb23 and n = 3 for OP50) of each of the three sample groups was used to generate a spectral library as well as to make protein identifications and relative quantifications.
Cell line and bacterial supernatant
All cell and bacteria culture media and reagents were purchased from Thermo Fisher Scientific (Roskilde, Denmark), unless otherwise stated. Pig intestinal epithelial cell line (IPEC-J2, ACC 701) was purchased from DSMZ (Braunschweig, Germany). Cells were maintained in DMEM supplemented with 20% FBS, at 37 °C with 5% CO2 atmosphere. Cell cultures were supplemented with antibiotics (Penicillin and Streptomycin, 100 ×). The working concentrations in medium was 1 U/mL penicillin and 1 μg/mL of streptomycin.
The bacterial supernatant used in this study were from Lb21 and Lb23. Both bacteria were grown on Tryptic Soy Broth (TSB) at 37 °C under an anaerobic atmosphere (Anaerocult, Merck, Darmstadt, Germany). The supernatant was then collected for cell culture assay. The concentration of the bacterial solution (CFU/mL) was measured by agar plate in parallel.
Culture conditions and RT-PCR
Pig epithelial cells (IPEC-J2) were cultured with bacterial supernatant (from 0, 0.5… till 2 × 107 CFU/mL) for 6 h and cell pellets were collected for RT-PCR analysis. RNA extraction, quality control and RT-qPCR was performed at Eurofins AROS as described previously68. Data are first normalized to two sets of house-keeping genes by following equation: Value = 2−(Ct sample − Ct house-keeping) × 103. The data represent 2 independent experiments with 6 replicates. A student paired t test was used for statistical analysis, assuming two-tail and unequal variance data distribution. The values with statistical significance are indicated. TaqMan assay ID’s are shown in Table S6.
References
Ventola, C. L. The antibiotic resistance crisis: Causes and threats. P T J. 40, 277–283 (2015).
Sikorska, H. & Smoragiewicz, W. Role of probiotics in the prevention and treatment of meticillin-resistant Staphylococcus aureus infections. Int. J. Antimicrob. Agents 42, 475–481 (2013).
Vesterlund, S., Karp, M., Salminen, S. & Ouwehand, A. C. Staphylococcus aureus adheres to human intestinal mucus but can be displaced by certain lactic acid bacteria. Microbiology 152, 1819–1826 (2006).
Stapleton, P. D. & Taylor, P. W. Europe PMC Funders Group Methicillin resistance in Staphylococcus aureus: Methicillin resistance. Sci. Prog. 85, 1–14 (2007).
Araya, M. et al. Guidelines for the evaluation of probiotics in food. Jt. FAO/WHO Work. Gr. Rep. Draft. Guidel. Eval. Probiotics Food 1–11. https://doi.org/10.1111/j.1469-0691.2012.03873 (2002).
Gueimonde, M., Margolles, A., de los Reyes-Gavilán, C. G. & Salminen, S. Competitive exclusion of enteropathogens from human intestinal mucus by Bifidobacterium strains with acquired resistance to bile—A preliminary study. Int. J. Food Microbiol. 113, 228–232 (2007).
Tsai, C. C. et al. Antagonistic activity against Salmonella infection in vitro and in vivo for two Lactobacillus strains from swine and poultry. Int. J. Food Microbiol. 102, 185–194 (2005).
Prince, T., McBain, A. J. & O’Neill, C. A. Lactobacillus reuteri protects epidermal keratinocytes from Staphylococcus aureus-induced cell death by competitive exclusion. Appl. Environ. Microbiol. 78, 5119–5126 (2012).
Piewngam, P. et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562, 532–537 (2018).
Zipperer, A. et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016).
O’Flaherty, S., Saulnier, D. M., Pot, B. & Versalovic, J. How can probiotics and prebiotics impact mucosal immunity?. Gut Microbes 1, 293–300 (2010).
Rhayat, L. et al. Effect of Bacillus subtilis strains on intestinal barrier function and inflammatory response. Front. Immunol. 10, 1–10 (2019).
Thomas, C. M. & Versalovic, J. Introduction: Probiotic modulation of host signaling pathways. Gut Microbes 13, 148–163 (2010).
