Abstract
A three-descriptor quantitative structure–activity/toxicity relationship (QSAR/QSTR) model was developed for the skin permeability of a sufficiently large data set consisting of 274 compounds, by applying support vector machine (SVM) together with genetic algorithm. The optimal SVM model possesses the coefficient of determination R2 of 0.946 and root mean square (rms) error of 0.253 for the training set of 139 compounds; and a R2 of 0.872 and rms of 0.302 for the test set of 135 compounds. Compared with other models reported in the literature, our SVM model shows better statistical performance in a model that deals with more samples in the test set. Therefore, applying a SVM algorithm to develop a nonlinear QSAR model for skin permeability was achieved.
Similar content being viewed by others
Introduction
Modeling the penetration of manmade and naturally derived chemicals through human skin is of great importance for pharmaceutical and cosmetic industries, as well as toxicology and risk assessment of environmental and occupational hazards. It is very time-consuming and expensive to estimate the skin permeability of chemicals. Further, there are many ethical challenges associated with human and animal testing for assessment of skin permeability1,2.
Quantitative structure–activity/toxicity relationship (QSAR/QSTR) models3,4,5,6 can be used for the prediction of physicochemical property of compounds, even for those that have not been synthesized. Some researchers have carried out QSAR studies for skin permeability of chemicals (the logarithm of the skin permeability coefficients, log Kp).
Patel et al. developed QSAR models for the skin permeability of 158 chemicals with multiple linear regression (MLR) analysis7. The model based on four descriptors has an excellent fit to the data with a coefficient of determination of R2 of = 0.90. Fujiwara et al. proposed MLR QSARs for the skin permeability of 94 structurally diverse compounds8. The models obtained from ten data sets of the skin permeability possess high R2 values with an average R2 of 0.815. Magnusson et al. introduced a regression model (R2 = 0.760) for the skin permeability of 269 compounds9. They found that molecular weight was the main determinant of log KP and QSAR model can be improved when other descriptors such as melting point and hydrogen bonding acceptor capability were added. Chauhan and Shakya built a QSAR model for the skin permeability from the training set of 150 compounds through partial least-squares regression10. The model with a R2 of 0.936 for the training set was validated by the test set of 53 compounds. The root mean square (rms) error and R2 from the test set were equal to 0.670 and 0.542. Xu et al. proposed an expanded version of a linear free-energy relationship model for the skin permeability of complex chemical mixtures11. The model (R2 = 0.70) showed a better fit and predictive power compared with the simple model (R2 = 0.21). Chen et al. generated a MLR model for the skin permeability with four molecular descriptors12. The model has a R2 of 0.858 for the training set (85 compounds), and 0.839 for the test set (21 compounds), which are accurate and acceptable. All these QSAR models referred to were obtained with the linear techniques.
Generally, nonlinear QSAR models possess better statistical performance than linear QSAR models because of the nonlinear correlation between molecular physicochemical properties and structure descriptors. Neely et al. constructed a nonlinear artificial neural network (ANN) model for the skin permeability of 160 molecular structures13. The ANN model (10-3-7-1) based on ten descriptor and two hidden layers had an absolute-average percentage deviation, rms error, and R of 8.0%, 0.34, and 0.93, respectively. Khajeh and Modarress introduced a novel nonlinear QSAR model for the skin permeability of 283 compounds with the hybrid of ANN and a fuzzy inference system, adaptive neuro-fuzzy inference system (ANFIS)14. The ANFIS model was based on a training set of 225 compounds and validated by a test set of 58 compounds. The R2 values for the two sets were 0.899 and 0.890, respectively. The model possesses good predictive ability, although there are nine compounds in duplicate in the data set.
ANN algorithm may easily fall into a local minimum value and possesses the disadvantages of slow convergence speed15. Support vector machine (SVM) algorithm is based on the principle of structural risk minimization. SVMs can effectively avoid local optimums and have unique advantages in solving practical problems such as limited training samples, high dimensional and nonlinear data. The aim of this study was to develop a nonlinear SVM QSAR model for the skin permeability of a sufficiently large data set consisting of 274 compounds.
Materials and methods
Khajeh and Modarress reported 283 compounds and their experimental log Kp values14. After careful investigation, we found that the sample, p-Chlorobenzene, should be 1-chloro-4-nitrobenzene and 4-Chloro-4-phenylenediamine should be 4-Chloro-m-phenylenediamine. There are no counterions or organometallics in the data set. The molecular weights of 283 compounds were calculated with ChemDraw Ultra 8.0 in ChemOffice 2004. These molecules possessing the same molecular weights were checked carefully to identify the duplicates. There are nine compounds in duplicate, including 4-phenylenediamine (1,4-benzenediamine), 4-hydroxynitrobenzene (4-nitrophenol), methylhydroxybenzoate (methyl 4-hydroxybenzoate), 1,2-benzenediamine (2-phenylenediamine), 2-naphthol (naphthalene-2-ol), 2-nitro-1,4-phenylenediamine (2-nitro-4-phenylenediamine), 1-nonanol (Nonanol), 4-chloro-1,3-phenylenediamine (4-Chloro-m-phenylenediamine), and 1-heptanol (Heptanol). After these duplicates were deleted, 274 compounds were obtained. Table S1 in “Supplementary Materials” shows their SMILES structures and the log Kp values. The units for skin permeability coefficients Kp are cm/h and these log Kp values ranged from − 6.10 to − 0.76. The Kennard-Stone algorithm16 was used to group the compounds in the training set (139 compounds) and test set (135 compounds). The training set was used to adjust model parameters and train QSAR models; and the test set was used to validate the models.
ChemDraw Ultra 8.0 in ChemOffice 2004 was adopted to generate the structures of 274 compounds, which were converted into three-dimensional structures with Chem3D Ultra 8.0 and optimized with a semi-empirical AM1 method in MOPAC. Dragon 6.017 was used to calculate 4885 molecular descriptors for each compound. After some molecular descriptors that equal a constant or their correlation coefficients are above 0.90 were deleted, 1820 descriptors (including Neoplastic-80) were obtained for descriptor selection. Stepwise MLR analysis in IBM SPSS Statistical 19 was performed to select the optimal subset of descriptors and develop MLR models.
For non-linear regression, SVM algorithms map input variables into high-dimensional feature space, from which linear regression analysis is carried out18,19. For sample data, \(\left( {y_{1} ,x_{1} } \right), \ldots ,\left( {y_{l} ,x_{l} } \right),\quad x \in R^{n} ,\;y \in R\), the regression function is expressed as follows:
The optimal regression function can be obtained by means of the following minimization problem:
In SVM regression, the ε-insensitive loss function is employed for minimizing the training error:
Thus, Eq. (1) is:
By applying a kernel function k(x, y), Eq. (6) can be expressed as:
Gaussian radial basis function (RBF) was used in this work:
For SVM models, their SVM parameters C and γ can affect greatly their prediction performance. Both C and γ were optimized with the genetic algorithm. In this study, the LibSVM toolbox20 working on Matlab platform was used to develop models, which can be downloaded freely from https://www.csie.ntu.edu.tw/~cjlin/libsvm/.
Results and discussion
After carrying out stepwise MLR analysis in IBM SPSS Statistical 19 for the skin permeability log Kp of 274 compounds and 1820 descriptors, a three-descriptor QSAR model was obtained, which includes A log P, X3v, and Neoplastic-80.
The Ghose–Crippen–Viswanadhan octanol–water partition coefficient (A log P) is based on the A log P model21 and calculated by:
where ni is the number of atom of type i and ai is the corresponding hydrophobicity constant. Previous works have shown that A log P is positively correlation with skin permeability log Kp. In this work, the descriptors were converted to a new descriptor cos2[(4.31 + A log P)/8.66]. An analysis of cos2[(4.31 + A log P)/8.66] with respect to the skin permeability log Kp of 274 compounds resulted in regression Eq. (10) and statistical parameters:
where n is the number of samples in the training set, R2 is the coefficient of determination, R2adj is the adjusted R square, se is the standard error of the estimate, and F is the Fischer ratio. Figure 1 shows the correlation between cos2[(4.31 + A log P)/8.66] and log Kp. The descriptor cos2[(4.31 + A log P)/8.66] (or A log P) describes the hydrophobic character of a compound and is related to log Kp.
Connectivity indices are used widely in QSARs. They are based on the H-depleted molecular graph whose vertexes belong to non-hydrogen atom and are correlated with the number of connected non-hydrogen atoms17. The general formula for calculating connectivity indices is:
where n is the number of vertices; k is an integer ranging from 0 to 5, denoting the total number of kth order paths present in the molecular graph; and δ is the vertex degrees. Valence connectivity indices (Xkv) can be used to account for the presence of heteroatoms in the molecule as well as of double and triple bonds, by means of replacing the vertex degree with the valence vertex degree. The valence connectivity index of order 3, X3v, describes molecular size and shape.