Bermudez-Brito, M., Plaza-Díaz, J., Muñoz-Quezada, S., Gómez-Llorente, C. & Gil, A. Probiotic mechanisms of action. Ann. Nutr. Metab. 61, 160–174 (2012).
Suez, J., Zmora, N., Segal, E. & Elinav, E. The pros, cons, and many unknowns of probiotics. Nat. Med. https://doi.org/10.1038/s41591-019-0439-x (2019).
Félix, M.-A. & Braendle, C. The natural history of Caenorhabditis elegans. Curr. Biol. 20, R965–R969 (2010).
Olsen, A. & Gill, M. S. (eds) Ageing: Lessons from C. elegans (Springer International Publishing, 2017).
Cohen, L. B. & Troemel, E. R. Microbial pathogenesis and host defense in the nematode C. elegans. Curr. Opin. Microbiol. https://doi.org/10.1016/j.mib.2014.11.009 (2015).
Ewbank, J. J. & Pujol, N. Local and long-range activation of innate immunity by infection and damage in C. elegans. Curr. Opin. Immunol. 38, 1–7 (2016).
Christensen, K., Mørch, M., Morthorst, T., Lykkemark, S. & Olsen, A. Microbiota, probiotic bacteria and ageing. In Ageing: Lessons from C. elegans (eds Olsen, A. & Gill, M.) 411–429 (Springer, Paris, 2017).
Ikeda, T., Yasui, C., Hoshino, K., Arikawa, K. & Nishikawa, Y. Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against Salmonella enterica serovar enteritidis. Appl. Environ. Microbiol. 73, 6404–6409 (2007).
Park, M. R. et al. Probiotic Lactobacillus fermentum strain JDFM216 stimulates the longevity and immune response of Caenorhabditis elegans through a nuclear hormone receptor. Sci. Rep. 8, 1–10 (2018).
Zhao, L. et al. The transcription factor DAF-16 is essential for increased longevity in C. elegans exposed to Bifidobacterium longum BB68. Sci. Rep. 7, 1–7 (2017).
Kamaladevi, A. & Balamurugan, K. Lactobacillus casei triggers a TLR mediated RACK-1 dependent p38 MAPK pathway in Caenorhabditis elegans to resist Klebsiella pneumoniae infection. Food Funct. 7, 3211–3223 (2016).
Rangan, et al. A bacterial secreted peptidoglycan hydrolase enhances host resistance to intestinal pathogens. Science 353, 1434–1437 (2016).
Nakagawa, H. et al. Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in Caenorhabditis elegans. Aging Cell 15, 227–236 (2016).
Komura, T., Ikeda, T., Yasui, C., Saeki, S. & Nishikawa, Y. Mechanism underlying prolongevity induced by bifidobacteria in Caenorhabditis elegans. Biogerontology 14, 73–87 (2013).
Kim, Y. & Mylonakis, E. Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain ncfm enhances gram-positive immune responses. Infect. Immun. 80, 2500–2508 (2012).
Gumienny, T. L. & Savage-Dunn, C. TGF-β signaling in C. elegans. Wormb. ed. C. elegans Res. Community, Wormb. July, 1–34 (2013).
Suzuki, Y. et al. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126, 241–250 (1999).
Mallo, G. V. et al. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12, 1209–1214 (2002).
Tenor, J. L. & Aballay, A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 9, 103–109 (2008).
Portal-Celhay, C., Bradley, E. R. & Blaser, M. J. Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12, 49 (2012).
Mochii, M., Yoshida, S., Morita, K., Kohara, Y. & Ueno, N. Identification of transforming growth factor-β-regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc. Natl. Acad. Sci. U.S.A. 96, 15020–15025 (1999).
Berg, M. et al. TGFβ/BMP immune signaling affects abundance and function of C. elegans gut commensals. Nat. Commun. 10, 604 (2019).
Portal-Celhay, C. & Blaser, M. J. Competition and resilience between founder and introduced bacteria in the Caenorhabditis elegans gut. Infect. Immun. 80, 1288–1299 (2012).
Cabreiro, F. & Gems, D. Worms need microbes too: Microbiota, health and aging in Caenorhabditis elegans. EMBO Mol. Med. 5, 1300–1310 (2013).
Brejning, J. et al. Loss of NDG-4 extends lifespan and stress resistance in Caenorhabditis elegans. Aging Cell 13, 156–164 (2014).
Lithgow, G. J., White, T. M., Hinerfeld, D. A. & Johnson, T. E. Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J. Gerontol. https://doi.org/10.1093/geronj/49.6.B270 (1994).
Hesselager, M. O., Everest-Dass, A. V., Thaysen-Andersen, M., Bendixen, E. & Packer, N. H. FUT1 genetic variants impact protein glycosylation of porcine intestinal mucosa. Glycobiology https://doi.org/10.1093/glycob/cww009 (2016).
Tullet, J. M. A. DAF-16 target identification in C. elegans: Past, present and future. Biogerontology 16, 221–234 (2015).
Bauché, D. & Marie, J. C. Transforming growth factor β: A master regulator of the gut microbiota and immune cell interactions. Clin. Transl. Immunol. 6, e136 (2017).
Kwon, G., Lee, J., Koh, J. H. & Lim, Y. H. Lifespan extension of Caenorhabditis elegans by Butyricicoccus pullicaecorum and Megasphaera elsdenii with probiotic potential. Curr. Microbiol. 75, 557–564 (2017).
Roberts, A. F., Gumienny, T. L., Gleason, R. J., Wang, H. & Padgett, R. W. Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans. BMC Dev. Biol. 10, 1–10 (2010).
Pukkila-Worley, R. & Ausubel, F. M. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Curr. Opin. Immunol. 24(1), 3–9 (2012).
Schultz, R. D., Bennett, E. E., Ellis, E. A. & Gumienny, T. L. Regulation of extracellular matrix organization by BMP signaling in Caenorhabditis elegans. PLoS ONE 9, e101929 (2014).
Zugasti, O. & Ewbank, J. J. Neuroimmune regulation of antimicrobial peptide expression by a noncanonical TGF-β signaling pathway in Caenorhabditis elegans epidermis. Nat. Immunol. 10, 249–256 (2009).
Madaan, U. et al. BMP signaling determines body size via transcriptional regulation of collagen genes in Caenorhabditis elegans. Genetics 210, 1355–1367 (2018).
Zhang, X. & Zhang, Y. DBL-1, a TGF-, is essential for Caenorhabditis elegans aversive olfactory learning. Proc. Natl. Acad. Sci. 109, 17081–17086 (2012).
Zhou, M. et al. Investigation into in vitro and in vivo models using intestinal epithelial IPEC-J2 cells and Caenorhabditis elegans for selecting probiotic candidates to control porcine enterotoxigenic Escherichia coli. J. Appl. Microbiol. 117, 217–226 (2014).
Perez, M. F. & Lehner, B. Vitellogenins—Yolk gene function and regulation in Caenorhabditis elegans. Front. Physiol. 10, 1067 (2019).
Baker, M. E. Is vitellogenin an ancestor of apolipoprotein B-100 of human low-density lipoprotein and human lipoprotein lipase?. Biochem. J. https://doi.org/10.1042/bj2551057 (1988).
Goszczynski, B., Captan, V. V., Danielson, A. M., Lancaster, B. R. & McGhee, J. D. A 44 bp intestine-specific hermaphrodite-specific enhancer from the C. elegans vit-2 vitellogenin gene is directly regulated by ELT-2, MAB-3, FKH-9 and DAF-16 and indirectly regulated by the germline, by daf-2/insulin signaling and by the TGF-β/Sma/Mab pa. Dev. Biol. 413, 112–127 (2016).
Fischer, M., Regitz, C., Kull, R., Boll, M. & Wenzel, U. Vitellogenins increase stress resistance of Caenorhabditis elegans after Photorhabdus luminescens infection depending on the steroid-signaling pathway. Microbes Infect. 15, 569–578 (2013).
Ezcurra, M. et al. C. elegans eats its own intestine to make yolk leading to multiple senescent pathologies. Curr. Biol. 28, 2544-2556.e5 (2018).
Murphy, C. T. et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–284 (2003).