By correlating log Kp to the two descriptors, cos2[(4.31 + A log P)/8.66] and X3v, we obtained the following regression equation:
Compared with Eq. (10), the quality of Eq. (12) improved noticeably when the descriptor X3v was added. Figure 2 shows the correlation between the experimental and calculated log Kp with Eq. (12). As illustrated in Fig. 2, there were two samples, ouabain (No. 5 in Table S1), and fluocinonide (No. 11) with larger prediction errors for log Kp. Thus, more molecular descriptors should be added.
Plot of experimental versus calculated log Kp with Eq. (12), generated by OriginPro 7.5 SR1.
The descriptor Ghose–Viswanadhan–Wendoloski antineoplastic-like index at the qualifying range that covers approximately 80% of the drugs studied, Neoplastic-80, depends on A log P and reflects molecular polarity and hydrophobicity17. The Neoplastic-80 value of a molecule that has a benzene ring, heterocyclic ring, aliphatic amine, carboxamide group, alcoholic hydroxyl group, carboxy ester and/or keto group, was equal to 1, when its A log P value is in the range of − 1.5 to 4.7, the molar refractivity of 43–128, the molecular weight of 180–470, and the total number of atoms of 21–63; otherwise Neoplastic-80 equals zero. A molecule with larger Neoplastic-80 might have a smaller log Kp value. Carrying out regression analysis between log Kp of 274 compounds and the three descriptors stated above resulted in Eq. (13):
The correlation coefficient R of 0.945 in Eq. (13) was slightly higher than the 0.942 of the model13. Moreover, Eq. (13) has accurate prediction for the skin permeability log Kp of compounds including the two samples (Nos. 5 and 11 in Table S1 in “Supplementary Materials”) stated above, since Fig. 3 shows that there are no samples with obvious larger errors. When the descriptor A log P, together with X3v and Neoplastic-80, was directly used to develop the MLR model, its correlation coefficient R was only 0.939, which was lower than the 0.945 of Eq. (13). Thus the three descriptors, cos2[(4.31 + A log P)/8.66], X3v, and Neoplastic-80 shown in Table S1 in “Supplementary Materials” were used to develop QSAR models.
Plot of experimental versus calculated log Kp with Eq. (13), generated by OriginPro 7.5 SR1.
A correlation analysis between the skin permeability log Kp of 139 compounds in the training set and the three descriptors resulted in Eq. (14) (i.e., MLR model):
The characteristics of molecular descriptors in MLR model are listed in Table 1. As can been observed in Table 1, the three descriptors, cos2[(4.31 + A log P)/8.66], X3v, and Neoplastic-80 descriptor all were significant and made a contribution to log Kp, because their significance values (or P values) are less than 0.05. In addition, their variance inflation factors (VIF) were far less than ten suggesting that the three descriptors describe different structure factors affecting skin permeability log Kp. The t-test can be used to measure the significance of descriptors in making a contribution to molecular physicochemical properties. The higher the absolute value of the t-test, the greater the significance of the descriptor. According to the t-test values in Table 1, the absolute values of t-test increased in the sequence: Neoplastic-80, X3v, and cos2[(4.31 + A log P)/8.66], the significance of descriptors increased in the same sequence.
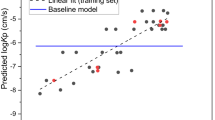
The MLR model was further used to predict the skin permeability log Kp of 135 compounds in the test set. The correlation coefficient R of the test set was 0.928. The rms errors for the training set, test set and total set were 0.343, 0.302, and 0.323, respectively. The prediction log Kp values are illustrated in Fig. 4 and listed in Table S1 in “Supplementary Materials”.
Plot of experimental versus predicted log Kp with Eq. (14), generated by OriginPro 7.5 SR1.
The three molecular descriptors used in Eq. (14) were used as input variables to develop SVM models for skin permeability log Kp from the training set of 139 compounds, by applying the LibSVM toolbox in the MATLAB R2014a software platform. A genetic algorithm was adopted to optimize the SVM parameters C and γ under the following conditions: the searching range of parameters C was [0, 1000], the searching range of γ was [0, 10], the m in m-fold-cross-validation was 5, the maximum generation was 200, the maximum population size was 20, and the ε in the ε-insensitive loss function was 0.001.