Lochnit, G., Grabitzki, J., Henkel, B., Tavernarakis, N. & Geyer, R. First identification of a phosphorylcholine-substituted protein from Caenorhabditis elegans: Isolation and characterization of the aspartyl protease ASP-6. Biol. Chem. 387, 1487–1493 (2006).
Wong, D., Bazopoulou, D., Pujol, N., Tavernarakis, N. & Ewbank, J. J. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 8, R194 (2007).
Kage-Nakadai, E. et al. Two very long chain fatty acid acyl-CoA synthetase genes, acs-20 and acs-22, have roles in the cuticle surface barrier in Caenorhabditis elegans. PLoS ONE 5, e8857 (2010).
Qu, M., Xu, K., Li, Y., Wong, G. & Wang, D. Using acs-22 mutant Caenorhabditis elegans to detect the toxicity of nanopolystyrene particles. Sci. Total Environ. 643, 119–126 (2018).
Zhao, Y. et al. Lactic acid bacteria protects caenorhabditis elegans from toxicity of graphene oxide by maintaining normal intestinal permeability under different genetic backgrounds. Sci. Rep. 5, 1–13 (2015).
Rourke, E. J. O., Soukas, A. A. & Carr, C. E. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell 10, 430–435 (2010).
Liang, J., Yu, L., Yin, J. & Savage-Dunn, C. Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Dev. Biol. 305, 714–725 (2007).
Clark, J. F., Meade, M., Ranepura, G., Hall, D. H. & Savage-Dunn, C. Caenorhabditis elegans DBL-1/BMP regulates lipid accumulation via interaction with insulin signaling. G3 Genes Genomes Genet. 8, 343–351 (2018).
Böttcher, Y. et al. Adipose tissue expression and genetic variants of the bone morphogenetic protein receptor 1A gene (BMPR1A) are associated with human obesity. Diabetes https://doi.org/10.2337/db08-1458 (2009).
Collado, M. C., Grześkowiak, Ł & Salminen, S. Probiotic strains and their combination inhibit in vitro adhesion of pathogens to pig intestinal mucosa. Curr. Microbiol. 55, 260–265 (2007).
Sniffen, J. C., McFarland, L. V., Evans, C. T. & Goldstein, E. J. C. Choosing an appropriate probiotic product for your patient: An evidence-based practical guide. PLoS ONE 13, 1–22 (2018).
Shen, C., Christensen, L. G., Bak, S. Y., Christensen, N. & Kragh, K. Immunomodulatory effects of thymol and cinnamaldehyde in chicken cell lines. J. Appl. Anim. Nutr. 8, 1–10 (2020).
Acknowledgements
The authors would like to thank Dr. Arthur Ouwehand (Dupont Nutrition and Health, Finland) for providing the lactobacilli strains and fruitful discussions, Prof. Karl Pedersen (National Veterinary Institute, Sweden) for providing the F18 strain, and Dr. Morten Krog Larsen (Aarhus University, Denmark) for constructive feedback. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs.
Funding
Funding was provided by Innovation Foundation Denmark grant number 4105-00019B.
Author information
Authors and Affiliations
Contributions
M.G.M., K.V.M., M.O.H., R.H.H., C.L.K., T.B., C.S., L.G.C. performed the experiments and analyzed the results. M.G.M., K.V.M., M.O.H. and A.O. wrote the manuscript. A.O., C.H.P., E.B., C.S., K.F. conceived the research idea. All authors reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mørch, M.G.M., Møller, K.V., Hesselager, M.O. et al. The TGF-β ligand DBL-1 is a key player in a multifaceted probiotic protection against MRSA in C. elegans. Sci Rep 11, 10717 (2021). https://doi.org/10.1038/s41598-021-89831-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-89831-y
This article is cited by
-
Bacterial vitamin B6 is required for post-embryonic development in C. elegans
Communications Biology (2024)
-
Geroprotective potential of microbiome modulators in the Caenorhabditis elegans model
GeroScience (2023)
-
Immunological pathogenesis of Bovine E. coli infection in a model of C. elegans
BMC Microbiology (2022)
-
Probiotic-mediated p38 MAPK immune signaling prolongs the survival of Caenorhabditis elegans exposed to pathogenic bacteria
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.