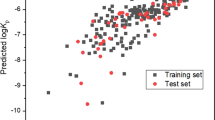
The optimization results for the SVM model were obtained: the parameters C being 7.2906 and γ being 1.7200, and the internal correlation coefficient based on leave-one-out (LOO) cross-validation method being 0.82. The optimal SVM model was further validated with the test set of 135 compounds. The SVM prediction results are listed in Table S1 in “Supplementary Materials” and illustrated in Fig. 5. The coefficient of determination R2 and rms error for the training set of 139 compounds were 0.946 and 0.253, respectively; R2 and rms for the test set of 135 compounds were 0.872 and 0.302, respectively; and R2 and rms error for the total set were 0.925 and 0.270, respectively. The rms errors of 0.253, 0.302, and 0.270, respectively, for the training set, test set and total set from the SVM model were lower than those (0.343, 0.302, and 0.323, respectively) of Eq. (14) (MLR model) in this study. Therefore, there were non-linear relationships between the skin permeability log Kp and molecular descriptors used.
The SVM model was further evaluated with the criteria by Golbraikh and Tropsha:22
where \(q_{ext}^{2}\) is external correlation coefficient; R02 and R0′2 are determination coefficients of the predicted vs. the observed values and of the observed vs. the predicted values, respectively; k and k′ are slopes of regression lines of the predicted vs. the observed values and of the observed values vs. the predicted values; \(\overline{y}_{{{\text{train}}}}\) is the average value of the training set; yi and \(\overline{y}_{i}\) are the observed and the predicted activities, respectively; \(y_{i}^{{r_{0} }} = k\widetilde{y}_{i}\) and \(\widetilde{y}_{i}^{{r_{0} }} = k^{\prime}y_{i}\). Obviously, our SVM model satisfied the validation criteria22,23.
The coefficient of determination R2 (= 0.946) in this study is higher than the R2 of 0.907, 0.8158, 0.7609, 0.93610, 0.7011, 0.85812, and 0.9313. In addition, the rms errors of the training set, test set and total set from the ANFIS model of Khajeh and Modarress that dealt with the 283 samples were 0.318, 0.308, and 0.316 respectively14, which were greater than the rms errors ( 0.253, 0.302, and 0.270, respectively) from our SVM model. Compared with results of other models reported in the literature9,10,11,12,13,14, our SVM model shows better statistical performance in a model that deals with more samples in the test set.
Conclusions
A three-descriptor SVM model with SVM parameters C of 7.2906 and γ of 1.7200 was successfully built for the skin permeability log Kp of a sufficiently large data set consisting of 274 compounds, by means of a genetic algorithm. The SVM model possesses rms errors of 0.253 for the training set (139 compounds), 0.302 for the test set (135 compounds), and 0.270 for the total set (274 compounds). Our SVM model shows better statistical performance in a model that deals with more samples in the test set, compared with other QSARs of the skin permeability of log Kp reported in the literature. There were non-linear relationships between the skin permeability log Kp and molecular descriptors used. It was reasonable applying a SVM algorithm to develop a nonlinear QSAR model for skin permeability.
Data availability
All data generated or analysed during this study are included in this published article (and its “Supplementary Information” files).
References
Fitzpatrick, D., Corish, J. & Hayes, B. Modelling skin permeability in risk assessment––The future. Chemosphere 55, 1309–1314 (2004).
Alves, V. M. et al. Predicting chemically-induced skin reactions. Part II: QSAR models of skin permeability and the relationships between skin permeability and skin sensitization. Toxicol. Appl. Pharmacol. 284, 273–280 (2015).
Varpe, B. D. et al. 3D-QSAR and Pharmacophore modeling of 3,5-disubstituted indole derivatives as Pim kinase inhibitors. Struct. Chem. 31, 1675–1690 (2020).
Heo, S. K., Safder, U. & Yoo, C. K. Deep learning driven QSAR model for environmental toxicology: Effects of endocrine disrupting chemicals on human health. Environ. Pollut. 253, 29–38 (2019).
Lotfi, S., Ahmadi, S. & Zohrabi, P. QSAR modeling of toxicities of ionic liquids toward Staphylococcus aureus using SMILES and graph invariants. Struct. Chem. 31, 2257–2270 (2020).
Rahmani, N., Abbasi-Radmoghaddam, Z., Riahi, S. & Mohammadi-Khanaposhtanai, M. Predictive QSAR models for the anti-cancer activity of topoisomerase IIα catalytic inhibitors against breast cancer cell line HCT15: GA-MLR and LS-SVM modeling. Struct. Chem. 31, 2129–2145 (2020).
Patel, H., ten Berge, W. & Cronin, M. T. D. Quantitative structure-activity relationships (QSARs) for the prediction of skin permeation of exogenous chemicals. Chemosphere 48, 603–613 (2002).
Fujiwara, S.-I., Yamashita, F. & Hashida, M. QSAR analysis of interstudy variable skin permeability based on the “latent membrane permeability” concept. J. Pharm. Sci. 92, 1939–1946 (2003).
Magnusson, B. M., Anissimov, Y. G., Cross, S. E. & Roberts, M. S. Molecular size as the main determinant of solute maximum flux across the skin. J. Invest. Dermatol. 122, 993–999 (2004).
Chauhan, P. & Shakya, M. Role of physicochemical properties in the estimation of skin permeability: In vitro data assessment by Partial Least-Squares Regression. SAR QSAR Environ. Res. 21, 481–494 (2010).
Xu, G., Hughes-Oliver, J. M., Brooks, J. D. & Baynes, R. E. Predicting skin permeability from complex chemical mixtures: Incorporation of an expanded QSAR model. SAR QSAR Environ. Res. 24, 711–731 (2013).
Chen, C.-P., Chen, C.-C., Huang, C.-W. & Chang, Y.-C. Evaluating molecular properties involved in transport of small molecules in stratum corneum: A quantitative structure-activity relationship for skin permeability. Molecules 23, 911 (2018).
Neely, B. J., Madihally, S. V., Robinson, R. L. & Gasem, K. A. M. Nonlinear quantitative structureproperty relationship modeling of skin permeation coefficient. J. Pharm. Sci. 98, 4069–4084 (2009).
Khajeh, A. & Modarress, H. Linear and nonlinear quantitative structure–property relationship modelling of skin permeability. SAR QSAR Environ. Res. 25, 35–50 (2014).
Zhou, T., Lu, H., Wang, W. & Yong, X. GA-SVM based feature selection and parameter optimization in hospitalization expense modeling. Appl. Soft. Comput. 75, 323–333 (2019).
Daszykowski, M. et al. TOMCAT: A MATLAB toolbox for multivariate calibration techniques. Chemom. Intell. Lab. Syst. 85, 269–277 (2007).
Talete srl. DRAGON (Software for Molecular Descriptor Calculation) Version 6.0. http://www.talete.mi.it/ (2012).
Yu, X., Xu, L., Zhu, Y., Lu, S. & Dang, L. Correlation between 13C NMR chemical shifts and complete sets of descriptors of natural coumarin derivatives. Chemom. Intell. Lab. Sys. 184, 167–174 (2019).
Yu, X. Prediction of depuration rate constants for polychlorinated biphenyl congeners. ACS Omega 4, 15615–156205 (2019).
Chang, C. C. & Lin, C. J. LIBSVM: A library for support vector machines. Acm. T. Intel. Syst. Tec. 2, 27 (2011).
Ghose, A. K., Viswanadhan, V. N. & Wendoloski, J. J. Prediction of hydrophobic (liphophilic) properties of small organic molecules using fragmental methods: An analysis of ALOGP and CLOGP methods. J. Phys. Chem. A 102, 3762–3772 (1998).
Golbraikh, A. & Tropsha, A. Beware of q2. J. Mol. Graph. Model. 20, 269–276 (2002).
Yu, X., Bing, Y., Yu, W. & Wang, X. DFT-based quantum theory QSPR studies of molar heat capacity and molar polarization of vinyl polymers. Chem. Pap. 62, 623–629 (2008).
Acknowledgements
This work was supported by the Provincial Natural Science Foundation of Hunan (No. 2020JJ6012) and the Open Project Program of Hunan Provincial Key Laboratory of Environmental Catalysis & Waste Regeneration (Hunan Institute of Engineering) (No. 2018KF11).
Author information
Authors and Affiliations
Contributions
R.Z., J.D. data curation, manuscript revision. L.D.: data collection and curation, descriptor calculation. X.Y.: conceptualization, methodology, software, model development, writing-original draft preparation, manuscript revision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zeng, R., Deng, J., Dang, L. et al. Correlation between the structure and skin permeability of compounds. Sci Rep 11, 10076 (2021). https://doi.org/10.1038/s41598-021-89587-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-89587-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.